Abstract
Small conductance calcium-activated potassium channels (SK, KCa) are a family of voltage-independent K+ channels with a distinct physiology and pharmacology. The bee venom toxin apamin inhibits exclusively the three cloned SK channel subtypes (SK1, SK2 and SK3) with different affinity, highest for SK2, lowest for SK1 and intermediate for SK3 channels. The high selectivity of apamin made it a valuable tool to study the molecular makeup and function of native SK channels. Three amino acids located in the outer vestibule of the pore are of particular importance for the different apamin sensitivities of SK channels. Chimeric SK1 channels, enabling the homomeric expression of the rat SK1 (rSK1) subunit and containing the core domain (S1-S6) of rSK1, are apamin insensitive. By contrast, channels formed by the human orthologue hSK1 are sensitive to apamin. This finding hinted to the involvement of regions beyond the pore as determinants of apamin sensitivity, since hSK1 and rSK1 have an identical amino acid sequence in the pore region. Here we investigated which parts of the channels outside the pore region are important for apamin sensitivity by constructing chimeras between apamin insensitive and sensitive SK channel subunits and by introducing point mutations. We demonstrate that a single amino acid situated in the extracellular loop between the transmembrane segments S3 and S4 has a major impact on apamin sensitivity. Our findings enabled us to convert the hSK1 channel into a channel that was as sensitive for apamin as SK2, the SK channel with the highest sensitivity.
Ca2+-activated K+ channels (KCa)1 are activated by rises in intracellular Ca2+. The KCa potassium channel family comprises of at least three subfamilies, KCa1-3 (1). Channels containing the KCa1.1 α-subunit (BK channels) have large single channel conductance and are maximally activated by micromolar concentrations of intracellular free calcium and concurrent depolarization (2). Their kinetic and pharmacological properties are modified upon assembly with membrane standing ß-subunits (3). The KCa2 subfamily of small-conductance Ca2+-activated K+ channels, also known as SK channels, has three closely related members SK1 (KCa2.1), SK2 (KCa2.2) and SK3 (KCa2.3), which are characterized by a small single channel conductance. The IK channel (KCa3.1) shows an intermediate single channel conductance. Both SK and IK channels are voltage independent and activated by submicromolar concentrations of intracellular free Ca2+. The gating of SK and IK channels is induced upon Ca2+ binding to calmodulin, which is constitutively bound to each channel subunit. Ca2+ binding to calmodulin induces a conformational change, which leads to the opening of these channels (4-6).
SK channels, predominantly of the SK2 type, have been identified in sensory systems, such as the retina (7) and the cochlear inner and outer hair cells (8, 9). SK channels have also been described in heart (10, 11), liver (12, 13), skeletal muscle (14, 15) and visceral smooth muscle (16), including the urinary bladder (17). All three SK channels are expressed in the brain and show a differential distribution (18). The SK channel mediated current (IAHP) has been studied in detail in various regions of the central nervous system (19-21). The IAHP regulates membrane excitability, increases the precision of neuronal firing (22-30), and modulates synaptic plasticity by regulating excitatory synaptic transmission in the amygdala (31) and the hippocampus (32). Inhibition of SK channels facilitates hippocampal-independent (33, 34) as well as dependent (35) learning and improves memory performance (33, 34, 36, 37).
The unequivocal, molecular correlation between native KCa currents with their corresponding channel subunit(s) was possible because of the availability of highly specific blockers, in particular peptide toxins, which selectively inhibit these channels (37, 38). The 18-amino acid bee venom toxin apamin has proven to be extremely valuable as a specific blocker for which the only known receptors are the SK channels. Besides apamin, the scorpion toxins scyllatoxin (39), P05 (39) and tamapin (40) have the ability to inhibit SK-mediated currents, whereas other scorpion toxins (maurotoxin, Pi1, P01 and Tsk) compete with 125I-apamin binding (reviewed in 37), but do not inhibit SK2 or SK3 channels (39).
The rat SK1 (rSK1) subunits, unlike the human SK1 (hSK1), do not form functional homomeric SK channels (41, 42), but form heteromeric channels together with the SK2 subunit in heterologous expression systems (41). The construction of chimeric rSK1 channels, comprising the amino- and/or carboxyl-terminal regions of rSK2 or hSK1, enabled the expression of homomeric rSK1-like channels displaying more than a 25 fold reduction in apamin sensitivity compared to hSK1 (42). This finding was rather surprising, as hSK1 and rSK1 share an identical primary sequence within the pore region, where three amino acids have been identified as determinants of the apamin sensitivity (43). In this study, to test the hypothesis that amino acids outside the pore region contribute to apamin binding, chimera were generated to identify new regions in the SK channels involved in determining sensitivity to apamin. The introduction of point mutations and their analysis revealed the existence of an amino acid beyond the pore region that crucially influences SK channel sensitivity for apamin. This finding provides a new insight into the molecular determinants for the apamin receptor and extends our understanding of how the difference in sensitivity of the different SK channel subunits is generated. Furthermore, this is to our knowledge the first description of a toxin that inhibits potassium channels by interacting simultaneously with amino acids inside and outside the pore region. A detailed understanding of the structural basis of the differences in the pharmacological profile of SK channel subtypes is essential for the rational development of subunit specific inhibitors.
MATERIAL AND METHODS
DNA constructs
The coding regions of hSK1, rSK1 and rSK2 (GenBankTM accession numbers: NM_002248, NM_019313, NM_019314) were cloned into pcDNA3 or pcDNA5/FRT (40, 42). Point mutations and chimera were generated by splicing of overlap extension (44). The range of amino acids combined is given in plain numbers for residues corresponding to rSK1, bold numbers for residues corresponding to hSK1 and underlined numbers for residues corresponding to rSK2. The chimera rSK1NSK2-CSK2 contains the transmembrane domains and the pore of rSK1 and the NH2 and COOH termini of rSK2. The generation of this chimera (1-121/89-372/406-580) has been described previously (42). L1-hSK1 (1-121/89-120/125-143/140-372/406-580) was generated by substituting the extracellular region between transmembrane segments S1 and S2 in rSK1NSK2-CSK2 for the corresponding region in hSK1. In L3-hSK1 (1-121/89-200/205-227/224-372/406-580) the extracellular region between transmembrane segments S3 and S4 of rSK1NSK2-CSK2 was exchanged for the corresponding region of hSK1. Finally, chimera hSK1-SK2-hSK1 (1-122/152-256/228-543) was created by exchanging the region from the end of transmembrane segment S1 until the beginning of transmembrane segment S4 of hSK1 for the corresponding region in rSK2. In the text, hSK1-QDN refers to point mutation hSK1-KEH310/312/339QDN, and hSK1-QDN+S refers to hSK1-KEH310/312/339QDN+T216S.
All modified sequences were verified by sequencing with the BigDye Terminator Cycle Sequencing Kit and an ABI3100 Avant Genetic-Analyzer (Applied Biosystems).
Maintenance and Transient Transfection of HEK293 and CHO Cells
Cell lines were cultured in a humidified atmosphere (5% CO2, 95% air) at 37°C. HEK293 cells were grown in Dulbecco's modified Eagle's medium/F12 with 2 mM L-glutamine. CHO cell lines were grown in F-12 Nutrient Mixture (Ham) with GlutaMAX™ I. Both media were supplemented with 10% fetal calf serum and penicillin/streptomycin. For transient transfections cells were transfected using Lipofectamine 2000 (Invitrogen). 2 μg of plasmid DNA encoding one of the SK channel constructs were co-transfected with 0.5 μg of pEGFP-C2. Recordings were performed 1 to 3 days after transfection.
Generation and Maintenance of Cell Lines Stably Expressing SK Channels
For SK channel constructs hSK1-T216S, hSK1 and hSK1(QDN+S) stable cell lines were generated. Constructs were cloned into the pcDNA5/FRT vector. HEK Flp-In cells (Invitrogen) were grown under the same conditions as HEK293, but in the presence of Zeocin (100 μg/ml). Cells were transfected using Lipofectamine 2000 or by CaPO4 precipitation. For Lipofectamine 2000 transfection cells were plated into 60 mm culture dishes and co-transfected with a mixture of 0.7 μg expression construct and 7.3 μg pOG44 plasmid according to the manufacturer's protocol. For the CaPO4 precipitation method cells were plated into 90 mm culture dishes and co-transfected with a mixture of 2 μg expression construct and 23 μg pOG44 plasmid. For polyclonal selection of cells, Zeocin was replaced by Hygromycin B (75 μg/ml). Hygromycin-resistent cells were pooled and maintained in the presence of Hygromycin B.
