Abstract
Breast cancers lacking estrogen and progesterone receptor expression and Her2 amplification exhibit distinct gene expression profiles and clinical features, and they comprise the majority of BRCA1-associated tumors. Here we demonstrated that the p53 family member p63 controls a pathway for p73-dependent cisplatin sensitivity specific to these “triple-negative” tumors. In vivo, ΔNp63 and TAp73 isoforms were coexpressed exclusively within a subset of triple-negative primary breast cancers that commonly exhibited mutational inactivation of p53. The ΔNp63α isoform promoted survival of breast cancer cells by binding TAp73 and thereby inhibiting its proapoptotic activity. Consequently, inhibition of p63 by RNA interference led to TAp73-dependent induction of proapoptotic Bcl-2 family members and apoptosis. Breast cancer cells expressing ΔNp63α and TAp73 exhibited cisplatin sensitivity that was uniquely dependent on TAp73. Thus, in response to treatment with cisplatin, but not other chemotherapeutic agents, TAp73 underwent c-Abl–dependent phosphorylation, which promoted dissociation of the ΔNp63α/TAp73 protein complex, TAp73-dependent transcription of proapoptotic Bcl-2 family members, and apoptosis. These findings define p63 as a survival factor in a subset of breast cancers; furthermore, they provide what we believe to be a novel mechanism for cisplatin sensitivity in these triple-negative cancers, and they suggest that such cancers may share the cisplatin sensitivity of BRCA1-associated tumors.
Introduction
The identification of novel cancer subtypes promises to provide more specific, more effective, and less toxic therapies. Global gene expression profiling has uncovered previously unrecognized subsets of human breast cancer, including the “triple-negative” or “basal-like” subset characterized by a lack of estrogen receptor (ER) and progesterone receptor (PR) expression, the absence of Her2 amplification, and the expression of basal epithelial markers (1, 2). Triple-negative breast cancers are the most common subtype arising in patients harboring germline mutations in the breast cancer predisposition gene breast cancer 1, early onset (BRCA1). Both BRCA1-associated and the more common sporadic triple-negative tumors share similar gene expression profiles (3, 4). More importantly, both are refractory to commonly used chemotherapeutic agents and as a result are associated with a relatively poor prognosis (1). Therefore, identification of new therapeutic options for patients with these tumors is an important goal. While some reports have suggested that BRCA1-associated triple-negative tumors exhibit particular sensitivity to cisplatin chemotherapy (5), it is unknown whether a common mechanism for cisplatin sensitivity is shared between sporadic and BRCA1-associated triple-negative tumors.
The p53-related transcription factor p63 is an essential regulator of mammary epithelial development. In both mice and humans, mutation or inactivation of p63 leads to failure of early mammary development as well as severe developmental abnormalities of the skin, limbs, and other ectoderm-derived tissues (6, 7). In the adult breast, p63 expression is restricted to the basal myoepithelial cell layer, which is known to contribute to proliferation, differentiation, and polarity of mammary epithelia (8, 9). However, expression of p63 is not detectable in the majority of invasive breast carcinomas, consistent with the fact that most breast tumors exhibit a luminal, rather than a basal, epithelial phenotype (2, 10). Nevertheless, previous studies have suggested that a fraction of invasive ductal breast carcinomas express p63 protein, with reports ranging from 0%–30% of cases (11–14). The importance of p63 in mammary development and its potential expression in a subset of breast cancers therefore suggest a possible role for p63 in breast cancer pathogenesis.
Like p53 and the related protein p73, p63 is a sequence-specific DNA-binding factor that regulates transcription of critical downstream target genes. All 3 p53 family members possess a highly homologous DNA-binding domain, through which they regulate both shared and distinct subsets of transcriptional targets (15). Expression from 2 distinct p63 promoters produces protein isoforms that either contain or lack the N-terminal transactivation domain (TAp63 or ΔNp63, respectively). Differential mRNA splicing also gives rise to multiple C-terminal variants (16). The predominant p63 isoform expressed in most epithelial cells, ΔNp63α, exhibits properties of both a transcriptional activator and a repressor (17). A tumor-specific role for ΔNp63α in epithelial cells is supported by the observation that ΔNp63α is overexpressed in up to 80% of primary squamous cell carcinomas (SCCs) of the head and neck, lung, and esophagus (18–22). Our recent work demonstrates that in head and neck SCC, ΔNp63α promotes tumor cell survival through repression of p73-dependent apoptosis (23, 24).
Through our studies seeking to determine the role of p63 in human breast cancer, we have uncovered a p63-dependent tumor survival pathway that directly mediated cisplatin sensitivity specifically in triple-negative tumors. We found that ΔNp63 and proapoptotic TAp73 isoforms were coexpressed exclusively within this tumor subset in vivo. Breast tumor cells coexpressing ΔNp63 and TAp73 exhibited sensitivity to cisplatin, but not to common chemotherapeutic agents used for breast cancer treatment, as a result of phosphorylation-dependent dissociation of TAp73 from ΔNp63α and subsequent transcription of proapoptotic effectors. Together these results identify a p73-dependent pathway for specific chemosensitivity in a biologically defined subset of refractory breast cancers.
Results
TAp73 and ΔNp63 are coexpressed in a biologically defined subset of primary invasive breast carcinomas.
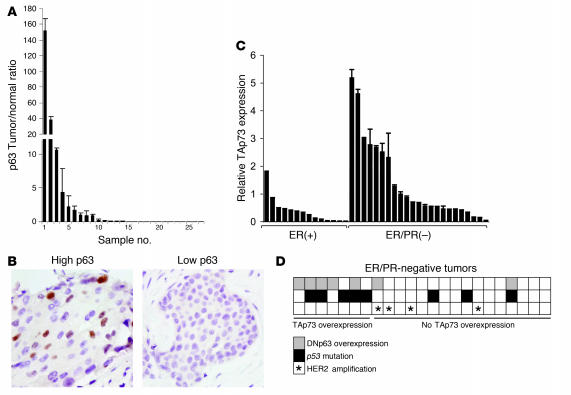
Previous studies of p63 expression in breast carcinoma have relied almost exclusively on immunohistochemistry, which is relatively insensitive and only roughly quantitative. In order to perform quantitative assessment of p63 levels, we initially used a panel of breast tumor specimens that have been extensively characterized by laser-capture microdissection and microarray-based gene expression profiling (25). These specimens allowed us to compare p63 expression levels in specimen-matched normal luminal epithelial cells and invasive carcinoma cells. We directly examined p63 expression by real-time quantitative RT-PCR (QRT-PCR) in 27 matched normal epithelium/invasive tumor pairs. While most tumors exhibited p63 expression levels that were equal to or lower than the matched normal epithelia, we found that p63 was expressed at more than 2-fold the level of matched normal epithelia in 5 of 27 specimens (Figure 1A). To confirm that these results reflected p63 expression within malignant cells, we performed immunohistochemistry for p63 in specimens exhibiting both low and high p63 levels. Consistent with the results from QRT-PCR, we observed nuclear expression of p63 protein specifically in malignant epithelial cells from specimens showing p63 mRNA overexpression, but not from those showing low or no expression (Figure 1B). Thus p63 mRNA and protein are expressed in a subset of primary breast tumor cells in vivo.
Figure 1. Coexpression of ΔNp63 and TAp73 in triple-negative primary breast tumors.
(A) Overexpression of p63 in primary microdissected invasive breast carcinomas relative to specimen-matched normal luminal epithelia. The ratio of tumor/normal p63 mRNA was determined by QRT-PCR. (B) Nuclear p63 protein correlated with p63 mRNA expression, as assessed by immunohistochemistry of representative tumors from A. Photomicrographs demonstrate low and high expression of p63 mRNA. Original magnification, ×100. (C) TAp73 was overexpressed in ER/PR-negative tumors. Shown is QRT-PCR for TAp73 in 14 ER-positive and 23 ER/PR-negative primary breast carcinomas. Statistical significance was analyzed using both a mean-value approach (ER-positive, 0.373 ± 0.126; ER/PR-negative, 1.381 ± 0.303; P = 0.0172, 2-way Student’s t test) and a binning approach whereby tumors exhibiting p73 expression more than 2-fold the mean of the sample set were categorized as high and the rest as low (P = 0.0309, Fisher’s exact test). (D) Expression of TAp73 correlated with ΔNp63 overexpression (P = 0.0107, Fisher’s exact test) and with p53 mutation (P = 0.0257, Fisher’s exact test) in ER/PR-negative primary breast carcinomas. Levels of ΔNp63 were determined by QRT-PCR, and p53 mutation was determined by cDNA sequencing. Note that TAp73/ΔNp63 coexpression was not observed in Her2-overexpressing tumors as assessed by QRT-PCR.
