Abstract
Understanding plant metabolism as an integrated system is essential for metabolic engineering aimed at the effective production of compounds useful to human life and the global environment. The “omics” approach integrates transcriptome and metabolome data into a single data set and can lead to the identification of unknown genes and their regulatory networks involved in metabolic pathways of interest. One of the intriguing, although poorly described metabolic pathways in plants is the biosynthesis of glucosinolates (GSLs), a group of bioactive secondary products derived from amino acids that are found in the family Brassicaceae. Here we report the discovery of two R2R3-Myb transcription factors that positively control the biosynthesis of GSLs in Arabidopsis thaliana by an integrated omics approach. Combined transcriptome coexpression analysis of publicly available, condition-independent data and the condition-specific (i.e., sulfur-deficiency) data identified Myb28 and Myb29 as candidate transcription factor genes specifically involved in the regulation of aliphatic GSL production. Analysis of a knockout mutant and ectopic expression of the gene demonstrated that Myb28 is a positive regulator for basal-level production of aliphatic GSLs. Myb29 presumably plays an accessory function for methyl jasmonate-mediated induction of a set of aliphatic GSL biosynthetic genes. Overexpression of Myb28 in Arabidopsis-cultured suspension cells, which do not normally synthesize GSLs, resulted in the production of large amounts of GSLs, suggesting the possibility of efficient industrial production of GSLs by manipulation of these transcription factors. A working model for regulation of GSL production involving these genes, renamed Production of Methionine-Derived Glucosinolate (PMG) 1 and 2, are postulated.
Keywords: coexpression, functional genomics, transcriptomics
Glucosinolates (GSLs) produced by vegetables in the family Brassicaceae, such as broccoli and cabbage, have recently attracted considerable attention, because they apparently provide anticarcinogenic, antioxidative, and antimicrobial activity (1–3). In addition, interest in rapeseed is increasing because of the growing demand for biodiesel fuel, which can be derived from its oil (4). GSLs function as defense compounds against pests (5, 6) that can potentially reduce oil production. In nature, there are ≈120 GSLs that differ in side-chain structures [supporting information (SI) Fig. 6]. GSLs are synthesized from several amino acids, including Met, Trp, and Phe (5, 6). Met-derived 4-methylsulfinylbutyl GSL (4MSOB) deserves special attention because its degraded product, sulforaphane, which was first isolated from broccoli, exhibits pronounced anticarcinogenic activity (1–3). The model plant Arabidopsis thaliana (L.) Heynh. also contains GSLs, including 4MSOB. For biotechnological applications that require increasing 4MSOB production, it is important to identify all of the genes involved in GSL biosynthesis and to elucidate the entire regulatory mechanism in Arabidopsis. To date, however, there has been no report on the genes regulating Met-derived aliphatic GSL biosynthesis.
Along with the massive accumulation of microarray data sets, transcriptome coexpression analysis has proven to be a powerful tool for identifying regulatory relationships in the transcriptional networks of model organisms, including Escherichia coli (7) and yeast (8). Assuming that a set of genes coexpressed under a given experimental regimen is involved in the same or related metabolic pathway, candidate genes involved in the regulation or synthesis steps of a particular metabolic pathway can be comprehensively identified, or at least predicted with some confidence, by using publicly available transcriptome databases (9–13). Although the strategy of coexpression analysis has great potential for versatility, its application has thus far been limited in the actual discovery of useful genes in important pathways in Arabidopsis (11). If a coexpression profile for a specific condition (e.g., nutrient-deficiency stress) is compared with the set of genes that is always coexpressed in all tissues under all experimental conditions that have been tested, or “condition-independent” profile derived from public data sets, the reliability and feasibility of predicting a gene function would greatly increase. In this study, we identified two previously unrecognized genes, Myb28 and Myb29, which encode R2R3-Myb transcription factors involved in the regulation of aliphatic GSL biosynthesis, by using an integrated strategy based on transcriptome coexpression analysis for both public data sets and our own data, along with metabolic profiling. We also concurrently predicted many of the unknown structural genes encoding enzymes of aliphatic GSL biosynthesis and clarified their regulatory network. Overexpression of Myb28 in Arabidopsis cell suspension cultures resulted in the production of large amounts of GSLs, indicating the usefulness of these transcription factors for the production of GSLs in biotechnological applications.
Results and Discussion
Discovery of Myb28 and Myb29 as Regulators of GSL Biosynthesis on the Basis of Transcriptome Coexpression Analysis.
The discovery of two transcription factors was made by the combined analyses of transcriptome coexpression profiles from a public domain database and our own data set. A data set of condition-independent coexpression profiles was generated by calculating Pearson's correlation coefficients between all combinations of 22,263 Arabidopsis genes by using the publicly available 1,388 microarray data of AtGenExpress (14). By using this correlation coefficient data, we exhaustively analyzed coexpression between metabolic pathway genes and transcription factor genes. Coexpression relationships were visualized as a graph in which a pair of genes (two vertices) with a high correlation coefficient (>0.65 in this case) was connected by a line (i.e., an “edge”) forming a module (Fig. 1). This analysis revealed that the genes involved in aliphatic GSL biosynthesis (SI Fig. 6) were clustered in a discrete module together with two uncharacterized transcription factor genes, Myb28 (At5g61420) and Myb29 (At5g07690) (Fig. 1). The known GSL biosynthetic genes were highly coexpressed with Myb28, and to a lesser extent with Myb29. Some of the putative Leu biosynthetic gene homologs (AtLeuC1, AtLeuD1 and D2, AtIMD1 and/or 3, AtBCAT-3, and AtBCAT-4; see below) were also clustered in this module, suggesting that aliphatic GSL biosynthesis and some putative Leu biosynthetic gene homologs are coordinately regulated. These coexpression analyses led to the hypothesis that Myb28 and Myb29 may be transcription factors that positively regulate aliphatic GSL biosynthesis and that some putative Leu biosynthetic gene homologs may be involved in aliphatic GSL biosynthesis rather than in Leu biosynthesis.