Electrophysiology
Whole cell patch clamp recordings of HEK cells transiently transfected or stably expressing SK channel constructs were performed. Transfected cells were grown on coverslips, placed in a recording chamber and perfused at a flow rate of 1 ml/min. Currents were recorded using an EPC-10 amplifier (HEKA, Lambrecht, Germany). All recordings were performed at room temperature (22°C). Data were acquired with Pulse/Pulsefit software (HEKA, Lambrecht, Germany). Pipettes were pulled from borosilicate glass with a horizontal patch electrode puller (Zeitz-Instruments GmbH, Muenchen, Germany) and had a resistance of 1.8-3 MΩ when filled with intracellular solution (see below). After gigaseal formation, the fast capacitative transients were automatically compensated.
SK channels were activated by whole-cell dialysis with an intracellular solution containing (in mM): 130 KCl, 10 HEPES, 10 EGTA at pH 7.2. MgCl2 and CaCl2 were added to obtain free magnesium and calcium concentrations of 1 mM and 1 μM, respectively using EqCal (Biosoft, Cambridge, UK). Recordings were performed in a high potassium extracellular solution containing (in mM): 144 KCl, 10 HEPES, 2 CaCl2, 1 MgCl2, 10 D-Glucose, pH 7.4 with KOH, or a low potassium extracellular solution containing (in mM): 4 KCl, 140 NaCl, 10 HEPES, 2 CaCl2, 1 MgCl2, 10 D-Glucose, pH 7.4 with NaOH. The high potassium solution was used in most experiments and inward currents were analyzed in order to avoid problems due to SK channel rectification and activation of endogenous voltage-gated K+ channels at positive potentials. In tetraethlyammonium (TEA) containing solutions, TEA-Cl replaced NaCl in the low potassium extracellular solution. Membrane currents were recorded upon application of 400-ms voltage ramps from −140 to +40 mV, repeated every 10 s. Given the large size of some currents, voltages were corrected off-line for the occurring voltage-clamp error. Cells that showed >20% residual current after application of 100 nM apamin were excluded from the analysis. Concentration-response curves were analyzed using a Langmuir-Hill equation of the type I/Imax = (1-a0)/[1+([Inhibitor]/IC50)h]+a0, where h corresponds to the Hill coefficient and IC50 to the concentration of inhibitor that produces a 50% inhibition. The value for curve minimum (a0) was not fixed. The data points for the concentration-response curves were obtained from different cells and therefore treated as independent replicates. The fraction of unblocked current (I/Imax) was calculated at −80 mV or −20 mV. Curve fitting was performed by using a least square fitting routine. All data were analyzed with Igor Pro 5.0 (WaveMetrics, Lake Oswego, OR) or Prism 4.0 (GraphPad Software Inc., San Diego, CA). Values are given as mean ± S.E. or 95% confidence interval (95% CI). Unpaired two-tailed Student's t test was used for statistical comparisons between groups.
RESULTS
Amino acids outside the pore affect apamin sensitivity
The three members of the SK channel family, SK1, SK2 and SK3, can be distinguished by their sensitivity to apamin. Homomeric SK2 channels from rat (rSK2), mouse (mSK2) and human (hSK2) are the most apamin sensitive and homomeric hSK1 channels are the least sensitive channels of the KCa2 subfamily (21, 37, 45-47). In heterologous expression systems, the rSK1 α-subunit does not form functional homomeric channels (41, 42), but forms functional heteromeric channel with SK2 (41). The chimeric rSK1 α-subunit (rSK1NSK2-CSK2), which contains the S1-S6 region of rSK1 with amino- and carboxyl-termini of rSK2, assembles into homomers (42). This chimera is ideally suited to obtain information on the pharmacological properties of the rSK1 subunit.
The hSK1 channel and the rSK1NSK2-CSK2 chimera were transiently expressed in HEK293 cells. In solutions with nearly identical intra- and extracellular potassium concentrations, a calcium activated potassium current was elicited by voltage ramps ranging from −140 to +40 mV in the presence of 1 μM intracellular free calcium. The observed current showed a reversal potential close to 0 mV and was shifted to hyperpolarized values upon lowering the extracellular potassium concentration to 4 mM (Fig. 1A). As expected the observed reversal potentials corresponded to values predicted by the Nernst equation for a potassium conductance.
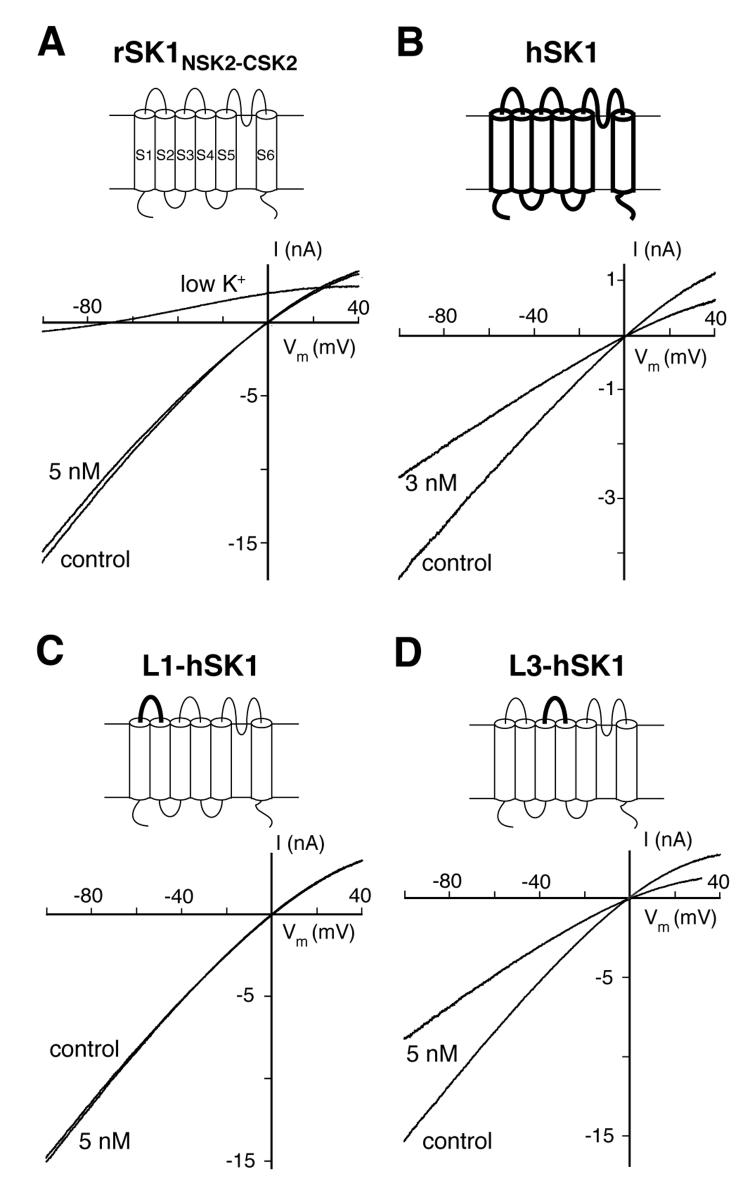
Fig. 1.
Amino acids outside the pore affect apamin sensitivity. Schematic drawings illustrate the SK α-subunit constructs. Thin lines indicate regions of the rSK1NSK2-CSK2 α-subunit and thick lines correspond to regions of the hSK1 α-subunit. HEK293 cells expressing SK channels and chimeras were measured in the whole-cell configuration in the presence of 1 μM intracellular Ca2+. Currents elicited by voltage ramps from −100 to 40 mV (duration: 400 ms) are shown. A, Representative current traces of HEK293 cells expressing rSK1NSK2-CSK2 before and after application of 5 nM apamin. The current was not inhibited by 5 nM apamin (residual current 99 ± 2%, n=3). An extracellular solution containing 4 mM K+ produced the expected left shift of the current-voltage relationship (low K+). B, for comparison, currents generated by hSK1 were inhibited by 42 ± 1% (n=6) by 3 nM apamin. C and D, L1-hSK1 and L3-hSK1 were generated by substituting the extracellular regions between transmembrane segments S1 and S2 (L1) or S3 and S4 (L3) in rSK1NSK2-CSK2 for the corresponding regions in hSK1. C, L1-hSK1 currents were unaffected by the application of 5 nM apamin (residual current 99 ± 1%, n=4), like rSK1NSK2-CSK2 currents. D, Currents generated by the chimera L3-hSK1 were inhibited by to 44 ± 5% (n=5) by 5 nM apamin.
The primary sequence of the hSK1 α-subunit and the rSK1NSK2-CSK2 chimera is identical between transmembrane segments S5 and S6. This region comprises the pore of potassium channels and has been ascribed the structural basis for the different apamin sensitivity of members of the KCa2 family (39, 43). However the current generated by the rSK1NSK2-CSK2 chimera was not inhibited by 5 nM apamin (Fig. 1A), whereas 3 nM apamin inhibited hSK1 by nearly 50% (Fig. 1B, 42 ± 1%, n=6). The identical pore regions of hSK1 and rSK1NSK2-CSK2 and the observed intriguing difference in apamin sensitivity (Fig. 1, A and B) strongly suggest the involvement of amino acids located outside the pore domain for apamin binding.