Given the distinct functional properties of N-terminal TAp63 and ΔNp63 isoforms (16), we next examined expression of these forms of p63, as well as the related gene p73, in primary breast carcinomas. We identified a group of primary invasive breast carcinoma specimens from which frozen tissue was available for preparation and analysis of unamplified RNA. Of note, all tumors were removed prior to any radiation, hormonal, or chemotherapy treatment. We used isoform-specific QRT-PCR to directly assess levels of TAp63, ΔNp63, TAp73, and N-terminally truncated p73 isoforms ΔNp73 and ΔN’p73 (hereafter referred to as ΔNp73) in these samples. In order to compare the level of each isoform relative to the others, we used cDNA templates to generate standard curves for each isoform (see Methods). To ensure that our findings reflected gene expression within malignant cells, samples underwent histopathologic analysis and were found to contain no more than 5% ductal carcinoma in situ (DCIS) or normal epithelia.
In a series of 37 such tumor specimens, the most striking finding of our examination was the overexpression of ΔNp63 (more than 2-fold the mean value of the sample set) and its statistically significant association (P < 0.05, Fisher’s exact test) with overexpression of TAp73 isoforms in a subset of breast carcinomas lacking ER and PR expression (Figure 1, C and D). Overexpression of TAp73 in particular was highly associated with ER/PR negativity (Figure 1C). To validate the correlation between mRNA and protein levels of TAp73 and ΔNp63, we compared QRT-PCR values to the respective protein levels determined by Western blot analysis in a panel of breast cancer cell lines. We obtained r2 values greater than 0.9 for both TAp73 (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI30866DS1) and ΔNp63 (data not shown). In contrast to the relatively high expression of TAp73, ΔNp73 isoforms were expressed at very low levels, with a mean fold ratio of TAp73/ΔNp73 mRNA in the sample set greater than 70:1, consistent with prior reports (26). In addition, TAp63 isoforms were expressed at substantially lower levels than ΔNp63 isoforms in most of the specimens and were not associated with TAp73 expression (data not shown). Thus, TAp73 and ΔNp63 are selectively co-overexpressed within a subset of ER/PR-negative primary breast tumors.
Given that p63 and p73 may also exhibit functional interactions with p53, we next examined the mutational status of p53 in these tumors by cDNA sequencing (Supplemental Table 1). Among the 7 tumors exhibiting overexpressed TAp73, 5 of 7 harbored missense or nonsense mutations in p53, while only 3 of 16 tumors lacking TAp73 overexpression exhibited mutant p53 (P = 0.0257, Fisher’s exact test; Figure 1D). This statistically significant finding suggests a link between p53 inactivation and TAp73 overexpression, which is in agreement with a prior report suggesting an association between p73 and mutant p53 expression in primary breast carcinomas (27).
As noted above, breast tumors characterized by the absence of both ER/PR expression and Her2 overexpression appear biologically and clinically distinct (1, 10). We therefore assessed Her2 mRNA expression in the tumor set by QRT-PCR, using a clinically validated cutoff value for Her2 overexpression of greater than 10-fold the mean normal level (28), and found that no tumors exhibiting co-overexpression of TAp73 and ΔNp63 overexpressed Her2 (Figure 1D). Together these findings demonstrate that overexpression of TAp73 and ΔNp63 is restricted to a subset of ER/PR/Her2-negative primary breast carcinomas that commonly exhibit mutation of p53. These tumors may therefore represent a distinct biologic subtype with particular phenotypic properties.
Requirement of p63 for survival in breast cancer cells.
To study the potential functional contribution of p63 and p73 in breast cancer cells, we next examined their expression in human tumor–derived breast carcinoma cell lines. Consistent with our findings in primary breast tumors, we found that a subset of breast cancer cell lines expressed ΔNp63α, the major p63 isoform expressed in normal epithelia (16). Several cell lines, including MCF-7, exhibited low levels of ΔNp63α mRNA and protein, while other lines, including HCC-1937, MDA-MB-468, and T47D, expressed substantially higher levels of ΔNp63α (Figure 2A and Supplemental Figure 1). As in primary breast tumors, TAp73 was coexpressed with ΔNp63 in these cell lines (Supplemental Figure 1). Also consistent with our findings in primary tumors, 2 of 3 tumor-derived cell lines we identified as coexpressing ΔNp63 and TAp73, HCC-1937 and MDA-MB-468, exhibited a triple-negative phenotype and mutational inactivation of p53. Furthermore, HCC-1937 is a BRCA1-associated triple-negative tumor cell line, while MDA-MB-468 is a triple-negative tumor cell line expressing wild-type BRCA1 (29).
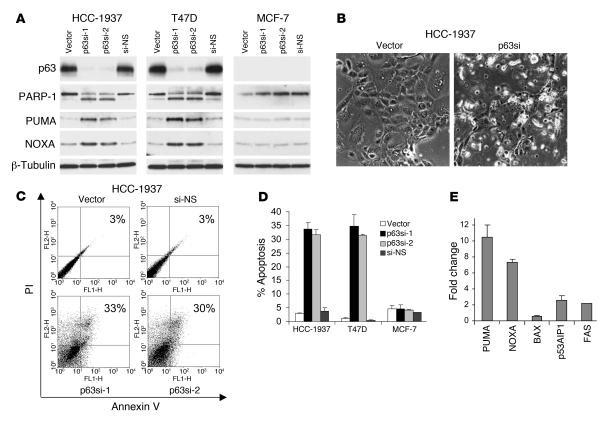
Figure 2. Endogenous p63 is required for survival in breast cancer cells.
(A) Knockdown of endogenous p63–induced Puma, Noxa, and PARP-1 cleavage, as assessed by immunoblot of the indicated cells 72 hours after infection with lentiviral shRNA vectors targeting 2 distinct p63 sequences (p63si-1 and p63si-2) or a nonspecific sequence (si-NS). None of these effects were observed in MCF-7 cells, which do not express abundant p63. (B) Apoptotic morphology following lentiviral p63 knockdown in HCC-1937 cells. Photomicrographs were taken 72 hours after lentiviral shRNA infection. Original magnification, ×100. (C) Apoptosis was observed following endogenous p63 knockdown, as assessed by annexin V/PI staining of unfixed cells 72 hours following infection with the indicated lentiviral shRNA vectors. Percentages indicate apoptotic cells (annexin V–positive and/or PI-positive). (D) Apoptosis following p63 inhibition was specific to breast cancer cells expressing abundant p63. Shown are results of annexin V/PI staining for 3 independent experiments. Error bars represent SD. (E) Puma and Noxa were induced following p63 knockdown. RNA was prepared 72 hours following lentiviral infection and assayed by QRT-PCR for the indicated genes. Error bars represent SD for 2 independent experiments, each performed in duplicate.
To study the function of endogenous p63, we recently developed an efficient system for lentiviral-based RNA interference (RNAi). These viruses express small hairpin RNA (shRNA) species that are processed to siRNAs. We tested the effect of endogenous p63 knockdown by lentiviral RNAi in breast cancer cells that express endogenous ΔNp63α. As a control for the specificity of our shRNA species, we tested the effect of 2 independent p63-directed shRNAs. As an additional control, we also examined the effects of expressing these shRNAs in MCF-7 (Figure 2A) and Saos-2 cells (data not shown), both of which express little or no endogenous p63.
