Abstract
Direct ionization of crystalline d(CGCGCGCG) and d(CGCGCGCGCG) oligomers produces 3’- and 5’-phosphate-terminated fragments as the main strand breakage products detectable by ion-exchange chromatography. The nature of the base has no effect on the probability of strand break at the given site. The yields of 3’-phosphates are systematically lower than of the 5’-phosphates originating from the same cleavage site, pointing to the possible presence of unidentified products with sugar remnants attached to the 3’-end. These results show that direct ionization is efficient at producing single strand breaks in DNA and its action is relatively indiscriminate with respect to base sequence.
Introduction
Direct-type effects arise from the direct ionization of the DNA molecule or from holes and electrons transferred to the DNA from its solvation shell (1–3). This pathway, resulting in direct-type damage, may be particularly important in chromosomes, where the concentration of “free” water is relatively low. Work on cellular DNA has implicated direct-type damage as a significant factor in causing strand breaks (4).
Recently we reported the yields of single strand breaks in DNA crystals formed by d(CGCG)2 and d(CGCGCG)2 duplexes, x-irradiated both at 4 K and room temperature (5). This study showed that the yield of strand breaks is fairly insensitive to the irradiation temperature and corresponds to about 10% of the free radical yield measured at 4 K. With respect to individual product yields, statistically sound conclusions were not possible because these two sequences were too short. One observation of particular interest was the difference in yield between products with 5’-phosphorylated ends compared to products with 3’-ends, the former being about1.8-fold greater than the latter. In order to further explore this and other observations, we have extended our study to the crystals formed by the d(CGCGCGCG) and d(CGCGCGCGCG) sequences.
Based on our previous results on unaltered base release from several DNA crystals we hypothesized that the precursor for the strand break in directly ionized DNA is the sugar radical cation, which further undergoes deprotonation to form a neutral sugar radical (6). This process competes with hole transfer to DNA bases. Since guanine is the most oxidizable DNA base, the possibility exists that guanine could “quench” the sugar radical cation more efficiently than any other bases (7) and, therefore, avert localization of radiation damage on the adjacent sugar. For highly exothermic processes, however, such a correlation between the free energy of electron transfer and the rate constant does not hold (8), and there are good reasons to believe that hole transfer from oxidized sugars to bases may proceed in this regime(9). Through the investigation of crystalline d(CGCGCGCG)2 and d(CGCGCGCGCG)2, the evidence continues to build toward the conclusion that base sequence has either a minor, or no influence, on the probability of strand breakage by direct ionization.
Methods
Materials
The oligodeoxynucleotides, d(CGCGCGCG) (CG8) and d(CGCGCGCGCG) (CG10), were purchased from Ransom Hill Bioscience and used as received for crystallization. Synthetic 3’- and 5’-phosphorylated oligodeoxynucleotides from Midland Certified Reagent Company were employed as reference compounds in the identification of DNA strand break products.
Crystals of CG8 and CG10 were grown following the published procedure for CG8 (10). Because the crystal structure of CG10 had not been determined previously, x-ray diffraction data was taken and analyzed by two of us (SBH and LDW). CG10 was found be nearly isomorphous to CG8 (10) and to the closely related crystal of d(CGCICICG)2 (11). In each case, the Z-DNA duplexes are packed so as to form parallel columns of continuously stacked CG base pairs. However, CG10 crystals differ by displaying disorder along the unit cell axis parallel to the columns; that is the CG base pairs of CG10 are in phase with one another but the helices termini are not. While the impact of such disorder on the radiation chemistry is not known, we believe it is likely to be negligible relative to other variables.
Product analysis
The protocol for sample preparation and irradiation has been described elsewhere (12). After irradiation, the crystals were transferred to 1 ml plastic vials and weighed to ±1 μg accuracy on a Cahn-60 microbalance. The crystals (samples ranged from 90–400 μg: a typical sample weighed ~ 200 μg) were then dissolved in a 50 mM Tris/ 10% acetonitrile medium (pH 10.2, 50 μL of the buffer per 100 μg of crystals) containing 50 μM thymidine-5’ -monophosphate (TMP) employed as an internal standard for quantification purposes, and held at 70° C for 30 minutes (6, 5). This preventive measure was taken to ensure the absence of any uncontrolled strand breakage originating from heat-labile lesions during subsequent sample processing.