The most obvious regions potentially harbouring these amino acids are the hydrophilic extracellular loop L1, located between transmembrane segments S1 and S2, and L3, located between transmembrane segments S3 and S4. Two chimera, L1-hSK1 and L3-hSK1, were created to analyse these regions. L1-hSK1 was generated by substituting the extracellular loop between the transmembrane segments S1 and S2 in rSK1NSK2-CSK2 with the corresponding region of hSK1. In L3-hSK1 the extracellular loop between S3 and S4 (S3-S4 loop) of rSK1NSK2-CSK2 was exchanged for the corresponding region of hSK1. The chimera were transiently expressed in HEK293 cells and currents were elicited as described above. Both chimera assembled into functional homomeric channels (Fig. 1, C and D). L1-hSK1 showed, like rSK1NSK2-CSK2, only a marginal inhibition upon application of 5 nM apamin (1 ± 1%, n=4, Fig. 1C). In contrast, 5 nM apamin suppressed around 50% of the current generated by L3-hSK1 (44 ± 5%, n=5, Fig. 1D), thereby demonstrating an apamin sensitivity similar to hSK1. These results show that the extracellular S3-S4 loop (L3) influences apamin sensitivity and is most likely sufficient to explain the observed difference in apamin sensitivity between hSK1 and rSK1.
The hydrophilic extracellular loop L3 between transmembrane segments S3 and S4 influences apamin binding
Since amino acids located outside the pore influence the apamin sensitivity of the rSK1NSK2-CSK2 chimera (Fig. 1, A-D), we wondered whether it is a general feature of SK channels that regions outside the pore influence their sensitivity towards apamin. To address this question, we generated a chimera between hSK1 and SK2, which differ more than 100 fold in apamin sensitivity (46, 47). In this chimera, hSK1-SK2-hSK1, the region from the end of the transmembrane segment S1 until the beginning of the transmembrane segment S4 of hSK1 was exchanged for the corresponding region of SK2 (Fig. 2A). If only amino acids in the pore region (between S5 and S6) are responsible for the apamin sensitivity of SK channels, the hSK1-SK2-hSK1 chimera should display an apamin sensitivity comparable to hSK1. If, however, amino acids outside the pore region influence the apamin sensitivity, hSK1-SK2-hSK1 should show an increased sensitivity when compared to hSK1. To test these predictions, hSK1-SK2-hSK1 was transiently expressed in HEK293 cells and calcium-activated potassium currents were elicited as described earlier (see also Material and Methods). Apamin at a concentration of 300 pM was applied and suppressed the current generated by the hSK1-SK2-hSK1 chimera by more than 50% (Fig. 2, A and D, 58 ± 3%, n=5). The concentration of 300 pM apamin was used because it does not affect hSK1 (Fig. 2, B and D) but inhibits SK2 channels nearly completely (Fig. 2, C and D). The IC50 value obtained for the hSK1-SK2-hSK1 chimera (Fig. 2E) is 124 pM (95% CI: 100 pM to 156 pM). The clear inhibition of the hSK1-SK2-hSK1 current by low concentrations of apamin shows that the exchanged region contributes to the difference in apamin sensitivity between hSK1 and SK2, and thereby supports the hypothesis that amino acids located beyond the pore region influence the apamin sensitivity in SK channels in general. The observations made for the L1-hSK1 and L3-hSK1 constructs were therefore not an oddity only observed within the rSK1NSK2-CSK2 chimera.
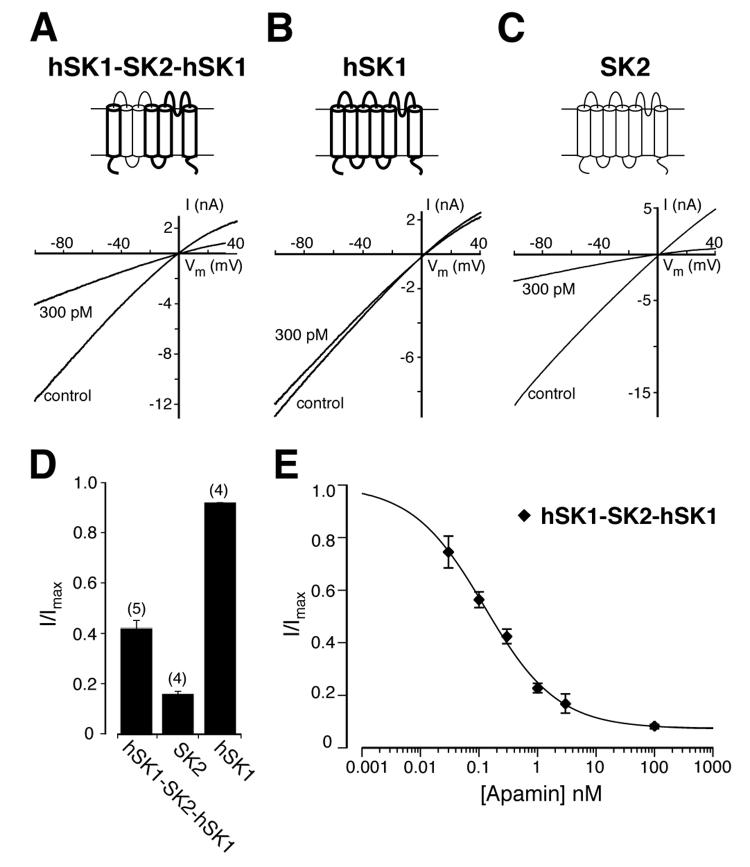
Fig. 2.
The apamin sensitivity of hSK1 is increased after exchange of the extracellular loops L2 and L3. Schematic drawings illustrate the SK α-subunit constructs. Thin lines indicate regions of the rSK2 α-subunit and thick lines indicate regions of the hSK1 α-subunit. Representative current traces of the hSK1-SK2-hSK1 chimera (A), hSK1 (B), and SK2 (C) before and after application of 300 pM apamin were obtained using symmetrical potassium concentrations. D, bar diagram indicating the fraction of residual current (I/Imax) in the presence of 300 pM apamin for hSK1-SK2-hSK1 (I/Imax = 0.42 ± 0.03, n=5), SK2 (I/Imax = 0.16 ± 0.01, n=4), and hSK1 (I/Imax = 0.92 ± 0.01, n=4) with P<0.0001 for the differences between hSK1-SK2-hSK1 and SK2 or hSK1. E, concentration-response curve for the inhibition of hSK1-SK2-hSK1 (◆) by apamin. Data points (n ≥ 4) were fitted with a Hill-Langmuir equation and yielded an IC50 value of 124 pM (95% CI: 100 pM to 156 pM).