We first demonstrated efficient knockdown of p63 expression by lentiviral RNAi in HCC-1937 and T47D cells, both of which express endogenous ΔNp63α. We routinely achieved greater than 75% knockdown of p63 mRNA and protein within 48–72 hours of lentiviral infection in these cells, as assessed by QRT-PCR and Western analysis, respectively (Figure 2A and Supplemental Figure 2). In contrast, infection with the control lentivirus or a nonspecific shRNA did not affect endogenous p63 levels. Inhibition of p63 in both these p63-expressing breast cancer lines induced obvious cell death associated with nuclear blebbing characteristic of apoptosis (Figure 2B). These morphologic changes were not observed following infection of these cells with either control lentivirus, nor were they observed following p63-directed RNAi in either MCF-7 or Saos-2 cells (Supplemental Figure 2 and ref. 24).
To confirm whether p63 loss induced apoptosis, we performed Western analysis for cleavage of poly(ADP-ribose) polymerase–1 (PARP-1), a specific hallmark of apoptotic cell death. PARP-1 cleavage was observed specifically in response to p63 knockdown, not following control lentiviral infection. Furthermore, PARP-1 cleavage was observed in HCC-1937 and T47D cells, but not in MCF-7 cells, which do not express endogenous p63 (Figure 2A). To quantitate apoptotic cell death, we assayed annexin V/propidium iodide (annexin V/PI) staining of unfixed cells by FACS analysis. This assay detects both early apoptotic (annexin V–positive, PI-negative) and late apoptotic (annexin V–positive, PI-positive) cells (30). Prominent annexin V/PI staining correlated with PARP-1 cleavage and with morphologic features of apoptosis and was induced specifically in response to p63 knockdown only in cell lines expressing endogenous ΔNp63α (Figure 2, C and D). Together these findings demonstrate that specific inhibition of endogenous p63 induces apoptosis in breast cancer cells in which it is expressed.
To begin to address the mechanism by which p63 inhibition induces apoptosis of breast cancer cells, we first assayed expression of proapoptotic genes known to be regulated by p53 family members. We observed substantial induction of Puma and Noxa, but not of Bax, p53AIP1, or Fas, coincident with p63 knockdown in HCC-1937 and T47D cells (Figure 2, A and E). Induction of both Puma and Noxa was a specific effect of p63 inhibition, as neither gene was induced following control shRNA lentiviral infection in these cells, nor were they induced by p63-directed shRNA in MCF-7 or Saos-2 cells, which do not express endogenous ΔNp63α (Figure 2A, Supplemental Figure 2, and data not shown).
Apoptosis following p63 inhibition depends on endogenous p73.
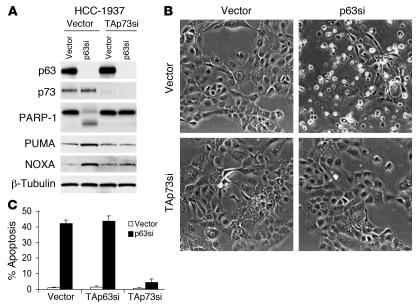
ΔNp63α has been hypothesized to function as an inhibitor of its proapoptotic paralogs p53 and TAp73 (16, 17, 24, 31). Neither HCC-1937 nor T47D cells express wild-type p53, demonstrating that the p63-dependent survival effect in these cells is not mediated through repression of p53 function (32). In contrast, both cell lines expressed TAp73 isoforms (Figure 3A and Supplemental Figure 3), and we and others have previously demonstrated that TAp73 is a direct transcriptional activator of Puma, implying a potential contribution of TAp73 to induction of Puma and cell death following inhibition of p63 (24, 33). In order to determine whether Puma induction and apoptosis following loss of p63 were mediated by TAp73, we inhibited TAp73 in HCC-1937 cells by expressing a lentiviral shRNA construct that we have previously shown to be a potent and specific inhibitor of this isoform, followed by brief drug selection (24). We confirmed knockdown of p73 mRNA and protein by QRT-PCR and IP/Western blot analysis, respectively, in these cells compared with control vector–infected cells (Figure 3A and Supplemental Figure 3). We then quantitated cell death following p63 knockdown in the TAp73-inhibited cells compared with that of control cells. As an additional control, we also generated cells expressing an shRNA directed against TAp63. Only inhibition of TAp73 consistently abrogated Puma induction, PARP cleavage, and cell death following knockdown of ΔNp63α in HCC-1937 cells (Figure 3). Similar results were observed in T47D cells (Supplemental Figure 3). These data suggest that ΔNp63α, when present in breast cancer cells, promotes cell survival by inhibiting the proapoptotic activity of transactivating isoforms of p73. As discussed below, this inhibition involves direct binding of ΔNp63α to TAp73 (see Dissociation of the p63/p73 complex is induced by cisplatin treatment).
Figure 3. Survival of breast cancer cells is promoted by p63 through repression of TAp73-dependent apoptosis.
(A) PARP-1 cleavage, Puma and Noxa induction, and apoptosis induced by p63 knockdown were TAp73 dependent. Pools of cells expressing a TAp73-targeted shRNA (TAp73si) or the control vector were then infected with a p63-directed lentiviral shRNA or control, and lysates were harvested at 72 hours for immunoblot and IP/immunoblot (for p73). (B) Morphologic features of apoptosis were TAp73 dependent. Shown are photomicrographs of representative fields of cells treated as in A and harvested 72 hours following p63 knockdown. Original magnification, ×100. (C) Rescue from apoptosis following ablation of TAp73 but not TAp63. Quantitation of apoptosis by annexin V/PI staining of cells treated as in A and harvested 72 hours following p63 knockdown. Error bars represent SD for 3 independent experiments.
TAp73 mediates specific cisplatin sensitivity in breast cancer cells exhibiting ΔNp63/TAp73 expression.
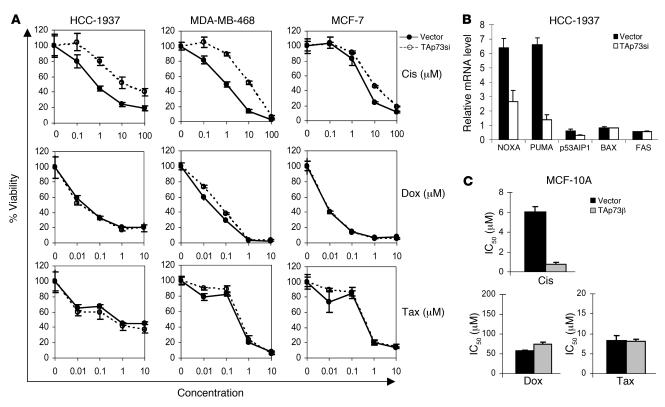
Expression of p73 has recently been identified as a potential contributor to chemosensitivity in a variety of contexts (34–38). Because TAp73 is expressed in triple-negative breast cancer cells but functionally repressed by ΔNp63α, we asked whether chemotherapy exposure might promote escape from this repression and thereby allow TAp73 to mediate chemosensitivity. To directly assess the contribution of TAp73 to chemosensitivity, we first tested the effect of ablating endogenous TAp73 by lentiviral RNAi prior to chemotherapy treatment. We examined HCC-1937 and MDA-MB-468, 2 triple negative cell lines that coexpress ΔNp63 and TAp73. As a control we examined MCF-7, an ER-positive, p63-negative breast cancer line that did not exhibit p63-dependent survival (Figure 2). Pools of cells stably expressing the TAp73-directed lentivirus or the vector control were generated (Supplemental Figure 4). We first tested chemosensitivity to doxorubicin or paclitaxel, 2 of the most commonly used agents for treatment of early-stage breast cancer (39). Knockdown of TAp73 had less than a 2-fold effect on the IC50 for either drug as assessed by quantitative cell viability assay (Figure 4A). In contrast, TAp73 ablation induced marked cisplatin resistance in both triple-negative cell lines: the IC50 for cisplatin was increased by more than 10-fold in each of these cell lines following TAp73 inhibition (Figure 4A and Supplemental Figure 4). In MCF-7 cells, TAp73 was not a mediator of sensitivity to cisplatin or other chemotherapeutic agents (Figure 4A). These findings suggest that TAp73 is a mediator of cisplatin sensitivity in breast cancer cells expressing ΔNp63 and TAp73.