The DNA cleavage products were analyzed by ion-exchange chromatography (IC) on a Waters Alliance™ system equipped with an autosampler, a 2690 solvent delivery module and a 996 PDA detector using a Dionex DNAPac PA-100 4.6×250 mm column operated at 60°C and a flow rate of 1 ml/min. The column was washed with 50 mM Tris buffer containing 10% acetonitrile by volume (pH 10.22), and the products separated by applying a linear Cl− gradient (typically from 0.1 M to 1 M NaCl over 30 min). The products were detected at 260 nm and identified by co-injection with authentic reference compounds.
Quantification
Areas under the identified product peaks (Spr) were converted into the product absorbencies (Apr) using the formula
where Ast and Sst are the absorbance and the peak areas of the internal standard TMP. The typical concentration and absorbance of TMP in the loading buffer was 50 μM, corresponding to 0.33 o.d.u. at 260 nm. The areas are further converted into concentrations using the nm/o.d.u. ratios provided by Midland Certified Reagent Company for each of the references, and then into the number of μmoles of the products formed in the crystalline DNA using the known volume of the buffer used to dissolve the DNA. The absorbed energy in Joules is calculated from the dose rate and mass of the crystal. The slope of the product amount plotted against dose is the radiation-chemical yield of the product expressed in μmol/J.
Results
There were two experimental constraints that we were unable to overcome. One was a very limited amount of sample, due to the cost of materials and difficulties in crystallization. The other was that the presence of impurities in the starting material and the strong overlap of the peak of the parent oligomer with the peaks of some of the products in IC chromatograms impeded quantification of the products more than 6–7 bases in length. As a consequence, the measured yields have significantly larger errors than those reported in our previous studies. But a major advantage of IC in studies of DNA damage is that it allows for the simultaneous detection of each strand cleavage products in one experiment. This is especially helpful when comparing product yields. Relative yields are the focus of this report.
Another advantage of IC is that the relative retention times of end-phosphorylated DNA fragments generated from alternating CG-sequences can be predicted from three simple rules. Looking at the chromatogram of CG8 in Figure 1A, five groups of product are readily identified. Each group consists of two peaks corresponding to the products of the same length in terms of the number of bases n. Rule 1 is that the total charge on the phosphates is a key factor controlling the retention times. Therefore, any n-mer always runs faster than any n+1-mer regardless of their composition. Therefore, the group of monomers come out before the group of dimers, dimers before trimers, etc. Rule 2 is that inside each group the cytosine-rich product always runs faster than the guanine-rich one owing to the presence of the extra negative charge on guanines at pH 10.2 (the pKa of guanine is ~9.4). Rule 3 is that in sequences of the same composition and length 3’ phosphates run slightly faster than 5’ phosphates. These groups with even n’s (such as CGp and pCG, CGCGp and pCGCG, etc.) appear as easily identifiable “doublets” in IC chromatograms, which separate the groups corresponding to odd n’s. These rules are useful when reflecting on the nature of the identified peaks, which we return to in the discussion.
Fig 1.