Fig. 1.
Coexpression analysis of aliphatic GSL biosynthetic genes and transcription factors. Yellow and red points indicate genes encoding enzymes and transcription factors, respectively. Transcripts from AtIMD1 and AtIMD3 and those from CYP79F1 and CYP79F2 were cross-hybridized to the same probe sets on an ATH1 microarray used in AtGenExpress and hence are indistinguishable. Lengths of the lines are valueless in these displays.
This prediction for candidate transcription factors deduced from a public database was reinforced by in-house sulfur-deficiency stress transcriptome data. Previous studies integrating transcriptomics and metabolomics (15, 16) indicated that the biosynthetic genes for both aliphatic and Trp-derived indole GSLs were coordinately down-regulated under sulfur-deficiency conditions and were thus placed in the same cluster by batch-learning self-organizing map analysis. Myb28 and Myb29 were located in this same cluster, in addition to ATR1/Myb34, a known positive regulator of indole GSL biosynthesis (17) (SI Fig. 7). A set of putative Leu biosynthetic genes, AtLeuC1, AtLeuD1 and D2, AtIMD1, and AtBCAT-4, was also clustered with the GSL biosynthetic genes (SI Fig. 7). These results from sulfur-deficiency coexpression profiles support the hypothesis that Myb28 and Myb29 are positive regulators of aliphatic GSL biosynthesis.
Functional Analysis of Myb28 and Myb29 by Gene-Knockout Plants.
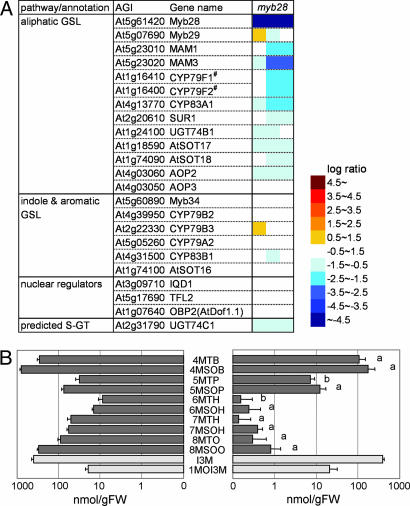
To confirm the predicted function of Myb28 and Myb29, we analyzed transcript (i.e., transcriptome analysis by microarray) and GSL accumulation in gene-knockout and -knockdown plants, designated as myb28 and myb29, respectively, both of which grew normally. As shown in Fig. 2A, the expression of most of the genes involved in aliphatic GSL biosynthesis was repressed in myb28, whereas there were no significant changes in indole and Phe-derived aromatic GSL biosynthetic gene expression or in the expression of three nuclear factor genes reported to be involved in GSL metabolism (18–20). The other genes whose expression changed in myb28 are shown in SI Table 1. Most of the genes down-regulated in myb28 other than GSL biosynthetic genes were sulfur-deficiency-inducible genes (15), suggesting that a decrease of GSL amount (see below) might result in perturbation of sulfur metabolism, i.e., a sulfur-surplus status in myb28, to repress these genes secondarily.
Fig. 2.
Expression of GSL biosynthetic genes and GSL contents in Myb28-knockout mutant myb28. (A) Heat map showing gene expression levels. Log ratio (base 2) of signal intensity in myb28 to wild type is shown by the color scale. The data represent three hybridizations. Transcripts of CYP79F1 and CYP79F2 were cross-hybridized to the same probe set on an ATH1 microarray because of similarity of their nucleotide sequences. (B) GSL contents of rosette leaves. The means and SD of three replicates are shown. Closed and open bars indicate aliphatic and indole GSLs, respectively. A statistically significant decrease in myb28 (Right) compared with wild type (Left) is shown as “a” (Welch's t test; P < 0.01) or “b” (P < 0.02) on the bar. 8MTO, 8-methylthiooctyl GSL; 8MSOO, 8-methylsulfinyloctyl GSL; I3M, indol-3-ylmethyl GSL; 1MOI3M, 1-methoxyindol-3-ylmethyl GSL. Note that the scale is logarithmic.