Figure 4. TAp73 mediates cisplatin sensitivity in breast cancer cells expressing TAp73 and ΔNp63.
(A) Inhibition of TAp73 induced resistance specifically to cisplatin. Dose-response curves (MTT cell viability assay) of cells expressing the control vector or a TAp73-directed lentiviral shRNA 5 days following treatment with cisplatin (Cis), doxorubicin (Dox), or paclitaxel (Tax). Little or no effect of TAp73 knockdown was observed in MCF-7 cells. Error bars show SD for 3 independent experiments. (B) TAp73 mediated selective proapoptotic target gene induction in response to cisplatin. QRT-PCR analysis of the indicated genes in HCC-1937 cells as in A 6 hours after cisplatin treatment (at IC70, 6.6 μM). (C) TAp73 expression conveyed specific cisplatin sensitivity to normal basal mammary epithelial cells. MCF-10A cells were infected with a retrovirus encoding TAp73β or the control vector, followed by quantitative dose-response analysis as shown in A (P < 0.01, 1-tailed Student’s t test). TAp73β increased sensitivity (i.e., decreased the IC50) only for cisplatin. Error bars show SD for 3 independent experiments.
To further explore the p73-dependent pathway induced by cisplatin, we next examined expression of candidate apoptotic effector genes following chemotherapy treatment. We observed a pattern of gene induction that was remarkably similar to that observed following p63 knockdown: Puma and Noxa, but not other proapoptotic effector genes, were robustly induced following cisplatin treatment. Most importantly, their induction was p73 dependent, as it was attenuated following treatment with TAp73-directed shRNA (Figure 4B). In contrast, doxorubicin treatment caused a 2-fold increase in Puma and no significant induction of other proapoptotic effectors at an equally toxic concentration (data not shown). Together these findings suggest that TAp73 induces a transcriptional program specifically in response to cisplatin treatment that leads to the death of breast cancer cells.
We previously reported that normal basal mammary epithelial cells express ΔNp63α but little or no TAp73 (9). The findings described above predict that ectopic TAp73 expression in such normal cells should enhance cellular sensitivity to cisplatin. To test this possibility we expressed TAp73 ectopically in MCF-10A, a nontransformed, nontumorigenic breast epithelial cell line that expresses relatively high levels of ΔNp63α but little or no TAp73 (9). MCF-10A cells were stably infected with a retrovirus encoding TAp73β, followed by brief drug selection (Supplemental Figure 4). Ectopic TAp73β expression significantly increased the sensitivity of MCF-10A cells to cisplatin (P < 0.01; Figure 4C and Supplemental Figure 4), even though it did not affect their baseline cell proliferation or viability (data not shown). This effect was highly specific, as TAp73β expression did not significantly enhance sensitivity to either doxorubicin or paclitaxel (Figure 4C). Together these data suggest that TAp73, which is normally inactive in breast cancer cells due to its coexpression with ΔNp63α, is activated to induce a cell death pathway specifically in response to cisplatin treatment, resulting in chemosensitivity to this agent.
Dissociation of the p63/p73 complex is induced by cisplatin treatment.
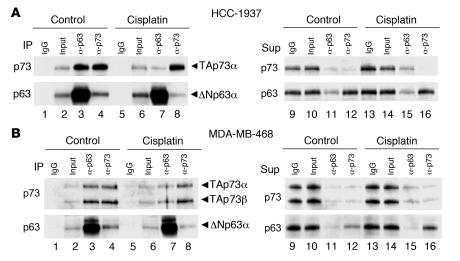
We next sought to determine the mechanism by which cisplatin treatment induced specific p73 activation in breast carcinoma cells. In SCCs endogenous ΔNp63α binds to TAp73, which we and others have shown to be sufficient for repression of TAp73-dependent transcription (24, 31). To determine whether this same mechanism was operative in breast carcinoma cells, we tested for ΔNp63α/TAp73 interaction by coimmunoprecipitation of the respective endogenous proteins. In both HCC-1937 and MDA-MB-468 cells, endogenous TAp73 was immunoprecipitated by p63-directed antisera; similarly, endogenous ΔNp63α was immunoprecipitated following IP for p73 (Figure 5, A and B). Furthermore, in both cell lines ΔNp63α was in molar excess relative to TAp73, and we observed that IP for either p63 or p73 yielded the same mass of p73 and essentially depleted p73 from the post-IP supernatant (Figure 5, A and B). These observations suggest that the vast majority of TAp73 is complexed to ΔNp63α in breast cancer cells under normal growth conditions.
Figure 5. Cisplatin induces dissociation of the ΔNp63α/TAp73 complex.
(A) Quantitative binding of TAp73 to ΔNp63α in HCC-1937 cells and dissociation following cisplatin treatment. Left, IPs of control or cisplatin-treated cultures (IC70 for 6 hours); right, corresponding post-IP supernatants (Sup). IP for either p63 or p73 resulted in complete immunodepletion of TAp73 (lanes 11 and 12). Following cisplatin treatment, less TAp73 was associated with ΔNp63α (lanes 3 and 7), resulting in detectable “free” TAp73 in the depleted post-IP supernatant (lanes 11 and 15). Note the absence of change in ΔNp63α or TAp73 protein levels following cisplatin treatment (lanes 2, 6, 10, and 14). Controls demonstrated these antibodies to be non–cross-reactive (ref. 24 and data not shown). (B) MDA-MB-468 cells showed quantitative ΔNp63α/TAp73 binding similar to that of HCC-1937 cells and partial dissociation following cisplatin treatment. Cells were treated as in A. Left, IP product; right, post-IP supernatant. Note the decrease in TAp73 associated with ΔNp63α following cisplatin treatment (lanes 3 and 7), despite no change in ΔNp63α or TAp73 protein levels (lanes 10 and 14). HCC-1937 cells expressed TAp73α (A), while MDA-MB-468 cells expressed both TAp73α and TAp73β (B).
As noted above, we observed TAp73-dependent transcription of proapoptotic effector genes within several hours of cisplatin treatment, absent any change in ΔNp63α or TAp73 protein levels (Figure 4B and Figure 5, A and B). We therefore sought to explain how cisplatin treatment allows TAp73 to escape ΔNp63α-mediated repression. Remarkably, we observed substantial dissociation of the ΔNp63α/TAp73 complex 6 hours following cisplatin treatment, as assessed by coimmunoprecipitation for either endogenous protein (Figure 5, A and B). These findings were confirmed by examining the immunodepleted post-IP supernatants: little or no residual p73 was detected following depletion for p63 in the basal state; in contrast, “free” TAp73 was readily detectable following cisplatin treatment (Figure 5A). Notably, these findings were highly reproducible in both HCC-1937 and MDA-MB-468 cells (Figure 5B). While we did observe a decline in ΔNp63α protein expression beginning at 12 hours, consistent with prior reports (40), our findings suggest that dissociation of the ΔNp63/TAp73 complex contributes to cisplatin-induced TAp73 activation.
TAp73 is phosphorylated in a c-Abl–dependent manner specifically following cisplatin treatment.
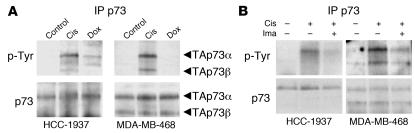
Previous studies have implicated phosphorylation of p73 in its activation following DNA damage (35, 37, 38). Therefore, to uncover the molecular events leading to TAp73 dissociation and activation, we examined p73 phosphorylation following chemotherapy treatment. In both HCC-1937 and MDA-MB-468 cells we observed tyrosine phosphorylation of p73 — which was maximal at 6 hours — in response to cisplatin treatment, but not doxorubicin treatment (Figure 6A). Of note, this phosphorylation coincided temporally with TAp73-dependent transcription (Figure 4B) and with dissociation of the ΔNp63/TAp73 complex (Figure 5). Given these previous studies linking c-Abl to activation of p73, we next tested whether phosphorylation was c-Abl dependent using the well-established c-Abl kinase inhibitor imatinib (41). Pretreatment of cells with imatinib resulted in substantial inhibition of p73 phosphorylation following cisplatin treatment (Figure 6B). These findings suggest that p73 is phosphorylated in a c-Abl–dependent manner specifically following cisplatin treatment.