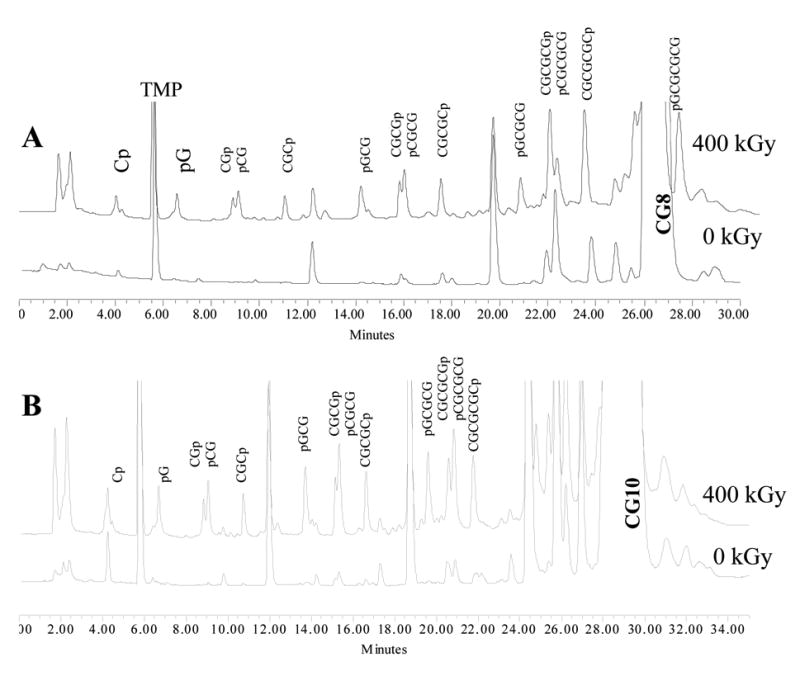
Ion-exchange chromatograms of CG8 (A) and CG10 (B) before and after irradiation to a 400 kGy dose.
CG8
An ion exchange chromatogram of irradiated CG8 is shown in Figure 1A. The large off-scale peaks belong to the TMP reference and the undamaged CG8. All the major peaks that grow in with increasing dose are 3’ - and 5’ -end phosphates; i.e., remnants of the sugar that sustained the initial damage are no longer covalently bound. None of the major products are 3’-phosphoglycolates, which are common when damage is initiated by OH radicals in oxygenated media (13).
All the major products were identified and quantified except for CGCGCGp and pCGCGCG. The latter two were problematic because these peaks overlap with a strong impurity peak, the intensity of which decreases with dose. These two hexamers, therefore, were excluded from further analysis. Concentrations of the other products, measured after dissolving the crystals in the loading buffer, are reported in Table 1 along with the yields calculated from the dose response curves.
Table 1.
Radiation-Chemical Yields of Product Formation in crystalline CG8
| Amount, μmol/g
|
|||||
|---|---|---|---|---|---|
| Product | 100 kGy | 200 kGy | 300 kGy | 400 kGy | Yield nmol/J |
| Cp | 1.4 | 1.2 | 1.3 | 4.7 | 9±4 |
| pG | 1.1 | 1.2 | 1.5 | 3.6 | 7±2 |
| CGp | 0.4 | 0.4 | 0.5 | 1.4 | 3±1 |
| pCG | 0.5 | 0.4 | 0.6 | 1.5 | 3±1 |
| CGCp | 0.3 | 0.3 | 0.4 | 1.0 | 2.0±0.5 |
| pGCG | 0.4 | 0.5 | 0.5 | 1.3 | 2.7±0.7 |
| CGCGp | 0.3 | 0.3 | 0.2 | 0.8 | 1.4±0.5 |
| pCGCG | 0.6 | 0.6 | 0.6 | 1.5 | 3.0±0.8 |
| CGCGCp | 0.2 | 0.2 | 0.2 | 0.9 | 1.6±0.6 |
| pGCGCG | 0.5 | 0.4 | 0.4 | 1.0 | 1.9±0.6 |
| CGCGCGCp | 0.8 | 0.1 | 0.1 | 1.2 | 2±2 |
| pGCGCGCG | 1.1 | 0.9 | 1.1 | 2.6 | 5±2 |
CG10
The ion exchange chromatogram of CG10 irradiated to the dose of 400 kGy is shown in Figure 1B along with the chromatogram of an unirradiated sample. Due to the superposition with intense peaks from contaminants and starting material, quantification of Cp, pGCGCGCG, the two 8-mers, and two 9-mers was not pursued. The results for the other cleavage products are reported in Table 2.
Table 2.