The aliphatic GSL contents of myb28 rosette leaves as determined by liquid chromatography–mass spectrometry were significantly decreased (Fig. 2B), whereas the indole GSLs did not change significantly. Plants in the myb28 line contained only 17–33% of GSLs with 4C or 5C Met side-chains [4-methylthiobutyl GSL (4MTB), 4-methylsulfinylbutyl GSL (4MSOB) 5-methylthiopentyl GSL (5MTP), and 5-methylsulfinylpentyl (5MSOP)], 1.7% of GSLs with a 6C chain [6-methylthiohexyl GSL (6MTH) and 6-methylsulfinylhexyl GSL (6MSOH)], and 0.3–0.7% of GSLs with 7C or 8C chains [7-methylsulfinylheptyl (7MTH), 7-methylsulfinylheptyl (7MSOH), 8-methylthiooctyl GSL (8MTH), and 8-methylsulfinyloctyl GSL (8MSOO)] as the wild-type plant. These differences could be due to the process of long-chain GSL formation through repetitive cycles of side-chain elongation reactions; thus, long side chains are more affected by the repression of enzyme activities than short-chain GSLs. GSL content also decreased in myb28 seeds (data not shown). The introduced defect of gene expression and GSL content in myb28 was genetically complemented by Agrobacterium-mediated stable transformation with a DNA fragment harboring the intact Myb28 gene (SI Fig. 8). All of these data indicated that Myb28 is a positively acting transcription factor specific for the expression of aliphatic GSL biosynthetic genes but not for indole or aromatic GSL biosynthetic genes.
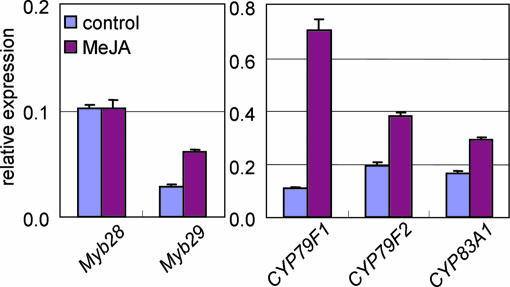
However, there was no apparent change in the expression of GSL biosynthetic genes or GSL content in myb29 gene-knockdown plants (SI Fig. 9), suggesting that Myb29 may not be essential for GSL biosynthesis. Because GSL biosynthesis is known to be enhanced by the plant hormone methyl jasmonate (MeJA) (21), we measured changes of gene expression in wild-type plants in response to MeJA application (Fig. 3). Under control conditions (without MeJA), Myb28 expression was higher than Myb29. MeJA application, however, induced expression of Myb29 but not Myb28. Expression of the aliphatic GSL biosynthetic genes CYP79F1, CYP79F2, and CYP83A1 was also up-regulated with the induction of Myb29. These results suggest that Myb28 is essential for basal-level synthesis of aliphatic GSLs, and Myb29 presumably has a function in the induction of aliphatic GSL biosynthetic genes in response to MeJA signaling.
Fig. 3.
Gene expression in response to MeJA. The relative expression levels of Myb28, Myb29, CYP79F1, CYP79F2, and CYP83A1 normalized to the constitutive ubiquitin gene UBC9 are shown as means and SD of three replicates. Detailed methods of quantitative RT-PCR are described in SI Methods. Note scalar differences between regulatory and structural gene expression levels.
GSL Content in Myb28-Overexpressing Lines.
To further confirm the regulatory function of Myb28 and to evaluate it for biotechnological applications, Myb28-overexpressing Arabidopsis plants and suspension cell cultures were investigated for their gene expression and GSL production. As shown in Fig. 4A, Myb28 transcript levels were increased by an average of 8.7-fold in transgenic suspension cells. The expression of every aliphatic GSL biosynthetic structural gene was markedly induced in the Myb28-overexpressing suspension cells, whereas there was no apparent increase in the expression of indole and aromatic GSL biosynthetic genes. Myb28-overexpressing whole plants, in which Myb28 transcript accumulated by 2-fold, overexpressed aliphatic GSL genes by ≈2-fold (data not shown). These results indicate that ectopic expression of Myb28 specifically up-regulates the expression of aliphatic GSL biosynthetic genes, but the indole and aromatic GSL biosynthetic genes, as well as Myb29, are unaffected. The other affected genes in Myb28-overexpressing cell cultures are shown in SI Table 2, which indicated that most of the genes up-regulated in Myb28-overexpressing cell cultures other than GSL biosynthetic genes were sulfur-deficiency-inducible genes.
Fig. 4.
Expression of GSL biosynthetic genes and GSL contents in Myb28-overexpressing suspension cell cultures. (A) Heat map showing gene expression levels. Log ratio (base 2) of signal intensity in Myb28-overexpressing T87 cell suspension cultures (T87OX) to control lines (empty-vector transformed) is shown by the color scale. The data are of nine hybridizations. (B) GSL contents. The data of three independent transformants of control (empty-vector transformed) (Left) and Myb28-overexpressing suspension cells (T87OX) (Right) are shown as three bars for each type of GSL. The means and SD of three replicates are shown. Closed and open bars indicate aliphatic and indole GSLs, respectively. nd, not detected in all three control cell culture lines; 8MTO, 8-methylthiooctyl GSL; 8MSOO, 8-methylsulfinyloctyl GSL; I3M, indol-3-ylmethyl GSL; 1MOI3M, 1-methoxyindol-3-ylmethyl GSL. Note that the scale is logarithmic.