Figure 6. Cisplatin treatment specifically induces c-Abl–dependent p73 phosphorylation.
(A) TAp73 is tyrosine phosphorylated in response to cisplatin but not doxorubicin. Immunoprecipitated p73 was probed for anti–phosphorylated tyrosine (p-Tyr) by immunoblot 6 hours following control or cisplatin (Cis) or doxorubicin (Dox) treatment (both at IC70). The same blot was then stripped and reprobed for total p73 protein. HCC-1937 cells expressed TAp73α, while MDA-MB-468 cells expressed both TAp73α and TAp73β. (B) Induction of c-Abl–dependent TAp73 phosphorylation following cisplatin treatment. Cells were pretreated with imatinib (Ima; 1 μM for 2 hours) or vehicle control as indicated and then treated with cisplatin and analyzed as in A.
Phosphorylation of TAp73 Y99 is essential for dissociation of the p63/p73 complex and for cisplatin sensitivity.
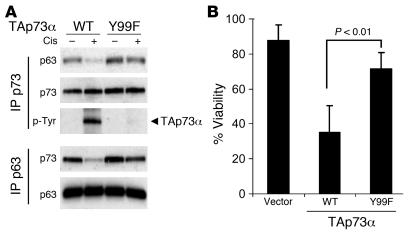
We next sought to determine whether c-Abl–dependent phosphorylation of p73 was required for p63/p73 dissociation and for the specific chemosensitivity to cisplatin mediated by TAp73. Previously published data indicate that Y99 is the major site of p73 phosphorylation by c-Abl and that phosphorylation at this site is critical for activation of p73 in response to DNA damage (38, 42, 43). Therefore, we first compared the effects of expressing either wild-type TAp73α or the site-specific Y99F mutant TAp73α in MCF-10A cells. As noted above, these cells express endogenous ΔNp63α but not TAp73. Stable retroviral expression of TAp73 did not affect baseline proliferation or viability of MCF-10A cells (data not shown). As predicted, ectopic wild-type TAp73 was strongly phosphorylated in response to cisplatin treatment in MCF-10A cells, while Y99F TAp73 underwent little detectable phosphorylation (Figure 7A). Furthermore, while IP for both wild-type and Y99F TAp73 coimmunoprecipitated abundant ΔNp63α at baseline, cisplatin induces substantial dissociation of ΔNp63α only from wild-type, but not Y99F, TAp73 (Figure 7A). The same result was evident following IP for p63 under these same conditions: dissociation of wild-type TAp73 from ΔNp63α was observed with 6 hours of cisplatin treatment, while essentially no dissociation of Y99F TAp73 was observed (Figure 7A). Finally, although wild-type TAp73 expression significantly increased sensitivity of MCF-10A cells to cisplatin treatment (P < 0.01), Y99F TAp73 had little, if any, effect on cisplatin sensitivity (Figure 7B). Of note, these findings are in contrast to another point mutant we tested, Y121F TAp73, which still underwent substantial phosphorylation and dissociation from ΔNp63α and still induced cellular sensitivity following cisplatin treatment (data not shown). Together these observations demonstrate that phosphorylation of TAp73 at Y99 is essential for dissociation of the ΔNp63α/TAp73 complex and for chemosensitivity following cisplatin treatment.
Figure 7. TAp73 phosphorylation at Y99 is required for cisplatin-induced ΔNp63α/TAp73 dissociation and cell death in MCF-10A cells.
(A) Wild-type or Y99F TAp73α were expressed in MCF-10A cells via retrovirus. Lysates from either cisplatin-treated or untreated cells were halved and subjected to IP for either p63 or p73, followed by immunoblots as shown. Wild-type TAp73α was tyrosine phosphorylated and dissociated from ΔNp63α following cisplatin treatment (1 μM, 6 hours), while Y99F TAp73α remained unphosphorylated and bound to ΔNp63α. Note there was no change in the total level of retroviral TAp73α or endogenous ΔNp63α following cisplatin treatment. (B) TAp73 Y99 phosphorylation was required to convey cisplatin sensitivity. MCF-10A cells described in A were treated with cisplatin (1 μM) for 5 days, and cell viability was assessed by MTT. Error bars show SD for 3 independent experiments.
TAp73 dissociation, proapoptotic transcription, and cell death induced by cisplatin are c-Abl dependent.
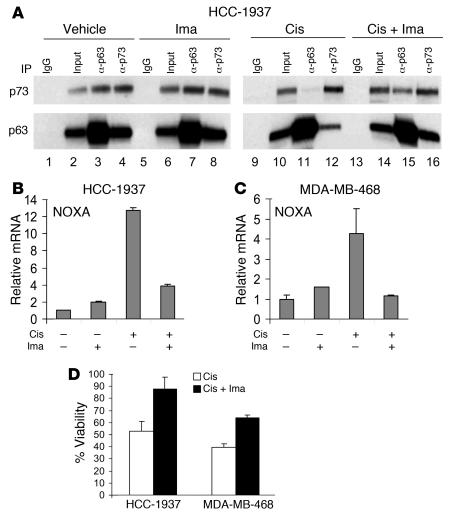
Given that Y99 is the major site for c-Abl phosphorylation of TAp73 (38, 42, 43), our data point to c-Abl as a critical kinase required for escape of TAp73 from ΔNp63α-mediated repression. To further substantiate this hypothesis, we asked whether imatinib could inhibit TAp73 activation and the resulting proapoptotic program induced by cisplatin. First, we examined dissociation of the ΔNp63α/TAp73 complex. Consistent with our model, pretreatment of cells with imatinib substantially blocked dissociation of the endogenous ΔNp63α/TAp73 complex induced by cisplatin in breast cancer cells (Figure 8A). Second, to determine whether imatinib could inhibit proapoptotic transcription induced by cisplatin, we assayed cisplatin-induced transcription of Noxa, which we had shown to be TAp73 dependent, with or without imatinib pretreatment. Indeed, pretreatment blocked induction of Noxa at 6 hours following cisplatin treatment in both HCC-1937 and MDA-MB-468 cells (Figure 8, B and C). This effect correlated with the ability of imatinib to block ΔNp63α/TAp73 dissociation at this same time point (Figure 8A). Of note, imatinib treatment had no effect on baseline Noxa expression in either cell line (data not shown). Finally, imatinib treatment led to a substantial rescue from cell death induced by cisplatin in both HCC-1937 and MDA-MB-468 cells (Figure 8D). Imatinib treatment did not affect cell proliferation in either cell line at the dose used in these assays (data not shown). Taken together, these results demonstrate that cisplatin induced death in breast cancer cells expressing ΔNp63α and TAp73 through a TAp73-mediated pathway involving c-Abl phosphorylation of TAp73. This phosphorylation was required for dissociation of the ΔNp63α/TAp73 complex, for TAp73-dependent transcription, and for apoptosis following cisplatin treatment.
Figure 8. Imatinib treatment blocks ΔNp63α/TAp73 dissociation, TAp73-dependent transcription, and cell death induced by cisplatin.
(A) Imatinib attenuated dissociation of ΔNp63α and TAp73. In left blots (control), cells were treated with imatinib (1 μM for 8 hours) or vehicle control; in right blots, cells were pretreated with imatinib (1 μM for 2 hours) or vehicle control, then treated with cisplatin (IC70 for 6 hours) prior to IP for p63 or p73. Dissociation of TAp73 and ΔNp63α following cisplatin treatment (compare lanes 3 and 11) was attenuated by imatinib treatment (lane 15). (B and C) TAp73-dependent proapoptotic transcription required c-Abl–mediated phosphorylation. HCC-1937 cells (B) or MDA-MB-468 cells (C) were pretreated with imatinib (1 μM for 2 hours) and/or treated with cisplatin (IC80 for 6 hours) as indicated, and mRNA was analyzed by QRT-PCR. (D) c-Abl–dependent phosphorylation is important for cisplatin sensitivity. Cells were pretreated with imatinib as in C, then treated with cisplatin (IC70) and analyzed for viability by MTT at 3 days. Error bars show SD for 3 independent experiments.