Radiation-Chemical Yields of Product Formation in crystalline CG10
| Product | Amount, μmol/g
|
||
|---|---|---|---|
| 100 kGy | 400 kGy | Approx. Yield nmol/J | |
| pG | 2.20 | 4.45 | 17 |
| CGp | 0.50 | 1.31 | 4 |
| pCG | 0.91 | 1.97 | 7 |
| CGCp | 0.46 | 1.17 | 4 |
| pGCG | 0.79 | 1.98 | 6 |
| CGCGp | 0.28 | 0.85 | 2 |
| pCGCG | 0.89 | 1.99 | 7 |
| CGCGCp | 0.43 | 1.00 | 3 |
| pGCGCG | 0.71 | 1.66 | 6 |
| CGCGCGp | 0.39 | 0.83 | 3 |
| pCGCGCG | 0.82 | 1.58 | 6 |
| CGCGCGCp | 0.59 | 0.95 | 4 |
Because the quantity of material allowed for only three samples (two irradiated + one control), the absolute values for the yields reported in Table 2 are rough estimates of the actual yields. Ratios of the yields, however, reflect relative efficiencies of product formation with reasonable accuracy.
Discussion
The spectrum of DNA cleavage products generated from CG8 and CG10 and detected by IC is fairly simple, consisting predominantly of 3’- and 5’phosphates. The presence of other products is likely, but they are either present in smaller quantities or have retention times comparable to that of the starting material. The latter is expected for many of the products with damage restricted to the bases. For example, the dihydropyrimidines are major products observed by Swarts in hydrated, solid-state, DNA (14, 15). It is, however, highly unlikely that dihydropyrimidine-containing oligomers would be resolved from the starting material by IC (given no difference in charge state). In addition, given the moderate alkalinity of the solutions (pH 10.2), the oligomers that sustain base damage are not likely to give rise to strand breaks. It is also notable that we do not detect oligomers containing the well known oxidation products of guanine even though one-electron oxidation of guanine most assuredly occurs at yields (15) higher than the sum of the strand break product yields.
The DNA fragmentation pattern induced by direct ionization is similar to the one induced by the indirect effect in the absence of oxygen, where hydroxyl radicals abstract hydrogen from the sugar-phosphate moiety (13). This similarity supports our model that strand breaks, produced by direct-type effects, arise from sugar radical precursors.
A notable difference between direct-type and indirect-type products is that the former contains a relative paucity of products containing a sugar remnant attached to the phosphate. Such products are well known in radiation chemistry of DNA in solution, both oxic and anoxic (16, 13). The structure of the sugar remnants vary: three such structures have been proposed for the remnants attached at the 3’-end, and two – for the 5’-end (16). While we find that the dominant reaction pathway for direct strand breakage is for the damaged sugar to degrade with the release of the 3’- and 5’- phosphates and the free base, we argue below that the yield of products with attached sugar remnants is likely to be significant as well.
The yield of a 5’-phosphate is systematically higher than the yield of a 3’-phosphate originating from the same cleavage site. This same phenomenon occurs in crystalline d(CGCG)2, d(CGCGCG)2, d(CACGCG):d(CGCGAT), and d(CGCACG):d(CGTGCG) (5, 9). (However, in these shorter sequences this effect is partially obscured by the increased probability of damage to the terminal sugars.) The number of reasonable explanations for this imbalance are limited. We suggest that products still carrying sugar remnants attached to the terminal phosphates are formed, with the yield of products containing 3’-attachments greater than those containing 5’-attachments. A number of factors make these products more difficult to identify and quantify: variability in remnant structures, lower yields, and lack of reference compounds. If this explanation is correct, the yields of such products containing remnants must be greater than or equal to the difference between the 5’-phosphate and 3’- phosphate products. The IC data is consistent with this explanation.