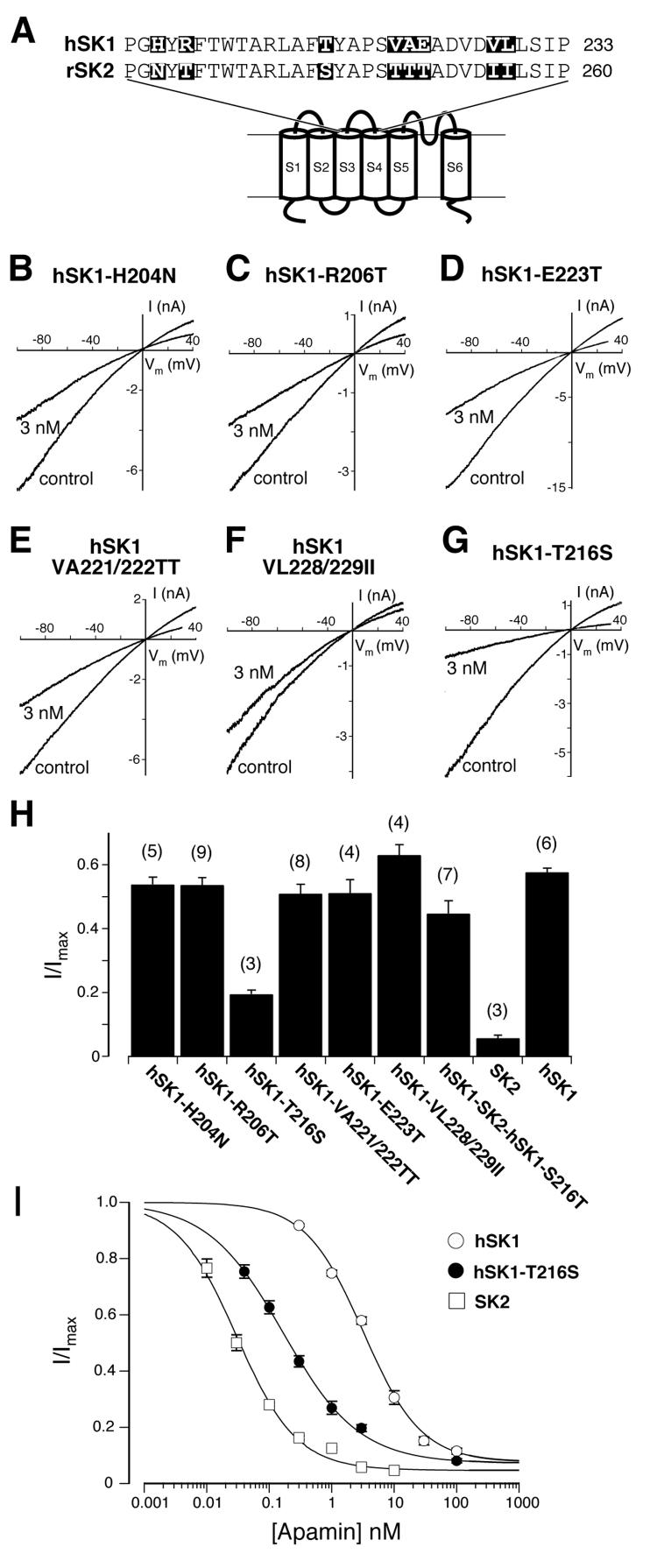
Next we were interested in identifying the amino acids located outside the pore region that influence the apamin sensitivity. Once more, we used chimeras of SK2 and hSK1, the most and the least apamin sensitive SK channel subunits, respectively. Based on the observations made with the chimeric SK channels (Fig. 1D and Fig. 2A), we decided to focus on differences in the amino acid sequences of hSK1 and SK2 in the hydrophilic extracellular loop L3 (Fig. 3A). The alignment shows differences between the amino acid sequences of hSK1 and SK2 in eight positions. By introducing single or double point mutations, the amino acids differing at these eight positions in hSK1 were replaced by the corresponding amino acids found in SK2. Consequently, if any of these amino acids influence the apamin sensitivity in SK2, the corresponding hSK1 mutant should show a higher sensitivity for apamin when compared to hSK1. At three positions, a charged amino acid in hSK1 was replaced for an uncharged amino acid present in SK2 (hSK1-H204N, hSK1-R206T, hSK1-E223T), or the two hydrophobic residues in hSK1 were replaced together for the two hydrophilic residues found at the corresponding positions in SK2 (hSK1-VA221/222TT). At the other three positions the exchanges were more subtle, as hydrophobic or hydrophilic amino acids were exchanged for similar ones (hSK1-T216S and hSK1-VL228/229II). Upon transfection into HEK293 cells, all point mutants expressed and generated calcium-activated potassium currents of comparable amplitude. Current densities at −80 mV for the lowest and highest expressing mutants ranged from 0.66 ± 0.17 nA/pF to 1.39 ± 0.36 nA/pF. To test whether the introduced mutations increased the apamin sensitivity of the mutant with respect to hSK1, 3 nM apamin was added to the extracellular solution. This concentration of apamin was chosen because it produces a half-maximal inhibition of the hSK1 current (Fig. 1B) and guarantees that even small changes in sensitivity of the mutated hSK1 channels are clearly measurable. Changing any of the three charged amino acids in hSK1 into the corresponding uncharged amino acid of SK2 did not change the sensitivity of the mutated hSK1. For all three mutations, hSK1-H204N (Fig. 3B and H), hSK1-R206T (Fig. 3, C and H) and hSK1-E223T (Fig. 3, D and H), 3 nM apamin still produced a half-maximal inhibition. Also in the two double mutants, hSK1-VA221/222TT (Fig. 3, E and H) and hSK1-VL228/229II (Fig. 3, F and H), the mutations did not render the channels more apamin sensitive. Only the substitution of threonine for serine in hSK1-T216S resulted in a channel that showed a large increase in apamin sensitivity. 80% of the current was inhibited by 3 nM apamin (Fig 3, G and H). The observed apamin sensitivity, reported as the relative amount of residual current (I/Imax) in the presence of 3 nM apamin, is shown in Fig. 3H for the six mutations in comparison to hSK1 and SK2. The differences between hSK1-T216S and hSK1, any of the six non affected point mutations or SK2 are all highly significant with P<0.01. Fig. 3I shows the concentration-response curve for hSK1-T216S in comparison with those for hSK1 and SK2. The IC50 value obtained for hSK1-T216S is 167 pM (95% CI: 145 pM to 193 pM). This makes hSK1-T216S 20 times more sensitive for apamin than hSK1, which shows an IC50 value of 3.2 nM (95% CI: 2.7 nM to 3.8 nM), and 5 times less sensitive for apamin than SK2, for which we measured an IC50 of 30 pM (95% CI: 27 pM to 33 pM). The IC50 value obtained for hSK1-T216S (Fig. 3I) is equivalent to that of the hSK1-SK2-hSK1 chimera (Fig. 2E). Furthermore, we replaced the serine at position 216 in the hSK1-SK2-hSK1 chimera by a threonine (hSK1-SK2-hSK1-S216T) to examine the importance of the threonine to serine switch, found in hSK1-T216S, with respect to the potential influence of all other amino acid residues in the S3-S4 loop. The apamin sensitivity of hSK1-SK2-hSK1-S216T is reduced and is comparable to that of hSK1 (Fig. 3H). These results show that a single amino acid located in the extracellular loop between S3 and S4 accounts for most of the difference in apamin sensitivity between hSK1 and SK2.
Fig. 3.
A single amino acid exchange in the extracelluar loop L3 increases the apamin sensitivity of hSK1 by one order of magnitude. A, sequence alignment of the extracellular loop L3 between the transmembrane segments S3 and S4 of hSK1 and SK2. Amino acids that differ in the two sequences are highlighted by black boxes. B - G, The indicated point mutations were introduced into hSK1, where amino acids were exchanged with those found at corresponding positions in SK2. Representative current traces of HEK293 cells expressing mutated channels before (control) and after application of 3 nM apamin for the various hSK1 mutants. H, The fraction of residual current (I/Imax) recorded for each mutant is summarized in a bar diagram. hSK1-H204N (I/Imax = 0.54 ± 0.02, n=5), hSK1-R206T (I/Imax = 0.54 ± 0.02, n=9), hSK1-T216S (I/Imax = 0.20 ± 0.01, n=3), hSK1-VA221/222TT (I/Imax = 0.51 ± 0.03, n=8), hSK1-E223T (I/Imax = 0.52 ± 0.04, n=4), hSK1-VL228/229II (I/Imax = 0.64 ± 0.03, n=4), hSK1-SK2-hSK1-S216T (I/Imax = 0.45 ± 0.04, n=7), as well as SK2 (I/Imax = 0.06 ± 0.01, n=3) and hSK1 (I/Imax = 0.58 ± 0.01, n=6). The differences between hSK1-T216S and all tested channels shown are highly significant (P<0.01). I, concentration-response curves for the inhibition of SK1 (○), hSK1-T216S (●), and SK2 (□) by apamin. Data points (n ≥ 3) were fitted with a Hill-Langmuir equation and yielded the following IC50 values: hSK1 (IC50 = 3.2 nM, 95% CI: 2.7 nM to 3.8 nM), hSK1-T216S (IC50 = 167 pM, 95% CI: 145 pM to 193 pM) and SK2 (IC50 = 30 pM, 95% CI: 27 pM to 33 pM).