Fig. 4B shows the GSL contents of the Myb28-overexpressing cell suspension cultures. Nontransformed and empty-vector-transformed control cell cultures accumulated no detectable levels of GSLs. All three independent Myb28-overexpressing cell lines accumulated 11 different GSLs at concentrations comparable to those found in wild-type whole Arabidopsis plants. These results clearly demonstrate that the overexpression of Myb28 is essential and sufficient to activate GSL biosynthetic genes leading to the production of GSLs even in dedifferentiated cells. This result is, as far as we know, a previously unreported example of metabolic engineering achieved by genetic engineering of a single transcription factor in Arabidopsis cell suspension cultures. In the Myb28-overexpressing whole plants, GSL contents did not significantly increase in leaves and seeds (data not shown) compared with those in the wild-type plants, supposedly because the expression of Myb28 was only approximately double.
Functional Description of the Genes Involved in GSL Biosynthesis.
On the basis of transcriptome analysis of the Myb28-knockout and -overexpressing lines, the genes involved in GSL biosynthesis can be described in some detail.
Genes involved in Met side-chain elongation.
Met is subjected to side-chain elongation cycles before entering the GSL core biosynthetic pathway. Elongation proceeds through the four-step reactions as in Leu biosynthesis (SI Fig. 10). Thus, the enzymes committed to Met side-chain elongation and Leu biosynthesis are presumably encoded by homologous genes belonging to the same gene families as follows: methylthioalkylmalate synthase (MAM) and isopropylmalate synthase (IPM-S) by four genes; MAM isomerase (MAM-I) and isopropylmalate isomerase (IPM-I) by three genes (AtLeuCs) for a large subunit and three genes (AtLeuDs) for a small subunit; MAM dehydrogenase (MAM-D) and isopropylmalate dehydrogenase (IPM-D) by three genes; and methionine-analog aminotransferase (MAAT) and branched-chain aminotransferase (BCAT) by six genes (SI Fig. 10). Of these 19 genes, only MAM1 and MAM3 have been functionally identified as coding for the methylthioalkylmalate synthase involved in Met side-chain elongation (22, 23). AtBCAT-1 has been shown to initiate degradation of the branched-chain amino acids Leu, Ile, and Val (24). Transcriptome analyses of the myb28-knockout and Myb28-overexpressing cell cultures indicated that MAM1, MAM3, AtLeuC1, AtLeuD1, AtLeuD2, AtIMD1, AtBCAT-3, and AtBCAT-4 were all positively regulated by Myb28 (SI Fig. 10), suggesting that these regulated genes are committed to aliphatic GSL biosynthesis; thus, the remainder of the 19 genes are not likely related to GSL but rather to Leu biosynthesis. AtBCAT-4 has recently been reported to be involved in Met side-chain elongation (25), confirming our methodology for predicting gene function.
Methionine biosynthetic genes.
Met is synthesized in plants from Cys by sequential reactions catalyzed by cystathionine γ-synthase (CGS), cystathionine β-lyase (CBL), and Met synthase (MS) (SI Fig. 10). Arabidopsis possesses three MS genes, AtMS1, AtMS2, and AtMS3. Of these genes, AtMS3, which encodes a chloroplastic isoform, is postulated to be responsible for the de novo synthesis of Met, whereas AtMS1 and AtMS2 are assumed to be involved in the recycling of cytosolic S-adenosyl-homoCys into Met (26). Transcriptome analyses of Myb28-engineered cells suggest that CGS, CBL, and AtMS2 are up-regulated by Myb28, which is indicative of the connection of these genes to GSL biosynthesis. These structural genes might be induced to compensate for the decrease in Met concentrations as the plant synthesizes aliphatic GSLs.
Other genes involved in GSL biosynthesis.
We suggested in ref. 15 that two GST genes, At1g78370 (ATGSTU20) and At3g03190 (ATGSTF11), are involved in GSL biosynthesis. Transcriptome analyses of myb28 and Myb28-overexpressing cell cultures indicated that both of them are regulated by Myb28, further supporting the specific involvement of these genes in aliphatic GSL biosynthesis.
We also assumed that At5g36160, which is annotated as a putative Tyr aminotransferase, is an additional C-S lyase gene functioning in GSL biosynthesis under certain conditions (15). However, the expression of this gene was not regulated by Myb28 (data not shown), suggesting that it may encode a C-S lyase involved in indole/aromatic GSL biosynthesis, or it encodes a Phe aminotransferase in side-chain elongation of Phe, directing the synthesis of 2-phenylethyl GSL derived from homoPhe (SI Fig. 6).
PMSR2, a gene encoding a putative peptide methionine sulfoxide reductase, was coexpressed with the GSL biosynthetic genes under sulfur deficiency, and was regulated by Myb28. Supposing that this enzyme could recognize the methylsulfinyl moiety of methylsulfinylalkyl GSL and that of methionine sulfoxide, this enzyme could be involved in side-chain conversion of aliphatic GSLs (Fig. 5).
Fig. 5.
Regulatory networks model of GSL biosynthetic pathway in Arabidopsis accession Columbia. PMG1/Myb28 and PMG2/Myb29 are positive regulators of aliphatic GSL biosynthesis, whereas ATR1/Myb34 positively regulates indole GSL formation. PMG1/Myb28 is the essential and sufficient master regulator and thus supports the basal production of aliphatic GSLs. PMG2/Myb29 is an accessory factor that plays a role in response to MeJA signaling. MeJA also induces the ATR1/Myb34 cascade. Sulfur deficiency represses the expression of all three Myb factors. Previously unidentified structural genes can be mapped in the aliphatic GSL biosynthetic pathway by the present study. MTG, methylthioalkyl GSL; MSOG, methylsulfinylalkyl GSL.