Material balance is expected between the products on one side of the cleavage site and those on the other side of the cleavage site. The IC data would be consistent with this expectation if it contains unidentified peaks with retention times in the ranges expected for products containing sugar remnants. The retention time of a product containing a sugar remnant can be estimated using the rules outlined above. As an example, consider damage at C5 of CG8; figure 2 shows that the yields of the two major products, CGCGp and pGCG, differ by about a factor of two. This difference would be balanced out if the yield of CGCGpR exceeds the yield of RpGCG (where R is a sugar remnant) by a compensating amount. Since the excess of CGCGpR would need to be comparable to the yield of CGCGp, one would expect to see peaks attributable to CGCGpR, if the structural variety of R is limited. The retention time of CGCGpR can be estimated using the rules outlined above. It should fall on the slow side of the 3-mer group because the sum of phosphate charges is -4. The expected retention time is, therefore, close to that of pGCG (14.3 min). Indeed at 14.4 minutes, there is an unidentified peak that grows in with dose. If one goes through this exercise with each of the product pairs shown in Figure 2, one finds unassigned product peaks that could correspond to the respective 3’-phoshpate product with a remnant attached. Hopefully, this hypothesis will be tested; this would be straight forward if sufficient product, in the unidentified peaks, can be isolated for further analysis.
Fig 2.
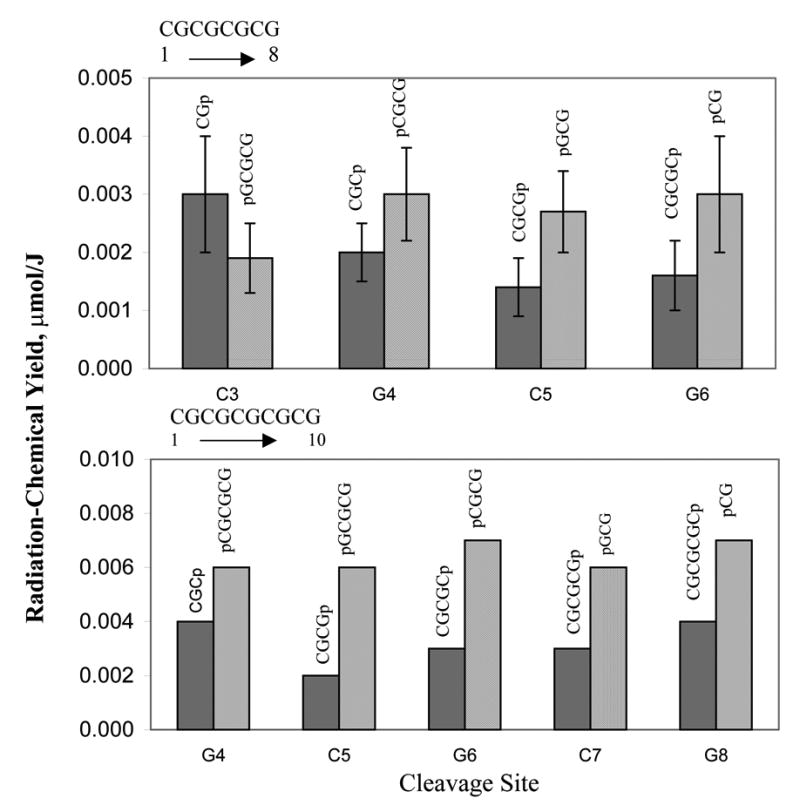
Product yields in CG8 and CG10 grouped by cleavage site. Site numbering starts at the 5’-end.
Another observation with respect to relative yields is that the yields for Cp and pG, in both CG8 and CG10, are 2–4 times that of the other products listed in Tables 1 and 2. The level of error in these two measurements, however, is large. The peak shapes indicate a superposition of each mononucleotide with unidentified products that have similar retention times and comparable intensity. The low extinction coefficients of Cp and pG, relative to the other cleavage products, make their quantification especially error-prone. For this reason, these two products were excluded from the analysis given in Tables 3–4.
Table 3.