The combination of mutations in the pore region with the amino acid identified in L3 reconstitutes the high affinity receptor of SK2 in hSK1
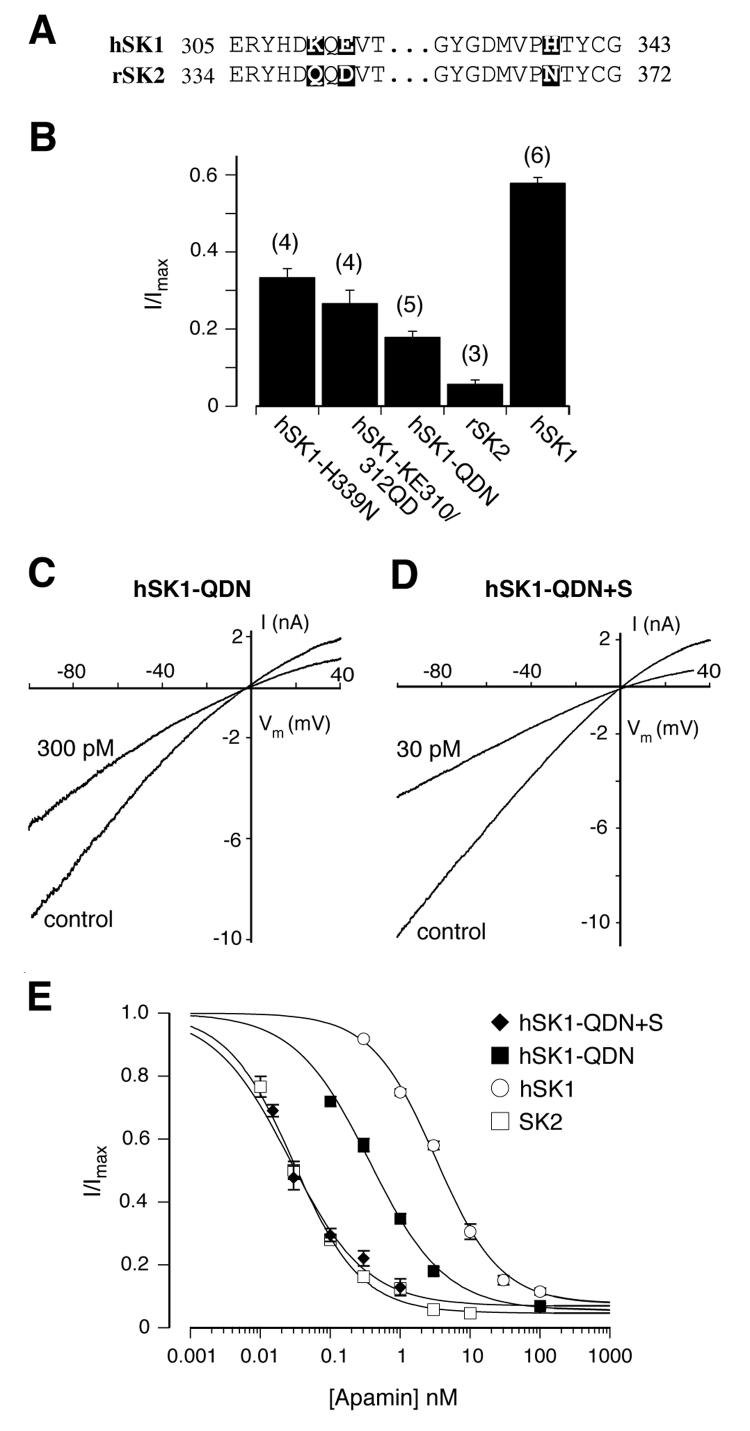
Electrophysiological recordings on chimeric SK channels and point mutations in Xenopus oocytes have demonstrated the importance of three amino acids in the outer vestibule of the pore for the differences in apamin sensitivity between hSK1 and SK2 (39, 43). We transiently expressed the same mutants in HEK293 cells and analyzed their apamin sensitivity under our recording conditions. In agreement with the observation made by Ishii and colleagues in X. oocytes (in ref. 43), both mutants, hSK1-H339N and hSK1-KE310/312QD, generated currents and showed a substantial increase in apamin sensitivity when compared to hSK1. 3 nM apamin inhibited 66 ± 2% (n=4) of the hSK1-H339N and 73 ± 3% (n=4) of the hSK1 KE310/312QD generated current (Fig. 4B). The combination of these mutations into a single hSK1 α-subunit, hSK1-QDN, showed a further increase in apamin sensitivity (Fig 4, B and C). The concentration-response curve for hSK1-QDN (Fig. 4E) gives an IC50 value of 366 pM (95% CI: 326 pM to 426 pM), which makes this mutant 9 times more sensitive to apamin than hSK1. However hSK1-QDN is still less sensitive than SK2 by more than one order of magnitude. Given our observation that the exchange of a single amino acid in the S3-S4 loop (L3) of hSK1 had a more prominent effect on its apamin sensitivity than the three mutations in the pore region, we wondered whether combining the pore and L3 mutations into one α-subunit would fully convert the apamin sensitivity of hSK1 into that of SK2. hSK1-QDN+S generated calcium activated potassium currents that were half maximally blocked by 30 pM apamin, indicating an apamin sensitivity comparable to SK2 (Fig. 4D). The concentration-response curve for hSK1-QDN+S (Fig. 4E) resulted in an IC50 of 26 pM (95% CI: 22 pM to 32 pM), very similar to the IC50 of 30 pM for SK2 (95% CI: 27 pM to 33 pM). The similarity of these IC50 values is supported by the superimposition of the concentration-response curves for hSK1-QDN+S and SK2, which are clearly distinct from the hSK1 and hSK1-QDN curves (Fig. 4E). This analysis demonstrates unequivocally that amino acids within the pore region as well as the identified amino acid in the extracellular S3-S4 loop (L3) are necessary to generate a high affinity receptor for apamin.
Fig. 4.
The combination of the point mutation T216S and the triple pore mutations KEH310/312/339QDN reconstitutes the apamin sensitivity of SK2 in hSK1. A, sequence alignment of the pore region of hSK1 and SK2. Amino acids from the middle part of the pore region are identical between hSK1 and SK2 and are omitted. Amino acids that differ in the two sequences are boxed in black. B, bar diagram summarizing the inhibitory effect of 3 nM apamin. The fraction of residual current (I/Imax) was 0.34 ± 0.02 for hSK1-H339N (n=4), 0.27 ± 0.03 for hSK1-KE310/312QD (n=4), and 0.18 ± 0.01 for hSK1-QDN (n=5). The differences between the triple pore mutant, hSK1-QDN, and hSK1 or SK2 are highly significant (P<0.001). C and D, representative current traces showing the degree of inhibition by 300 pM apamin of hSK1-QDN (I/Imax = 0.58 ± 0.02, n=11) and 30 pM apamin of hSK1-QDN+S (I/Imax =0.48 ± 0.04, n=7). E, concentration-response curves for the inhibition of hSK1-QDN+S (◆) and SK1-QDN (■) by apamin. The concentration-response curves for the effect of apamin on hSK1 (○, Fig. 3I) and SK2 (□, Fig. 3I) have been included for comparison. Data points (n ≥ 3) were fitted with a Hill-Langmuir equation and gave the following IC50 values: hSK1-QDN+S (IC50 = 26 pM, 95% CI: 22 pM to 32 pM), and SK1-QDN (IC50 = 366 pM, 95% CI: 326 pM to 426 pM).
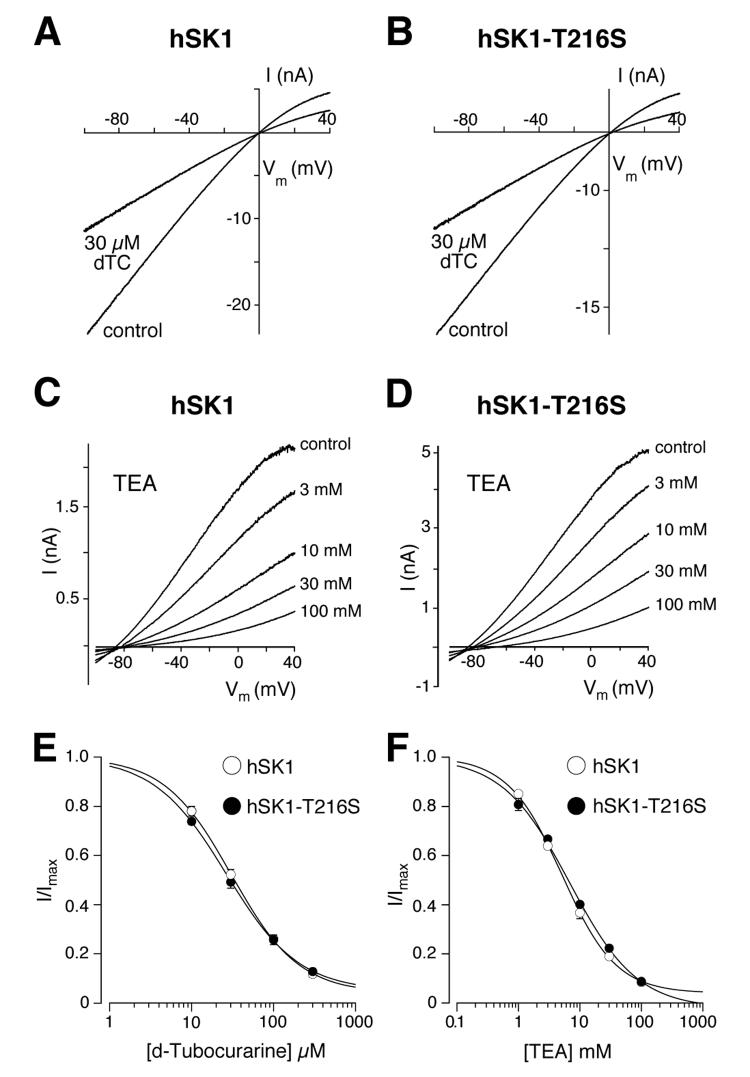
T216S does not influence the sensitivity of hSK1 to d-tubocurarine and TEA
Beside the peptide inhibitor apamin, the SK currents are also suppressed by small organic compounds like TEA and d-tubocurarine (7, 37, 40, 43, 46-48). The amino acids identified in the pore region that influence the sensitivity of the SK channels to apamin also affect the sensitivity for d-tubocurarine (43). We therefore wondered whether the d-tubocurarine sensitivity would be affected by the mutation hSK1-T216S. As illustrated in Fig. 5A and B, currents mediated by hSK1 and hSK1-T216S were both inhibited after application of 30 μM d-tubocurarine. A concentration-response curve gave a d-tubocurarine IC50 value for hSK1-T216S of 26.6 μM (95% CI: 20.4 μM to 34.6 μM). This value is not different from the IC50 of 30.8 μM for hSK1 (95% CI: 24.7 μM to 38.4 μM, Fig. 5E). Thus, in contrast to apamin, the sensitivity of SK channels to d-tubocurarine is not affected by the mutation T216S. The unchanged IC50 for d-tubocurarine of hSK1-T216S, when compared with hSK1, supports the view that the T216S mutation does not induce a major conformational rearrangement of the extracellular region of the SK channel, which would unspecifically affect the sensitivity to any SK channel inhibitor.