AOPs encoding 2-oxoglutarate-dependent dioxygenases are involved in the modification of Met side chains (27). Gene expression profiles of the Myb28-engineered cells suggest that Myb28 regulates AOP2 but not AOP3. Analysis of the expression profile of AOP3 in the Arabidopsis thaliana Trans-Factor and Cis-Element Prediction Database (ATTED-II) transcriptome expression database (www.atted.bio.titech.ac.jp/locus/At4g03050.html) revealed that this gene is expressed specifically in fruit, suggesting AOP3 as a fruit-specific protein in accession Columbia, and is thus independent of Myb28 regulation (Fig. 5).
A Working Model for Regulation of GSL Biosynthesis.
The present study identified Myb28 and Myb29 as positive regulatory factors for aliphatic GSL biosynthesis under certain conditions. A regulatory network for this pathway could thus be proposed, as summarized in Fig. 5. Myb28 and Myb29 belong to the R2R3-Myb gene family, clustered into a small subgroup with Myb34 and Myb76 (see below) in a molecular phylogenetic tree drawn by the relationship of their amino acid sequences (28). The subtle differences in hormonal or stress-signal recognition by these regulatory elements suggest an evolutionary conservation of their primary structures and functions with fine tuning of their distinct roles in GSL metabolism. On the basis of the results discussed above, we rename these genes Production of Methionine-Derived Glucosinolate (PMG) 1 and 2, that is, PMG1/Myb28 and PMG2/Myb29.
Because GSLs play important roles as storage forms of sulfur and defense compounds against herbivores and microorganisms, Arabidopsis has evolved sophisticated regulatory mechanisms to control GSL biosynthesis responding to changes in nutritional status and biotic/abiotic stresses. PMG1/Myb28 is apparently a master transcription factor, generally regulating the pathway from Met to aliphatic GSLs, and is necessary and sufficient for the biosynthesis of aliphatic GSLs at a basal level in Arabidopsis. In contrast, PMG2/Myb29 plays an accessory role in MeJA-mediated induction of aliphatic GSL biosynthesis. Because the expression of ATR1/Myb34, together with the indole GSL biosynthetic genes, was also induced by MeJA application (29), ATR1/Myb34 participates in the MeJA-mediated induction of indole GSL biosynthesis, as PMG2/Myb29 does for aliphatic GSLs.
GSLs allow the nontoxic storage of sulfur in plants, because GSLs contain two or three sulfur atoms per molecule. Under sulfur-deficiency conditions, the expression of PMG1/Myb28, PMG2/Myb29, and ATR1/Myb34 was down-regulated, presumably to shut down GSL biosynthesis, so that the limited available sulfur would not be diverted from the production of sulfur-containing primary metabolites such as Cys, Met, and glutathione (30). In this context, the relation of sulfur-deficiency gene regulation to the recently identified SLIM1 factor that regulates sulfate uptake and assimilation is particularly intriguing (31). Although ATR1/Myb34 is apparently negatively regulated by SLIM1 in roots (31), the effect of SLIM1 on PMG1/Myb28 expression is unclear, probably due to the relatively minor importance of PMG1/Myb28 in roots compared with ATR1/Myb34 (SI Fig. 11). ATR1/Myb34 was repressed in the PMG1/Myb28-overexpressing plants (Fig. 4A). In an ATR1 loss-of-function mutant, the expression of CYP79B2, CYP79B3, and CYP83B1 as part of indole GSL biosynthesis and thus presumed to be controlled by ATR1/Myb34 was repressed in adult leaves but not in seedlings (17), suggesting the presence of a complicated regulatory mechanism for indole GSL biosynthesis. Biosynthesis of indole GSL could be finely regulated by several regulatory networks in response to environmental stimuli and the developmental stage, because indole GSL metabolism is closely related to the biosynthesis of indole-3-acetic acid, an important plant hormone.
A gene expression database analysis revealed that PMG1/Myb28 and PMG2/Myb29 are expressed preferentially in leaves and nodes along with AtBCAT-4 and MAM-1, whereas ATR1/Myb34 and CYP79B2 are expressed more strongly in roots than in leaves (SI Fig. 11). Interestingly, SUR1, which is involved in both aliphatic and indole GSL biosyntheses, seems to be positively regulated by both PMG1/Myb28 and ATR1/Myb34 in a tissue-specific manner (i.e., in leaves and inflorescences by PMG1/Myb28 and in root by ATR1/Myb34) (SI Fig. 11).
Perspectives on the Application of PMG1/Myb28 and PMG2/Myb29 and Coexpression Strategy.
In terms of biotechnological application for GSL metabolic engineering, PMG1/Myb28 and PMG2/Myb29 are quite promising targets of genetic engineering for improved production of aliphatic GSLs on an industrial scale, because the ectopic expression of PMG1/Myb28 resulted in the production of GSLs at levels comparable to differentiated plants, even in dedifferentiated suspension cells. This case study using Arabidopsis leads to further applicable studies aimed at industrial production of human health-beneficial GSLs and at engineering potent pest-resistant rapeseed for biodiesel production.