Radiation-Chemical Yields of Product Formation in CG8 Grouped by the Cleavage Site (site numbering starts at the 5’-end)
| Site | Radiation-Chemical Yields (μmol/J)
|
||||||
|---|---|---|---|---|---|---|---|
| 5’-side
|
3'-side
|
Average | |||||
| Product | Yield | Error | Product | Yield | Error | ||
| C3 | CGp | 0.0030 | 0.0010 | pGCGCG | 0.0019 | 0.0006 | 0.0025 |
| G4 | CGCp | 0.0020 | 0.0005 | pCGCG | 0.0030 | 0.0008 | 0.0025 |
| C5 | CGCGp | 0.0014 | 0.0005 | pGCG | 0.0027 | 0.0007 | 0.0020 |
| G6 | CGCGCp | 0.0016 | 0.0006 | pCG | 0.0030 | 0.0010 | 0.0023 |
Table 4.
Radiation-Chemical Yields of Product Formation in CG10 Grouped by the Cleavage Site (site numbering starts at the 5’-end)
| Site | Radiation-Chemical Yield (μ mol/J)
|
||||
|---|---|---|---|---|---|
| 5'-side
|
3'-side
|
Average | |||
| Product | Yield | Product | Yield | ||
| G4 | CGCp | 0.004 | pCGCGCG | 0.006 | 0.005 |
| C5 | CGCGp | 0.002 | pGCGCG | 0.006 | 0.004 |
| G6 | CGCGCp | 0.003 | pCGCG | 0.007 | 0.005 |
| C7 | CGCGCGp | 0.003 | pGCG | 0.006 | 0.004 |
| G8 | CGCGCGCp | 0.004 | pCG | 0.007 | 0.006 |
Tables 3–4 show radiation-chemical yields for products in CG8 and CG10 grouped according to the cleavage site they originate from. Figure 2 shows the product yields from these tables plotted as a function of cleavage site for all more or less reliably quantifiable products in CG8 and CG10. Based on average yields, the cleavage probabilities appear to be fairly uniform for all internal sites regardless of the base. This is consistent with the model in which the rate of hole transfer from the sugar to the base is not controlled to any significant extent by oxidation potential of the base as it should be for a highly energetic process (9). It is also consistent with our earlier results on unaltered base release from crystalline DNA that showed statistically equal probability of damage for all sugars in the sequence.
A quantity of considerable interest is the yields of immediate strand breaks. In our previous studies of strand break products in crystalline DNA (5, 9), the yields (in μmol/J) are 0.14± 0.03 for d(CGCG)2, 0.06–0.11 for d(CGCGCG)2, 0.16±0.03 for d(CGCACG):d(CGTGCG) and 0.07±.02 for d(CACGCG):d(CGCGTG). Stressing the above caveats regarding absolute accuracy and using extrapolated values for the products that could not be quantified, the strand break yields for CG8 and CG10 are in the ranges, 0.02–0.04 μmol/J and 0.03–0.08 μmol/J, respectively. These approximate yields continue to build the case for ~10% of the direct ionizations resulting in immediate strand breaks and that the break is initiated by one-electron oxidation of the sugar phosphate backbone.
Conclusions
The rate of electron transfer from DNA bases to the holes generated on the sugar-phosphate backbone in directly ionized DNA is not controlled by the nature of the base, in agreement with the large negative G expected for the process. The sugar damage results in a strand break with release of a pair of 3’- and 5’-end phosphates per each cleavage site. The observed preference for the production of 5’-phosphorylated products suggests that some sugar remnants remain attached to the terminal phosphates primarily at the 3’-end. Products containing sugar remnants may account for IC peaks that have not yet been identified.
Acknowledgments
We thank Kermit R. Mercer for his invaluable technical assistance. The investigation was supported by PHS Grant 2-R01-CA32546, awarded by the National Cancer Institute, DHHS. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Cancer Institute.