Fig. 5.
Comparison of TEA and d-tubocurarine effects on hSK1 and hSK1-T216S. A and B, inhibition of hSK1 (A) and hSK1-T216S (B) by 30 μM d-tubocurarine (dTC). C and D, inhibition of hSK1 (C) and hSK1-T216S (D) by various concentrations (1 mM, 3 mM, 10 mM, 30 mM and 100 mM) of TEA. These recordings were performed using asymmetric potassium concentrations inside and outside the cell, to enable the replacement of NaCl for TEA-Cl without changing the driving force for potassium. E and F, concentration-response curves for d-tubocurarine (E) and TEA (F) on hSK1 (○) and hSK1-T216S (●). Data points (n = 3–6) were fitted with a Hill-Langmuir equation and gave the following IC50 values for d-tubocurarine: hSK1-T216S (IC50 = 26.6 μM, 95% CI: 20.4 μM to 34.6 μM) and hSK1 (IC50 = 30.8 μM, 95% CI: 24.7 μM to 38.4 μM); for TEA: hSK1-T216S (IC50 = 6.8 mM, 95% CI: 5.5 to 8.5) and hSK1 (IC50 = 5.2 mM, 95% CI: 4.5 to 5.9 mM.
Next, we analyzed whether the sensitivity to TEA, a classical pore blocker, was influenced by the mutation in the extracellular S3-S4 loop (L3). Increasing concentrations of TEA were bath applied to the HEK293 cells and progressively inhibited either hSK1 or hSK1-T216S (Fig. 5, C and D). The concentration-response curves obtained for the inhibition of the outward current (Fig. 5F) gave similar IC50 values for hSK1 (IC50: 5.2 mM, 95% CI: 4.5 to 5.9 mM) and hSK1-T216S (IC50: 6.8 mM, 95% CI: 5.5 to 8.5). The inhibition by TEA displayed a slight voltage dependence with stronger block at negative potentials for both hSK1 (0.21 ± 0.04 mM/10 mV) and hSK1-T216S (0.32 ± 0.12 mM/10 mV). The similarity of the IC50 values for the inhibition of hSK1 and hSK1-T216S by TEA demonstrates that the T216S mutation does not influence the TEA binding, and indicates that the general architecture of the SK channel pore is not changed. This underscores the observations made for d-tubocurarine, in that T216S influences the sensitivity of apamin specifically by a localized effect on the apamin receptor site.
DISCUSSION
Peptide toxins that bind to ion channels have been very valuable tools for structure-function studies and for elucidating the physiological roles of particular channel families or subtypes (2, 21, 30, 37, 39, 49-53). Apamin, one of the components of the bee venom, is one of the smallest peptide neurotoxins (54). It crosses the blood-brain barrier and induces hyperexcitability (55). Apamin has long been known as a highly selective inhibitor of Ca2+-activated K+ channels (16). The cloning of the SK channels (45, 56) allowed the molecular analysis of the apamin-receptor interaction, and showed the importance of the pore region (43), a region to which many scorpion and snake toxins bind on voltage-gated K+ channels (57-59).
In this study we show that, beside the residues in the pore region (Fig. 4, B, C, and E, 43), a threonine in the middle of the extracellular S3-S4 loop (L3; position 216 in hSK1) is of crucial importance for apamin binding. This result was unexpected in the light of the insensitivity to apamin of the SK2-SK1 chimera containing the S3-S4 loop shown by Ishii et al. (1997, Fig. 1A). The substitution of threonine 216 in hSK1 by serine, the amino acid found in SK2 at the corresponding position, leads to a ∼20 fold higher apamin sensitivity (Fig. 3I). Furthermore, the exchange of threonine together with the three amino acids in the pore region of hSK1 for the corresponding residues of SK2 leads to the reconstitution of the high apamin sensitivity of SK2 in the two orders of magnitude less sensitive SK1 (Fig. 4, D and E). This result underscores the importance of T216S for generating a high affinity apamin receptor, and is underpinned by the evidence that the apamin concentration-inhibition curves of the SK2 and the hSK1-QDN+S channels are superimposed, and the resulting half-maximal inhibitions are indistinguishable (Fig. 4E).
Hanatoxin, a tarantula toxin, inhibits Kv channels by binding to the voltage-sensor (53), which is in close proximity to the amino acid identified in this study that influence apamin binding in the SK channel family. Hanatoxin inhibits Kv channels by a voltage dependent mechanism, where binding to the outer ends of S3 and S4 stabilizes the resting conformation of the voltage sensor (53). However, it is not likely that apamin inhibits SK channels by a similar mechanism, because SK channels are not voltage dependent (45). Instead SK channel gating results from a conformational rearrangement at the carboxyl-terminus, induced by binding of Ca2+ to the constitutively bound calmodulin (4-6). Furthermore, apamin is much less hydrophobic than hanatoxin and therefore not likely to partition into the plasma membrane (60). Additionally, amino acids in the pore have been identified to influence inhibition by apamin (43, this study), whereas hanatoxin binding to Kv channels is not influenced by mutations in the pore region (61).
This study and the available evidence make it more likely that apamin inhibits SK channels by a mechanism elucidated in detail for scorpion toxins on Kv channels, by which the toxin occludes the channel pore (58). Mutagenesis studies that rendered SK channels sensitive for the Kv channel blockers charybdotoxin and kaliotoxin suggest that the pore region of these two channel families have a comparable architecture and that the inhibition of mutated SK channels by scorpion toxins may be described as a pore block (62). The affinity of the most subtype-selective SK channel inhibitor, Lei-Dab7, generated by introducing an unnatural, positively charged amino acid at position 7 in leiurotoxin, is also influenced by at least one amino acid in the pore region (39), in accordance with a pore blocking mechanism. Furthermore, a change in the structure of the turret is the most likely reason for the lack of sensitivity to apamin and scyllatoxin of the SK3 splice variant hSK3_ex4, presenting the insertion of an additional exon between S5 and the pore (63). The mechanism of pore block for scorpion toxins is also supported by molecular modelling and docking simulations (64-66). However, although there is a growing consensus that scorpion toxins occlude the ion pathway in SK channels, decisive experimental evidence is presently not available for any of the SK channel inhibitors (58).
How can the view that apamin occludes the conduction pathway be reconciled with the finding that the mutation of a single threonine into a serine in the extracellular S3-S4 loop (L3) has a bigger influence on apamin inhibition (Fig. 3, SK1-T216S IC50: 167 pM) than the three mutations in the pore outer vestibule (Fig. 4, SK1-QDN IC50: 366 pM)? The answer might lie simply in the fact that the extracellular loop in question is particularly long when compared with other members of the Kv channel superfamily (67), and that the end of S4 and the amino acids identified in the pore region that influence apamin sensitivity might lie in close proximity. In the absence of any structural or functional data on the localisation or movement of the S4 segment during the opening of SK channels, we have to base our arguments on the available data for the S4 of voltage-gated K+ channels. Unfortunately, as a consequence of the low electron density for the extracellular loops in the Kv1.2 channel crystals, no structural information for the S3-S4 loop (L3) is available (68). However, the recently solved crystal structure of the Kv1.2 channel shows the voltage sensing domain in close proximity to the pore domain of the neighbouring subunit, which would bring the extracellular S3-S4 loop (L3) close to the outer pore vestibule (68). Additionally, the proximity between the initial S4 segment and the pore domain has been demonstrated by the formation of disulfide bonds (69), and by analysing the reaction time of introduced cysteine residues in the pore loop with charged cysteine modifiying agents depending on the position of S4 (70). The influence of a region outside the pore on toxin binding has been observed for the Ca2+- and voltage-activated potassium channel complex, which consists of α- and ß-subunits. Here, the extracellular regions of the ß-subunits influence the interaction of charybdotoxin and iberiotoxin with the channel complex (71). In particular, the ß4-subunit renders the channel complex insensitive to charybdotoxin and iberiotoxin (71).
There may be more amino acids involved in creating the apamin binding site on the SK channels. Our analysis was guided by the difference in apamin sensitivity between hSK1 and rSK1NSK2-CSK2. This led to the identification of the threonine at position 216 in hSK1 and the corresponding serine in SK2 and SK3 described here. However, when we aligned the amino acids for rSK1NSK2-CSK2 and the hSK1-SK2-hSK1 chimera in the pore region and in the S3-S4 loop, we were surprised about the result. The resulting channels show a more than 150 fold difference in their sensitivity for apamin (Fig. 1, 2 and in Ref. 42), in spite of both having a serine at the corresponding amino acid position 216 in hSK1 and identical pores. This strongly suggests the involvement of further amino acids that render rSK1 apamin insensitive.