Besides PMG1/Myb28 and PMG2/Myb29, our strategy of combining omics analyses of public transcriptome coexpression data sets with condition-specific (i.e., sulfur deficiency) transcriptome and metabolome profiles in-house, lead to the prediction of additional transcription factors, Myb76 and Myb59, which exhibit weaker correlation with GSL biosynthetic genes, and a number of structural genes presumably encoding the enzymes involved in GSL biosynthesis. This result indicates that a strategy based on transcriptome coexpression analysis is highly versatile for the comprehensive identification of genes involved in plant metabolism. Especially when condition-independent coexpression profiles from public databases are combined with condition-specific transcriptome and metabolome profiles, whole regulatory frameworks can be outlined (15, 32), leading to a greater understanding of metabolic systems and subsequent biotechnological applications in plant production.
Methods
Coexpression Analysis.
Pearson's correlation coefficients between all combinations of 22,263 Arabidopsis genes were obtained from ATTED-II (14) (www.atted.bio.titech.ac.jp), which is based on the publicly available 1,388 microarray data of AtGenExpress. The cut-off value for Pearson's correlation coefficient was 0.65. The genes assigned to the Met side-chain elongation pathway and Leu biosynthetic pathway in KaPPA-View (map Ath00403 and Ath00009, respectively; http://kpv.kazusa.or.jp/kappa-view) (33), known aliphatic GSL biosynthetic genes, and transcription factor genes obtained from the Arabidopsis Gene Regulatory Information Server (AGRIS) (34) (http://arabidopsis.med.ohio-state.edu/RGNet), the Database of Arabidopsis Transcription Factors (DATF) (35) (http://attfdb.cbi.pku.edu.cn), and the RIKEN Arabidopsis Transcription Factor Database (RARTF) (36, 37) (http://rarge.gsc.riken.jp/rartf) were used for Fig. 1. The coexpression pattern of these genes was illustrated by using the Pajek program (38). Condition-specific microarray data for sulfur deficiency was reported in ref. 15, and the genes were clustered by batch-learning self-organizing map analysis according to their expression patterns as reported in ref. 16.
Vector Construction and Plant Materials.
For overexpression lines, full-length Myb28 cDNA was amplified by PCR using Arabidopsis leaf cDNA as a template. The cDNA was introduced into binary vector pGWB2 by TOPO and the Gateway system (Invitrogen, Carlsbad, CA), in which the expression of cDNA is under the control of the CaMV35S promoter. For the genetic complementation study, an ≈4-kb fragment spanning the upstream sequence and coding region of Myb28 was amplified by PCR using Arabidopsis leaf DNA as a template. This genomic fragment was introduced in pGWB1 by TOPO and the Gateway system. The resulting vectors were introduced into Agrobacterium tumefaciens EHA101 by the method of An et al. (39).
Wild-type Arabidopsis accession Columbia was transformed with full-length Myb28 cDNA by the floral dip method (40) to obtain Myb28-overexpressing plants. The T-DNA insertion mutant myb28 (see below) was complemented with a genomic fragment containing an intact copy of Myb28. Arabidopsis T87 cultured suspension cells (41) were transformed with the fusion construct of CaMV35S promoter linked to Myb28 cDNA to obtain Myb28 overexpressing suspension cell lines. Details of suspension cell culture and transformation are described in SI Methods.
Myb28-knockout plants, in which T-DNA was inserted into the 5′ UTR of Myb28 (SALK_136312) (42), was obtained from the Arabidopsis Biological Resource Center (Ohio State University, Columbus, OH). Homozygous lines of the T-DNA insertion mutant were selected and designated as myb28. A homozygous T-DNA-inserted line of Myb29, designated as myb29, in which T-DNA is inserted in the 5′ UTR of Myb29 (CS121027), was a kind gift from Mitsuhiro Aida (Nara Institute of Science and Technology, Ikoma, Japan).
T2 and T3 generations of mutants and transgenic plants were used for analysis. Plants were grown for ≈3 weeks on soil [PRO-MIX BX (Premier Horticulture Inc., Quakertown, PA): vermiculite = 2:1, supplemented with fertilizer] in a greenhouse at 22°C under natural and fluorescent light (16 h light/8 h dark cycle). Rosette leaves were harvested, immediately frozen in liquid nitrogen, and stored at −80°C.
MeJA Treatment.
Wild-type Arabidopsis plants were grown for 7 days in liquid culture (43). Plants were treated with MeJA for 3 h by direct addition to the liquid medium (final concentration of 10 μM). DMSO was used for mock-treatments (final concentration of 0.1%, vol/vol). The seedlings were harvested, immediately frozen in liquid nitrogen, and stored at −80°C until use.
Transcriptome Analysis.
Total RNA was extracted with an RNeasy Plant mini kit (Qiagen, Valencia, CA). In the case of myb28 and myb29, three independent hybridizations with an Affymetrix (Santa Clara, CA) ATH1 microarray using two biological replicates were conducted according to the manufacturer's instructions. In the case of Myb28-overexprssing plants, four hybridizations with an Affymetrix ATH1 microarray were conducted by using T2 hygromycin-resistant individuals and T3 homozygous lines of two independent transformants. To compare expression levels in myb28 and overexpressing plants with wild-type plants, comparison analysis of GeneChip Operating Software (GCOS) ver1.4 (Affymetrix) was conducted. For the transcriptome analysis of cell suspension cultures, nine hybridizations were conducted with Agilent Arabidopsis2 Oligo DNA Microarray (Agilent Technologies, Palo Alto, CA) using nine combinations of one of three overexpressing lines (Cy5 label) and one of three control lines transformed with an empty vector (Cy3 label).