Discussion
We have defined a specific pathway that mediates cisplatin sensitivity through activation of the proapoptotic p53 family member TAp73 in breast cancer cells. Under normal growth conditions, survival of these cells depends upon the presence of ΔNp63α, which functions to repress the proapoptotic activity of TAp73 through direct physical interaction. Either inhibition of ΔNp63α expression by RNAi or dissociation of TAp73 from ΔNp63α by cisplatin treatment led to TAp73-dependent transcription of the same proapoptotic effector genes, followed by apoptosis. The specificity of this pathway for cisplatin, but not other chemotherapeutic agents, involved phosphorylation of p73 in a c-Abl–dependent manner, which promoted dissociation of the TAp73/ΔNp63α complex and was required for activation of the p73-dependent apoptotic program. The presence of this pathway in breast cancers in vivo is strongly supported by our observation TAp73 was co-overexpressed with inhibitory ΔNp63 exclusively in a subset of triple-negative primary breast carcinomas.
Breast tumors that lack ER/PR expression and Her2 amplification are among the most refractory of human breast cancers, because they cannot be treated with effective hormonal or Her2-targeted therapies. Microarray-based gene expression profiling has revealed that most triple-negative breast carcinomas express proteins associated with mammary basal and/or myoepithelial cells, including basal cytokeratins 5 and 6 (4, 10). The fact that ΔNp63 is highly expressed exclusively in basal cells of the normal mammary gland is therefore consistent with our data and those of others (44), that ΔNp63 was expressed in a subset of triple-negative primary tumors. Notably, we also assayed p63 expression in premalignant DCIS specimens. We found that p63 was overexpressed relative to normal luminal epithelia in only 2 of 39 specimens (data not shown). Thus, p63 is expressed in a subset of primary breast carcinomas, and its expression is restricted largely to invasive disease. While ΔNp63 is abundantly expressed in normal basal epithelia, we reported previously that p73 is expressed at very low levels in normal basal and luminal epithelia of the epidermis and mammary gland (9, 23). In breast cancers, we show here that proapoptotic TAp73 isoforms were overexpressed preferentially in tumors that exhibited overexpression of inhibitory ΔNp63. Specifically, 32% (6 of 19) of the triple-negative tumors we examined exhibited ΔNp63 overexpression and detectable TAp73 expression, with all but one of these showing high-level TAp73 expression. One implication of these findings is that tumor-specific expression of TAp73 in triple-negative breast cancers might contribute to particular sensitivity of these tumor cells rather than normal cells to cisplatin chemotherapy.
Previous studies have suggested that p73 may be an important mediator of the response to chemotherapy and other forms of DNA damage in tumor cells (34–38). We found that TAp73 was expressed but was physically associated with ΔNp63α in triple-negative breast tumor cells, and we and others have demonstrated that ΔNp63α potently inhibits the transcriptional activity of TAp73 (24, 31). However, despite the presence of ΔNp63α, we found that these cells exhibited cisplatin sensitivity that was mediated through TAp73-dependent transcription of proapoptotic effector genes. Furthermore, we observed that TAp73-dependent transcription was initiated in the absence of any change in the levels of TAp73 or ΔNp63α proteins. The resolution of this seeming paradox was provided by our finding that phosphorylation of p73 functioned to promote dissociation of the ΔNp63/TAp73 complex, leading to activation of TAp73-dependent transcription.
Recent studies have uncovered several kinase pathways that might contribute to activation of p73 following cisplatin treatment. These include AKT-dependent coactivator recruitment (45, 46), Chk1/2-dependent induction of p73 mRNA (47), and c-Abl–dependent phosphorylation (35, 37, 38). We provide several lines of evidence to suggest that the phosphorylation of p73 we observed is required for transcription and cell death induced by cisplatin and is mediated by the c-Abl kinase. First, previous reports have documented that TAp73 Y99 is the major site of p73 phosphorylation by c-Abl and that phosphorylation at this site is critical for activation of p73 in response to DNA damage (38, 42, 43). We found that the Y99F TAp73 mutant was virtually inactive in undergoing phosphorylation, in dissociating from ΔNp63α, and in sensitizing cells to death following cisplatin treatment. Second, we find that imatinib, a specific inhibitor of c-Abl activity, blocks cisplatin-induced ΔNp63α/TAp73 dissociation, TAp73-dependent target gene induction, and cell death. Finally, our findings are supported by previous work showing the ability of imatinib to inhibit the p73-dependent response to DNA damage (48). Of note, our data also imply that in breast cancer cells this TAp73-dependent mechanism is specific for platinum-based chemotherapy, which is unique in triggering phosphorylation required for ΔNp63/TAp73 dissociation. In addition, this cisplatin-sensitivity pathway appears restricted to breast tumors that exhibit a triple-negative phenotype, as only these tumors co-overexpress ΔNp63 and TAp73.
While a detailed understanding of the molecular basis of triple-negative tumors is lacking, it is now recognized that these tumors share clinical features and gene expression profiles with tumors in patients who inherit germline mutations in the breast cancer predisposition gene BRCA1 (2, 3). Notably, of the 2 triple-negative cells used in this study, HCC-1937 cells express only mutationally inactivated BRCA1, while MDA-MB-468 cells express wild-type BRCA1 (29). Thus, expression of ΔNp63 and TAp73 appears to reflect a functional pathway shared by BRCA1-associated and sporadic triple-negative tumors. Our finding that this pathway mediates cisplatin sensitivity is consistent with recent reports suggesting that breast carcinoma cells derived from BRCA1 mutation carriers exhibit particular sensitivity to cisplatin (5). The potential benefit of cisplatin treatment for patients presenting with non–BRCA1-associated triple-negative tumors, who greatly outnumber BRCA1-associated cases, is of substantial clinical interest. Whether in vitro sensitivity to cisplatin will portend a favorable clinical response in vivo remains to be determined (49). It is clear, however, that the efficacy of cisplatin as a breast cancer treatment is very low in unselected patients who have received prior therapy (50), underscoring the need to establish a means to identify those patients who will experience a favorable response.
In summary, this work has identified a molecular pathway that mediates specific cellular sensitivity of breast cancer cells to cisplatin chemotherapy. This ΔNp63/TAp73 pathway appeared to be restricted to a subset of primary breast cancers that are thought to be phenotypically similar to BRCA1-associated cancers, which themselves are thought to exhibit cisplatin sensitivity. These findings suggest that a subset of sporadic triple-negative cancers may exhibit clinical sensitivity to cisplatin treatment. In addition, it is likely that the ΔNp63/TAp73 pathway is relevant in other tumor types that express these particular proteins. As noted above, ΔNp63α is overexpressed in SCCs of the head and neck, lung, and esophagus (18–22), and we previously demonstrated that ΔNp63α functions to repress apoptosis mediated by TAp73 in SCC cells (23, 24). Consistent with a role for the ΔNp63/TAp73 pathway in mediating chemosensitivity, it has been recently reported that the level of ΔNp63α in primary head and neck SCCs correlates with the clinical response of patients to cisplatin chemotherapy (51). Our findings predict that the presence of an active ΔNp63/TAp73 pathway may prove to be a useful clinical predictor of cisplatin sensitivity in treatment-refractory breast cancers and other tumors.
Methods
Primary breast carcinoma specimens.
Discarded tissue samples were collected and processed anonymously under human subjects research protocol 2002-P-002059/10. This protocol was reviewed and approved by the Human Research Committee, Partners Healthcare Inc., Boston, Massachusetts, USA. All samples underwent pathologic review. Microdissected tumor/normal paired samples were processed as described previously (25). For isoform-specific analysis, we performed pilot studies on a smaller tumor group, then selected a larger group of tumors enriched for ER/PR-negative cases. Initially, 39 primary invasive tumors were evaluated in total, and 2 were excluded due to greater than 5% DCIS and normal epithelia. RNA was prepared as described below.