Our data show that the apamin sensitivity of SK channels is influenced by the primary sequence in the pore region as well as by the extracellular S3-S4 loop (L3). The involvement of the pore region and simultaneously the S3-S4 loop (L3) in the binding of a blocker has to our knowledge, so far not been demonstrated for any other K+ channel. Interestingly, serine, the amino acid in SK2 and SK3, and theronine, the amino acid in SK1, differ only by one methyl group. Therefore, we think it is very likely that the identified amino acid residue in the S3-S4 loop (L3) is in close contact with apamin and might interact with residues on the toxin surface. Alternatively, the mutation T216S might have an effect on the three-dimensional structure of the apamin receptor, thereby influencing the apamin sensitivity of the SK channels. Small organic molecules such as TEA and d-tubocurarine are not affected by the mutation (Fig. 5), suggesting that changes in the S3-S4 loop do not induce major conformational changes in the pore region.
However, at the present time we do not know which apamin residues may face the S3-S4 loop (L3). The available molecular modelling and docking simulations for scorpion toxins (64-66), cannot answer this question in the absence of structural data for the S3-S4 loop (68). Our study might serve as a starting point to identify amino acids in close contact between toxin and channel to complete our understanding of the architecture of the SK channel vestibule and of how apamin sits in it. Furthermore, taken into consideration the larger size of the scorpion toxins when compared with apamin, we would expect that the amino acids identified in the extracellular S3-S4 loop (L3) also affect the binding of the scorpion toxins to SK channels.
A detailed understanding of the molecular determinants of SK channel inhibition by toxins is essential for the design of novel, subunit specific blockers that may constitute useful tools for the elucidation of the SK channel normal function and the treatment of neurological or psychiatric disorders by selectively targeting SK channel subtypes.
Acknowledgements
We thank Dr Paola Pedarzani for scientific discussion, and Dr Mala Shah, Hannah Morgan and Dr Paola Pedarzani for critical reading of the manuscript.
Footnotes
This work was supported by a Wellcome Prize studentship (T.F.) and a Wellcome Trust Senior Research fellowship (M.S.).
IAHP, afterhyperpolarizing current mediated by SK channels; CHO cells, Chinese hamster ovary cells; CI, confidence interval; EGFP, enhanced green fluorescent protein; HEK293 cells, human embryonic kidney 293 cells; KCa, Ca2+-activated potassium channel; SK, small conductance Ca2+-activated potassium channel; TEA tetraethlyammonium;
REFERENCES
- 1.Wei AD, Gutman GA, Aldrich R, Chandy KG, Grissmer S, Wulff H. Pharmacol Rev. 2005;57:463–472. doi: 10.1124/pr.57.4.9. [DOI] [PubMed] [Google Scholar]
- 2.Hille B. Ion Channels of Excitable Membranes. Sunderland, MA: Sinauer Associates Inc.; 2001. [Google Scholar]
- 3.Orio P, Rojas P, Ferreira G, Latorre R. News Physiol Sci. 2002;17:156–161. doi: 10.1152/nips.01387.2002. [DOI] [PubMed] [Google Scholar]
- 4.Fanger CM, Ghanshani S, Logsdon NJ, Rauer H, Kalman K, Zhou J, Beckingham K, Chandy KG, Cahalan MD, Aiyar J. J Biol Chem. 1999;274:5746–5754. doi: 10.1074/jbc.274.9.5746. [DOI] [PubMed] [Google Scholar]
- 5.Xia XM, Fakler B, Rivard A, Wayman G, Johnson-Pais T, Keen JE, Ishii T, Hirschberg B, Bond CT, Lutsenko S, Maylie J, Adelman JP. Nature. 1998;395:503–507. doi: 10.1038/26758. [DOI] [PubMed] [Google Scholar]
- 6.Schumacher MA, Rivard AF, Bachinger HP, Adelman JP. Nature. 2001;410:1120–1124. doi: 10.1038/35074145. [DOI] [PubMed] [Google Scholar]
- 7.Klocker N, Oliver D, Ruppersberg JP, Knaus HG, Fakler B. Mol Cell Neurosci. 2001;17:514–520. doi: 10.1006/mcne.2000.0956. [DOI] [PubMed] [Google Scholar]
- 8.Glowatzki E, Fuchs PA. Science. 2000;288:2366–2368. doi: 10.1126/science.288.5475.2366. [DOI] [PubMed] [Google Scholar]
- 9.Oliver D, Klocker N, Schuck J, Baukrowitz T, Ruppersberg JP, Fakler B. Neuron. 2000;26:595–601. doi: 10.1016/s0896-6273(00)81197-6. [DOI] [PubMed] [Google Scholar]
- 10.Schetz JA, Anderson PA. Cardiovasc Res. 1995;30:755–762. [PubMed] [Google Scholar]
- 11.Xu Y, Tuteja D, Zhang Z, Xu D, Zhang Y, Rodriguez J, Nie L, Tuxson HR, Young JN, Glatter KA, Vazquez AE, Yamoah EN, Chiamvimonvat N. J Biol Chem. 2003;278:49085–49094. doi: 10.1074/jbc.M307508200. [DOI] [PubMed] [Google Scholar]
- 12.Barfod ET, Moore AL, Lidofsky SD. Am J Physiol Cell Physiol. 2001;280:C836–842. doi: 10.1152/ajpcell.2001.280.4.C836. [DOI] [PubMed] [Google Scholar]
- 13.Capiod T, Ogden DC. J Physiol (Lond) 1989;409:285–295. doi: 10.1113/jphysiol.1989.sp017497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hugues M, Schmid H, Romey G, Duval D, Frelin C, Lazdunski M. EMBO J. 1982;1:1039–1042. doi: 10.1002/j.1460-2075.1982.tb01293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pribnow D, Johnson-Pais T, Bond CT, Keen J, Johnson RA, Janowsky A, Silvia C, Thayer M, Maylie J, Adelman JP. Muscle Nerve. 1999;22:742–750. doi: 10.1002/(sici)1097-4598(199906)22:6<742::aid-mus11>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 16.Banks BE, Brown C, Burgess GM, Burnstock G, Claret M, Cocks TM, Jenkinson DH. Nature. 1979;282:415–417. doi: 10.1038/282415a0. [DOI] [PubMed] [Google Scholar]
- 17.Herrera GM, Nelson MT. J Physiol (Lond) 2002;541:483–492. doi: 10.1113/jphysiol.2002.017707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stocker M, Pedarzani P. Mol Cell Neurosci. 2000;15:476–493. doi: 10.1006/mcne.2000.0842. [DOI] [PubMed] [Google Scholar]
- 19.Bond CT, Maylie J, Adelman JP. Curr Opin Neurobiol. 2005;15:305–311. doi: 10.1016/j.conb.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 20.Sah P. Trends Neurosci. 1996;19:150–154. doi: 10.1016/s0166-2236(96)80026-9. [DOI] [PubMed] [Google Scholar]
- 21.Stocker M. Nat Rev Neurosci. 2004;5:758–770. doi: 10.1038/nrn1516. [DOI] [PubMed] [Google Scholar]
- 22.Schwindt PC, Spain WJ, Foehring RC, Stafstrom CE, Chubb MC, Crill WE. J Neurophysiol. 1988;59:424–449. doi: 10.1152/jn.1988.59.2.424. [DOI] [PubMed] [Google Scholar]
- 23.Stocker M, Krause M, Pedarzani P. Proc Natl Acad Sci U S A. 1999;96:4662–4667. doi: 10.1073/pnas.96.8.4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pedarzani P, Kulik A, Muller M, Ballanyi K, Stocker M. J Physiol (Lond) 2000;527:283–290. doi: 10.1111/j.1469-7793.2000.t01-1-00283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolfart J, Neuhoff H, Franz O, Roeper J. J Neurosci. 2001;21:3443–3456. doi: 10.1523/JNEUROSCI.21-10-03443.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cingolani LA, Gymnopoulos M, Boccaccio A, Stocker M, Pedarzani P. J Neurosci. 2002;22:4456–4467. doi: 10.1523/JNEUROSCI.22-11-04456.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edgerton JR, Reinhart PH. J Physiol (Lond) 2003;548:53–69. doi: 10.1113/jphysiol.2002.027854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hallworth NE, Wilson CJ, Bevan MD. J Neurosci. 2003;23(20):7525–7542. doi: 10.1523/JNEUROSCI.23-20-07525.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Womack MD, Khodakhah K. J Neurosci. 2003;23:2600–2607. doi: 10.1523/JNEUROSCI.23-07-02600.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bond CT, Herson PS, Strassmaier T, Hammond R, Stackman R, Maylie J, Adelman JP. J Neurosci. 2004;24:5301–5306. doi: 10.1523/JNEUROSCI.0182-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Faber ES, Delaney AJ, Sah P. Nat Neurosci. 2005;8:635–641. doi: 10.1038/nn1450. [DOI] [PubMed] [Google Scholar]