GSL Analysis.
GSLs were analyzed by liquid chromatography–mass spectrometry using sinigrin as an internal standard for quantification (31).
Supplementary Material
Acknowledgments
We thank Dr. Yoshiyuki Ogata (Kazusa DNA Research Institute) for useful suggestions about gene coexpression analysis; Dr. Jonathan Gershenzon (Max Planck Institute for Chemical Ecology, Jena, Germany) for the kind gift of GSL standards; Dr. Tsuyoshi Nakagawa (Shimane University, Matsue, Japan) for providing binary vectors pGWB1 and pGWB2; Dr. Mitsuhiro Aida for providing a homozygous Myb29-knockdown line; and the Arabidopsis Biological Resource Center for distributing Arabidopsis seeds that are crucial to the advancement of work in this field. This work was supported, in part, by the New Energy and Industrial Technology Development Organization as part of a project called, “Development of Fundamental Technologies for Controlling the Material Production Process of Plants” and by grants-in-aid from the Ministry of Education, Culture, Sports, Science, and Technology, Japan.
Abbreviations
- GSL
glucosinolate
- MAM
methylthioalkylmalate synthase
- MeJA
methyl jasmonate.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Microarray data have been deposited in ArrayExpress database (accession nos. E-ATMX-6, E-ATMX-7, and E-ATMX-8).
This article contains supporting information online at www.pnas.org/cgi/content/full/0611629104/DC1.
References
- 1.Fahey JW, Haristoy X, Dolan PM, Kensler TW, Scholtus I, Stephenson KK, Talalay P, Lozniewski A. Proc Natl Acad Sci USA. 2002;99:7610–7615. doi: 10.1073/pnas.112203099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Talalay P, Fahey JW. J Nutr. 2001;131:3027S–3033S. doi: 10.1093/jn/131.11.3027S. [DOI] [PubMed] [Google Scholar]
- 3.Fahey JW, Zhang Y, Talalay P. Proc Natl Acad Sci USA. 1997;94:10367–10372. doi: 10.1073/pnas.94.19.10367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hill J, Nelson E, Tilman D, Polasky S, Tiffany D. Proc Natl Acad Sci USA. 2006;103:11206–11210. doi: 10.1073/pnas.0604600103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grubb CD, Abel S. Trends Plants Sci. 2006;11:89–100. doi: 10.1016/j.tplants.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 6.Halkier BA, Gershenzon J. Annu Rev Plant Biol. 2006;57:303–333. doi: 10.1146/annurev.arplant.57.032905.105228. [DOI] [PubMed] [Google Scholar]
- 7.Balazsi G, Barabasi AL, Oltvai ZN. Proc Natl Acad Sci USA. 2005;102:7841–7846. doi: 10.1073/pnas.0500365102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ihmels J, Levy R, Barkai N. Nat Biotechnol. 2004;22:86–92. doi: 10.1038/nbt918. [DOI] [PubMed] [Google Scholar]
- 9.Gachon CM, Langlois-Meurinne M, Henry Y, Saindrenan P. Plant Mol Biol. 2005;58:229–245. doi: 10.1007/s11103-005-5346-5. [DOI] [PubMed] [Google Scholar]
- 10.Lisso J, Steinhauser D, Altmann T, Kopka J, Mussig C. Nucleic Acids Res. 2005;33:2685–2696. doi: 10.1093/nar/gki566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Persson S, Wei H, Milne J, Page GP, Somerville CR. Proc Natl Acad Sci USA. 2005;102:8633–8638. doi: 10.1073/pnas.0503392102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rautengarten C, Steinhauser D, Bussis D, Stintzi A, Schaller A, Kopka J, Altmann T. PLoS Comput Biol. 2005;1:e40. doi: 10.1371/journal.pcbi.0010040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei H, Persson S, Mehta T, Srinivasasainagendra V, Chen L, Page GP, Somerville C, Loraine A. Plant Physiol. 2006;142:762–774. doi: 10.1104/pp.106.080358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Obayashi T, Kinoshita K, Nakai K, Shibaoka M, Hayashi S, Saeki M, Shibata D, Saito K, Ohta H. Nucleic Acids Res. 2007;35:D863–D869. doi: 10.1093/nar/gkl783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirai MY, Klein M, Fujikawa Y, Yano M, Goodenowe DB, Yamazaki Y, Kanaya S, Nakamura Y, Kitayama M, Suzuki H, et al. J Biol Chem. 2005;280:25590–25595. doi: 10.1074/jbc.M502332200. [DOI] [PubMed] [Google Scholar]
- 16.Hirai MY, Yano M, Goodenowe DB, Kanaya S, Kimura T, Awazuhara M, Arita M, Fujiwara T, Saito K. Proc Natl Acad Sci USA. 2004;101:10205–10210. doi: 10.1073/pnas.0403218101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Celenza JL, Quiel JA, Smolen GA, Merrikh H, Silvestro AR, Normanly J, Bender J. Plant Physiol. 2005;137:253–262. doi: 10.1104/pp.104.054395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levy M, Wang Q, Kaspi R, Parrella MP, Abel S. Plant J. 2005;43:79–96. doi: 10.1111/j.1365-313X.2005.02435.x. [DOI] [PubMed] [Google Scholar]