Cell lines and plasmids.
The human breast carcinoma cell lines MCF-7, HCC-1937, MDA-MB-468, and T47D were maintained in RPMI 1640 containing 10% FBS, 100 IU/ml penicillin, and 100 μg/ml streptomycin (Invitrogen). MCF-10A cells were grown in DMEM-F12 (Invitrogen) supplemented with 5% horse serum, 20 ng/ml EGF, 0.5 μg/ml hydrocortisone, 100 ng/ml cholera toxin, 10 μg/ml insulin, 100 IU/ml penicillin, and 100 μg/ml streptomycin. Wild-type and mutant TAp73α plasmids were generously provided by Z.-M. Yuan (Harvard School of Public Health, Boston, Massachusetts, USA).
Lentiviral and retroviral production and infection.
The shRNA lentiviral constructs were created by transferring the U6 promoter-shRNA cassette into a lentiviral backbone, and high-titer amphotrophic retroviral and lentiviral stocks were generated by cotransfection with packaging vectors into 293T cells as described previously (24). The targeted sequences for p63 were 5′-GGGTGAGCGTGTTATTGATGCT-3′ and 5′-GAGTGGAATGACTTCAACTTT-3′. The targeted sequence for TAp73, 5′-GGATTCCAGCATGGACGTCTT-3′, is found within p73 exon 3. Therefore, this shRNA does not target ΔNp73 (Supplemental Figure 3) (24).
QRT-PCR analysis.
Total RNA from cells was extracted using STAT-60 RNA isolation solution (Tel-Test Inc.) according to the manufacturer’s protocol. First-strand cDNA was synthesized from total RNA using random hexamer primers and the SuperScript II system for RT-PCR (Invitrogen). Gene expression levels were measured by QRT-PCR using the iQ SYBR Green Supermix reagent (Bio-Rad) and an Opticon real-time PCR detector system (MJ Research). Data analysis was performed using Opticon Monitor Analysis Software V1.08 (MJ Research). The expression of each gene was normalized to GAPDH or β2M as a reference. The relative copy numbers were calculated from an 8-point standard curve generated from a 10-fold serial dilution of full-length cDNA constructs as described previously (23). Specific forward and reverse primer sequences are provided in Supplemental Table 2. The conditions for all QRT-PCR reactions were as follows: 3 minutes at 94°C followed by 40 seconds at 94°C, 40 seconds at 60°C, and 25 seconds at 72°C for 40 cycles. All PCR products were confirmed by the presence of a single peak upon melting curve analysis and by gel electrophoresis. No-template (water) reaction mixtures and no-RT mixtures were performed on all samples as negative controls. All experiments were performed in duplicate.
Apoptosis assays.
Quantitation of apoptosis by annexin V/PI staining was performed as described previously (24). Briefly, both floating and attached cells were collected 72 hours after p63-directed shRNA lentiviral infection. Apoptotic cell death was determined using the BD ApoAlert annexin V–FITC Apoptosis Kit (BD Biosciences) according to the manufacturer’s instructions, and cells were analyzed on a FACSCalibur flow cytometer using CellQuest Pro software (version 5.1.1; BD Biosciences).
IP and immunoblot analysis.
Protein lysates from cells were extracted in ice-cold lysis buffer (0.75% NP-40, 1 mM DTT, and protease inhibitors in PBS) with or without phosphate inhibitor I and II (Sigma-Aldrich). For IP experiments, precleared lysates (2.5 mg) from confluent cells were incubated with either 2.0 μg anti-p63 polyclonal antibody (H-129; Santa Cruz Biotechnology Inc.) or 1.0 μg anti-p73 monoclonal antibody (Ab-2; CalBiochem) for 2 hours at 4°C. The immunocomplexes were precipitated using protein A or protein G sepharose (Amersham Biosciences), washed 4 times with lysis buffer, and analyzed by SDS-PAGE. Immunoblots were probed with the following antibodies: p63 (diluted 1:5,000, 4A4; Sigma-Aldrich); mouse monoclonal p73 (diluted 1:1,000, Ab-2; CalBiochem); PARP (diluted 1:1,000; Cell Signaling Technology); Puma (diluted 1:1,000, Ab9645; Abcam); β-tubulin (diluted 1:2,500, D-10; Santa Cruz Biotechnology); Noxa (diluted 1:1,000, Ab13654; Abcam); and phosphorylated tyrosine (diluted 1:1,000, PY99; Santa Cruz Biotechnology Inc.).
Chemosensitivity assay.
Dose-response curves and IC50 values were determined using the methyl thiazolyl tetrazolium (MTT) cell viability assay as described previously (52). Cells were seeded into 96-well microtiter plates for 24 hours at a density of 5 × 103 per well. Serial drug dilutions were prepared in medium immediately before each assay, and viable cell masses following 3 or 5 days of drug exposure were determined by cell-mediated MTT reduction. Cell growth as well as drug activity was determined by measuring absorbance at 550 nm using an Anthos systems plate reader.
Statistics.
For gene expression levels measured as continuous variables (Figure 1), unpaired 2-tailed Student’s t tests were used to evaluate differences between sample means, and all values were reported as mean ± SEM. No significant deviations from normality were observed. Levels of ΔNp63 and TAp73 greater than 2 times the sample mean for each gene (n = 37) were considered elevated. In order to ensure that our conclusions were robust to small sample sizes, 2-tailed Fisher’s exact tests were used to derive P values from the 2 × 2 contingency tables. Unless indicated, multiple comparisons were not performed, and thus no corresponding compensation was made. In all cases, a P value less than 0.05 was considered significant.
Supplementary Material
Acknowledgments
The authors would like to thank Avi Sofer for technical assistance, James Rocco for helpful discussion, and Daniel Haber and Nick Dyson for critical review of the manuscript. This work was supported by the Mary Kay Ash Charitable Foundation, by grant DE-15945 of the National Institute of Dental and Craniofacial Research, NIH, by the Tracey Davis Memorial Fund, and by the Avon Foundation (to L.W. Ellisen). No sponsors of this work had any role in the design or conduct of the study, in the collection, analysis, and interpretation of the data, or in the preparation, review, or approval of the manuscript.