- 32.Ngo-Anh TJ, Bloodgood BL, Lin M, Sabatini BL, Maylie J, Adelman JP. Nat Neurosci. 2005;8:642–649. doi: 10.1038/nn1449. [DOI] [PubMed] [Google Scholar]
- 33.Fournier C, Kourrich S, Soumireu-Mourat B, Mourre C. Behavioral Brain Research. 2001;121:81–93. doi: 10.1016/s0166-4328(00)00387-9. [DOI] [PubMed] [Google Scholar]
- 34.Messier C, Mourre C, Bontempi B, Sif J, Lazdunski M, Destrade C. Brain Res. 1991;551:322–326. doi: 10.1016/0006-8993(91)90950-z. [DOI] [PubMed] [Google Scholar]
- 35.Stackman RW, Hammond RS, Linardatos E, Gerlach A, Maylie J, Adelman JP, Tzounopoulos T. J Neurosci. 2002;22:10163–10171. doi: 10.1523/JNEUROSCI.22-23-10163.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hammond RS, Bond CT, Strassmaier T, Ngo-Anh TJ, Adelman JP, Maylie J, Stackman RW. J Neurosci. 2006;26:1844–1853. doi: 10.1523/JNEUROSCI.4106-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stocker M, Hirzel K, D'Hoedt D, Pedarzani P. Toxicon. 2004;43:933–949. doi: 10.1016/j.toxicon.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 38.Liegeois JF, Mercier F, Graulich A, Graulich-Lorge F, Scuvee-Moreau J, Seutin V. Curr Med Chem. 2003;10:625–647. doi: 10.2174/0929867033457908. [DOI] [PubMed] [Google Scholar]
- 39.Shakkottai VG, Regaya I, Wulff H, Fajloun Z, Tomita H, Fathallah M, Cahalan MD, Gargus JJ, Sabatier JM, Chandy KG. J Biol Chem. 2001;276:43145–43151. doi: 10.1074/jbc.M106981200. [DOI] [PubMed] [Google Scholar]
- 40.Pedarzani P, D'Hoedt D, Doorty KB, Wadsworth JD, Joseph JS, Jeyaseelan K, Kini RM, Gadre SV, Sapatnekar SM, Stocker M, Strong PN. J Biol Chem. 2002;277:46101–46109. doi: 10.1074/jbc.M206465200. [DOI] [PubMed] [Google Scholar]
- 41.Benton DC, Monaghan AS, Hosseini R, Bahia PK, Haylett DG, Moss GW. J Physiol (Lond) 2003;553:13–19. doi: 10.1113/jphysiol.2003.054551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.D'Hoedt D, Hirzel K, Pedarzani P, Stocker M. J Biol Chem. 2004;279:12088–12092. doi: 10.1074/jbc.C300382200. [DOI] [PubMed] [Google Scholar]
- 43.Ishii TM, Maylie J, Adelman JP. J Biol Chem. 1997;272:23195–23200. doi: 10.1074/jbc.272.37.23195. [DOI] [PubMed] [Google Scholar]
- 44.Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 45.Kohler M, Hirschberg B, Bond CT, Kinzie JM, Marrion NV, Maylie J, Adelman JP. Science. 1996;273:1709–1714. doi: 10.1126/science.273.5282.1709. [DOI] [PubMed] [Google Scholar]
- 46.Shah M, Haylett DG. Br J Pharmacol. 2000;129:627–630. doi: 10.1038/sj.bjp.0703111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strobaek D, Jorgensen TD, Christophersen P, Ahring PK, Olesen SP. Br J Pharmacol. 2000;129:991–999. doi: 10.1038/sj.bjp.0703120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Monaghan AS, Benton DC, Bahia PK, Hosseini R, Shah YA, Haylett DG, Moss GW. J Biol Chem. 2004;279:1003–1009. doi: 10.1074/jbc.M308070200. [DOI] [PubMed] [Google Scholar]
- 49.Harvey AL. Gen Pharmacol. 1997;28:7–12. doi: 10.1016/s0306-3623(96)00173-5. [DOI] [PubMed] [Google Scholar]
- 50.Terlau H, Olivera BM. Physiol Rev. 2004;84:41–68. doi: 10.1152/physrev.00020.2003. [DOI] [PubMed] [Google Scholar]
- 51.Villalobos C, Shakkottai VG, Chandy KG, Michelhaugh SK, Andrade R. J Neurosci. 2004;24:3537–3542. doi: 10.1523/JNEUROSCI.0380-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Garcia ML, Gao Y, McManus OB, Kaczorowski GJ. Toxicon. 2001;39:739–748. doi: 10.1016/s0041-0101(00)00214-2. [DOI] [PubMed] [Google Scholar]
- 53.Swartz KJ. Nat Rev Neurosci. 2004;5:905–916. doi: 10.1038/nrn1559. [DOI] [PubMed] [Google Scholar]
- 54.Strong PN. Pharmacol Ther. 1990;46:137–162. doi: 10.1016/0163-7258(90)90040-9. [DOI] [PubMed] [Google Scholar]
- 55.Habermann E. Science. 1972;177:314–322. doi: 10.1126/science.177.4046.314. [DOI] [PubMed] [Google Scholar]
- 56.Joiner WJ, Wang LY, Tang MD, Kaczmarek LK. Proc Natl Acad Sci U S A. 1997;94:11013–11018. doi: 10.1073/pnas.94.20.11013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Giangiacomo KM, Ceralde Y, Mullmann TJ. Toxicon. 2004;43:877–886. doi: 10.1016/j.toxicon.2003.11.029. [DOI] [PubMed] [Google Scholar]
- 58.Miller C. Neuron. 1995;15:5–10. doi: 10.1016/0896-6273(95)90057-8. [DOI] [PubMed] [Google Scholar]
- 59.Stocker M, Miller C. Proc Natl Acad Sci U S A. 1994;91:9509–9513. doi: 10.1073/pnas.91.20.9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Phillips LR, Milescu M, Li-Smerin Y, Mindell JA, Kim JI, Swartz KJ. Nature. 2005;436:857–860. doi: 10.1038/nature03873. [DOI] [PubMed] [Google Scholar]
- 61.Swartz KJ, MacKinnon R. Neuron. 1997;18:675–682. doi: 10.1016/s0896-6273(00)80307-4. [DOI] [PubMed] [Google Scholar]
- 62.Jager H, Grissmer S. Toxicon. 2004;43:951–960. doi: 10.1016/j.toxicon.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 63.Wittekindt OH, Visan V, Tomita H, Imtiaz F, Gargus JJ, Lehmann-Horn F, Grissmer S, Morris-Rosendahl DJ. Mol Pharmacol. 2004;65:788–801. doi: 10.1124/mol.65.3.788. [DOI] [PubMed] [Google Scholar]
- 64.Andreotti N, di Luccio E, Sampieri F, De Waard M, Sabatier JM. Peptides. 2005;26:1095–1108. doi: 10.1016/j.peptides.2005.01.022. [DOI] [PubMed] [Google Scholar]
- 65.Cui M, Shen J, Briggs JM, Fu W, Wu J, Zhang Y, Luo X, Chi Z, Ji R, Jiang H, Chen K. J Mol Biol. 2002;318:417–428. doi: 10.1016/S0022-2836(02)00095-5. [DOI] [PubMed] [Google Scholar]
- 66.Wu Y, Cao Z, Yi H, Jiang D, Mao X, Liu H, Li W. Biophys J. 2004;87:105–112. doi: 10.1529/biophysj.103.039156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Coetzee WA, Amarillo Y, Chiu J, Chow A, Lau D, McCormack T, Moreno H, Nadal MS, Ozaita A, Pountney D, Saganich M, Vega-Saenz de Miera E, Rudy B. Ann N Y Acad Sci. 1999;868:233–285. doi: 10.1111/j.1749-6632.1999.tb11293.x. [DOI] [PubMed] [Google Scholar]
- 68.Long SB, Campbell EB, MacKinnon R. Science. 2005;309:903–908. doi: 10.1126/science.1116270. [DOI] [PubMed] [Google Scholar]
- 69.Laine M, Lin MC, Bannister JP, Silverman WR, Mock AF, Roux B, Papazian DM. Neuron. 2003;39:467–481. doi: 10.1016/s0896-6273(03)00468-9. [DOI] [PubMed] [Google Scholar]
- 70.Elinder F, Mannikko R, Larsson HP. J Gen Physiol. 2001;118:1–10. doi: 10.1085/jgp.118.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Meera P, Wallner M, Toro L. Proc Natl Acad Sci U S A. 2000;97:5562–5567. doi: 10.1073/pnas.100118597. [DOI] [PMC free article] [PubMed] [Google Scholar]