References
- 1.LaVere T, Becker D, Sevilla MD. Yields of OH· in γ-irradaited DNA as a function of DNA hydration: Hole transfer vs. OH· formation. Radiation Research. 1996;145:673–680. [PubMed] [Google Scholar]
- 2.Mroczka NE, Mercer KR, Bernhard WA. The effects of lattice water on free radical yields in x-irradiated crystalline pyrimidines and purines: a low-temperature electron paramagnetic resonance investigation. Radiat Res. 1997;147:560–568. [PubMed] [Google Scholar]
- 3.Debije MG, Strickler MD, Bernhard WA. On the efficiency of hole and electron transfer from the hydration layer to DNA: an EPR study of crystalline DNA X-irradiated at 4 K. Radiat Res. 2000;154:163–170. doi: 10.1667/0033-7587(2000)154[0163:oteoha]2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baverstock KF, Will S. Evidence for the dominance of direct excitation of DNA in the formation of strand breaks in cells following irradiation. Int J Radiat Biol. 1989;55:563–568. doi: 10.1080/09553008914550611. [DOI] [PubMed] [Google Scholar]
- 5.Debije MG, Razskazovskiy Y, Bernhard WA. The Yield of Strand Breaks Resulting from Direct-Type Effects in Crystalline DNA X-Irradiated at 4 K and Room Temperature. J Am Chem Soc. 2001;123:2917–2918. doi: 10.1021/ja005790r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Razskazovskiy Y, Debije MG, Bernhard WA. Direct radiation damage to crystalline DNA: what is the source of unaltered base release? Radiat Res. 2000;153:436–441. doi: 10.1667/0033-7587(2000)153[0436:drdtcd]2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meggers E, Dussy A, Schafer T, Giese B. Electron transfer in DNA from guanine and 8-oxoguanine to a radical cation of the carbohydrate backbone. Chem--Eur J. 2000;6:485–492. doi: 10.1002/(sici)1521-3765(20000204)6:3<485::aid-chem485>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 8.Marcus R, Sutin N. Electron transfer in chemistry and biology. Biochim Biophys Acta. 1985;811:265–322. [Google Scholar]
- 9.Razskazovskiy Y, Debije MG, Bernhard WA. Strand Breaks Produced in X-irradiated Crystalline DNA: Influence of Base Sequence. Radiation Research. doi: 10.1667/0033-7587(2003)159[0663:sbpixc]2.0.co;2. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujii S, Wang AHJ, Quigley GJ, Westerink H, Van Der Marel GA, Van Boom JH, Rich A. The Octamers d(CGCGCGCG) and d(CGCATGCG) Both Crystallize as Z-DNA in the Same Hexagonal Lattice. Biopolymers. 1985;24:243–250. doi: 10.1002/bip.360240118. [DOI] [PubMed] [Google Scholar]
- 11.Kumar VD, Harrison RW, Andrews LC, Weber IT. Crystal Structure at 1.5 Angstroms Resolution of d(CGCICICG), an Octanucleotide Containing Inosine, and Its Comparison with d(CGCG) and d(CGCGCG) Structures. Biochemistry. 1992;31:1541–1550. doi: 10.1021/bi00120a035. [DOI] [PubMed] [Google Scholar]
- 12.Debije MG, Bernhard WA. Free radical yields in crystalline DNA X-irradiated at 4 K. Radiat Res. 1999;152:583–589. [PMC free article] [PubMed] [Google Scholar]
- 13.Pogozelski WK, Tullius TD. Oxidative Strand Scission of Nucleic Acids: Routes Initiated by Hydrogen Abstraction from the Sugar Moiety. Chem Rev. 1998;98:1089–1107. doi: 10.1021/cr960437i. [DOI] [PubMed] [Google Scholar]
- 14.Swarts SG, Miao L, Wheeler KT, Sevilla MD, Becker D. Radiation-induced DNA damage as a function of DNA hydration. In: Zimbrick AFF, Zimbrick JD, editors. Radiation Damage in DNA: Structure/Function Relationships at Early Times. Battelle Press; Columbus, OH: 1995. pp. 131–138. [Google Scholar]
- 15.Swarts SG, Becker D, Sevilla M, Wheeler KT. Radiation-induced DNA damage as a function of hydration. II. Base damage from electron-loss centers. Radiat Res. 1996;145:304–314. [PubMed] [Google Scholar]
- 16.von Sonntag C. The Chemical Basis of Radiation Biology. Taylor and Francis; New York: 1987. [Google Scholar]