- 19.Kim JH, Durrett TP, Last RL, Jander G. Plant Mol Biol. 2004;54:671–682. doi: 10.1023/B:PLAN.0000040897.49151.98. [DOI] [PubMed] [Google Scholar]
- 20.Skirycz A, Reichelt M, Burow M, Birkemeyer C, Rolcik J, Kopka J, Zanor MI, Gershenzon J, Strnad M, Szopa J, et al. Plant J. 2006;47:10–24. doi: 10.1111/j.1365-313X.2006.02767.x. [DOI] [PubMed] [Google Scholar]
- 21.Mikkelsen MD, Petersen BL, Glawischnig E, Jensen AB, Andreasson E, Halkier BA. Plant Physiol. 2003;131:298–308. doi: 10.1104/pp.011015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kroymann J, Textor S, Tokuhisa JG, Falk KL, Bartram S, Gershenzon J, Mitchell-Olds T. Plant Physiol. 2001;127:1077–1088. [PMC free article] [PubMed] [Google Scholar]
- 23.Field B, Cardon G, Traka M, Botterman J, Vancanneyt G, Mithen R. Plant Physiol. 2004;135:828–839. doi: 10.1104/pp.104.039347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schuster J, Binder S. Plant Mol Biol. 2005;57:241–254. doi: 10.1007/s11103-004-7533-1. [DOI] [PubMed] [Google Scholar]
- 25.Schuster J, Knill T, Reichelt M, Gershenzon J, Binder S. Plant Cell. 2006;18:2664–2679. doi: 10.1105/tpc.105.039339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ravanel S, Block MA, Rippert P, Jabrin S, Curien G, Rebeille F, Douce R. J Biol Chem. 2004;279:22548–22557. doi: 10.1074/jbc.M313250200. [DOI] [PubMed] [Google Scholar]
- 27.Kliebenstein DJ, Lambrix VM, Reichelt M, Gershenzon J, Mitchell-Olds T. Plant Cell. 2001;13:681–693. doi: 10.1105/tpc.13.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stracke R, Werber M, Weisshaar B. Curr Opin Plant Biol. 2001;4:447–456. doi: 10.1016/s1369-5266(00)00199-0. [DOI] [PubMed] [Google Scholar]
- 29.Taki N, Sasaki-Sekimoto Y, Obayashi T, Kikuta A, Kobayashi K, Ainai T, Yagi K, Sakurai N, Suzuki H, Masuda T, et al. Plant Physiol. 2005;139:1268–1283. doi: 10.1104/pp.105.067058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saito K. Plant Physiol. 2004;136:2443–2450. doi: 10.1104/pp.104.046755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maruyama-Nakashita A, Nakamura Y, Tohge T, Saito K, Takahashi H. Plant Cell. 2006;18:3235–3251. doi: 10.1105/tpc.106.046458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rischer H, Oresic M, Seppanen-Laakso T, Katajamaa M, Lammertyn F, Ardiles-Diaz W, Van Montagu MC, Inze D, Oksman-Caldentey KM, Goossens A. Proc Natl Acad Sci USA. 2006;103:5614–5619. doi: 10.1073/pnas.0601027103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tokimatsu T, Sakurai N, Suzuki H, Ohta H, Nishitani K, Koyama T, Umezawa T, Misawa N, Saito K, Shibata D. Plant Physiol. 2005;138:1289–1300. doi: 10.1104/pp.105.060525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davuluri RV, Sun H, Palaniswamy SK, Matthews N, Molina C, Kurtz M, Grotewold E. BMC Bioinformatics. 2003;4:25. doi: 10.1186/1471-2105-4-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo A, He K, Liu D, Bai S, Gu X, Wei L, Luo J. Bioinformatics. 2005;21:2568–2569. doi: 10.1093/bioinformatics/bti334. [DOI] [PubMed] [Google Scholar]
- 36.Iida K, Seki M, Sakurai T, Satou M, Akiyama K, Toyoda T, Konagaya A, Shinozaki K. DNA Res. 2005;12:247–256. doi: 10.1093/dnares/dsi011. [DOI] [PubMed] [Google Scholar]
- 37.Sakurai T, Satou M, Akiyama K, Iida K, Seki M, Kuromori T, Ito T, Konagaya A, Toyoda T, Shinozaki K. Nucleic Acids Res. 2005;33:D647–650. doi: 10.1093/nar/gki014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Batagelj V, Mrvar A. Connections. 1998;21:47–57. [Google Scholar]
- 39.An G, Ebert PR, Mitra A, Ha SB. Binary Vectors. Dordrecht, Germany: Kluwer; 1988. [Google Scholar]
- 40.Clough SJ, Bent AF. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 41.Axelos M, Curie C, Mazzolini L, Bardet C, Lescure B. Plant Physiol Biochem. 1992;30:123–128. [Google Scholar]
- 42.Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 43.Goda H, Shimada Y, Asami T, Fujioka S, Yoshida S. Plant Physiol. 2002;130:1319–1334. doi: 10.1104/pp.011254. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.