Footnotes
Nonstandard abbreviations used: BRCA1, breast cancer 1, early onset; ER, estrogen receptor; MTT, methyl thiazolyl tetrazolium; ΔNp63, p63 lacking the N-terminal transactivation domain; PARP-1, poly(ADP-ribose) polymerase–1; PI, propidium iodide; PR, progesterone receptor; QRT-PCR, real-time quantitative RT-PCR; RNAi, RNA interference; SCC, squamous cell carcinoma; shRNA, small hairpin RNA; TAp63, p63 containing the N-terminal transactivation domain.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 117:1370–1380 (2007). doi:10.1172/JCI30866
References
- 1.Sorlie T., et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. U. S. A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Z.C., et al. Loss of heterozygosity and its correlation with expression profiles in subclasses of invasive breast cancers. Cancer Res. 2004;64:64–71. doi: 10.1158/0008-5472.can-03-2570. [DOI] [PubMed] [Google Scholar]
- 3.Sorlie T., et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc. Natl. Acad. Sci. U. S. A. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Foulkes W.D., et al. Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. J. Natl. Cancer Inst. 2003;95:1482–1485. doi: 10.1093/jnci/djg050. [DOI] [PubMed] [Google Scholar]
- 5.Tassone P., et al. BRCA1 expression modulates chemosensitivity of BRCA1-defective HCC1937 human breast cancer cells. Br. J. Cancer. 2003;88:1285–1291. doi: 10.1038/sj.bjc.6600859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang A., et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- 7.Mills A.A., et al. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- 8.Adriance M.C., Inman J.L., Petersen O.W., Bissell M.J. Myoepithelial cells: good fences make good neighbors. Breast Cancer Res. 2005;7:190–197. doi: 10.1186/bcr1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carroll D.K., et al. p63 regulates an adhesion programme and cell survival in epithelial cells. Nat. Cell Biol. 2006;8:551–561. doi: 10.1038/ncb1420. [DOI] [PubMed] [Google Scholar]
- 10.Perou C.M., et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 11.Wang X., et al. p63 expression in normal, hyperplastic and malignant breast tissues. Breast Cancer. 2002;9:216–219. doi: 10.1007/BF02967592. [DOI] [PubMed] [Google Scholar]
- 12.Reis-Filho J.S., Milanezi F., Amendoeira I., Albergaria A., Schmitt F.C. Distribution of p63, a novel myoepithelial marker, in fine-needle aspiration biopsies of the breast: an analysis of 82 samples. Cancer. 2003;99:172–179. doi: 10.1002/cncr.11061. [DOI] [PubMed] [Google Scholar]
- 13.Ribeiro-Silva A., et al. p63 correlates with both BRCA1 and cytokeratin 5 in invasive breast carcinomas: further evidence for the pathogenesis of the basal phenotype of breast cancer. Histopathology. 2005;47:458–466. doi: 10.1111/j.1365-2559.2005.02249.x. [DOI] [PubMed] [Google Scholar]
- 14.Koker M.M., Kleer C.G. p63 expression in breast cancer: a highly sensitive and specific marker of metaplastic carcinoma. Am. J. Surg. Pathol. 2004;28:1506–1512. doi: 10.1097/01.pas.0000138183.97366.fd. [DOI] [PubMed] [Google Scholar]
- 15.Harms K., Nozell S., Chen X. The common and distinct target genes of the p53 family transcription factors. Cell. Mol. Life Sci. 2004;61:822–842. doi: 10.1007/s00018-003-3304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang A., et al. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death inducing, and dominant negative activities. Mol. Cell. 1998;2:305–316. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- 17.King K.E., et al. DeltaNp63alpha functions as both a positive and a negative transcriptional regulator and blocks in vitro differentiation of murine keratinocytes. Oncogene. 2003;22:3635–3644. doi: 10.1038/sj.onc.1206536. [DOI] [PubMed] [Google Scholar]
- 18.Thurfjell N., et al. Function and importance of p63 in normal oral mucosa and squamous cell carcinoma of the head and neck. Adv. Otorhinolaryngol. 2005;62:49–57. doi: 10.1159/000082464. [DOI] [PubMed] [Google Scholar]
- 19.Weber A., Bellmann U., Bootz F., Wittekind C., Tannapfel A. Expression of p53 and its homologues in primary and recurrent squamous cell carcinomas of the head and neck. Int. J. Cancer. 2002;99:22–28. doi: 10.1002/ijc.10296. [DOI] [PubMed] [Google Scholar]
- 20.Sniezek J.C., Matheny K.E., Westfall M.D., Pietenpol J.A. Dominant negative p63 isoform expression in head and neck squamous cell carcinoma. Laryngoscope. 2004;114:2063–2072. doi: 10.1097/01.mlg.0000149437.35855.4b. [DOI] [PubMed] [Google Scholar]
- 21.Massion P.P., et al. Significance of p63 amplification and overexpression in lung cancer development and prognosis. Cancer Res. 2003;63:7113–7121. [PubMed] [Google Scholar]
- 22.Hu H., et al. Elevated expression of p63 protein in human esophageal squamous cell carcinomas. Int. J. Cancer. 2002;102:580–583. doi: 10.1002/ijc.10739. [DOI] [PubMed] [Google Scholar]
- 23.Deyoung M.P., et al. Tumor-specific p73 up-regulation mediates p63 dependence in squamous cell carcinoma. Cancer Res. 2006;66:9362–9368. doi: 10.1158/0008-5472.CAN-06-1619. [DOI] [PubMed] [Google Scholar]
- 24.Rocco J.W., Leong C.O., Kuperwasser N., DeYoung M.P., Ellisen L.W. p63 mediates survival in squamous cell carcinoma by suppression of p73-dependent apoptosis. Cancer Cell. 2006;9:45–56. doi: 10.1016/j.ccr.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 25.Ma X.J., et al. Gene expression profiles of human breast cancer progression. Proc. Natl. Acad. Sci. U. S. A. 2003;100:5974–5979. doi: 10.1073/pnas.0931261100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dominguez G., et al. DeltaTAp73 upregulation correlates with poor prognosis in human tumors: putative in vivo network involving p73 isoforms, p53, and E2F-1. J. Clin. Oncol. 2006;24:805–815. doi: 10.1200/JCO.2005.02.2350. [DOI] [PubMed] [Google Scholar]
- 27.Dominguez G., et al. Wild type p73 overexpression and high-grade malignancy in breast cancer. Breast Cancer Res. Treat. 2001;66:183–190. doi: 10.1023/a:1010624717311. [DOI] [PubMed] [Google Scholar]
- 28.Saxby A.J., et al. Assessment of HER-2 status in pancreatic adenocarcinoma: correlation of immunohistochemistry, quantitative real-time RT-PCR, and FISH with aneuploidy and survival. Am. J. Surg. Pathol. 2005;29:1125–1134. doi: 10.1097/01.pas.0000160979.85457.73. [DOI] [PubMed] [Google Scholar]
- 29.Elstrodt F., et al. BRCA1 mutation analysis of 41 human breast cancer cell lines reveals three new deleterious mutants. Cancer Res. 2006;66:41–45. doi: 10.1158/0008-5472.CAN-05-2853. [DOI] [PubMed] [Google Scholar]
- 30.Martin S.J., et al. Early redistribution of plasma membrane phosphatidylserine is a general feature of apoptosis regardless of the initiating stimulus: inhibition by overexpression of Bcl-2 and Abl. J. Exp. Med. 1995;182:1545–1556. doi: 10.1084/jem.182.5.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan W.M., Siu W.Y., Lau A., Poon R.Y. How many mutant p53 molecules are needed to inactivate a tetramer? Mol. Cell. Biol. 2004;24:3536–3551. doi: 10.1128/MCB.24.8.3536-3551.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wistuba I.I., et al. Comparison of features of human breast cancer cell lines and their corresponding tumors. Clin. Cancer Res. 1998;4:2931–2938. [PubMed] [Google Scholar]
- 33.Melino G., et al. p73 Induces apoptosis via PUMA transactivation and Bax mitochondrial translocation. J. Biol. Chem. 2004;279:8076–8083. doi: 10.1074/jbc.M307469200. [DOI] [PubMed] [Google Scholar]
- 34.Bergamaschi D., et al. p53 polymorphism influences response in cancer chemotherapy via modulation of p73-dependent apoptosis. Cancer Cell. 2003;3:387–402. doi: 10.1016/s1535-6108(03)00079-5. [DOI] [PubMed] [Google Scholar]
- 35.Gong J.G., et al. The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature. 1999;399:806–809. doi: 10.1038/21690. [DOI] [PubMed] [Google Scholar]
- 36.Irwin M.S., et al. Chemosensitivity linked to p73 function. Cancer Cell. 2003;3:403–410. doi: 10.1016/s1535-6108(03)00078-3. [DOI] [PubMed] [Google Scholar]
- 37.Agami R., Blandino G., Oren M., Shaul Y. Interaction of c-Abl and p73alpha and their collaboration to induce apoptosis. Nature. 1999;399:809–813. doi: 10.1038/21697. [DOI] [PubMed] [Google Scholar]
- 38.Yuan Z.M., et al. p73 is regulated by tyrosine kinase c-Abl in the apoptotic response to DNA damage. Nature. 1999;399:814–817. doi: 10.1038/21704. [DOI] [PubMed] [Google Scholar]
- 39.Berry D.A., et al. Estrogen-receptor status and outcomes of modern chemotherapy for patients with node-positive breast cancer. JAMA. 2006;295:1658–1667. doi: 10.1001/jama.295.14.1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fomenkov A., et al. RACK1 and stratifin target DeltaNp63alpha for a proteasome degradation in head and neck squamous cell carcinoma cells upon DNA damage. Cell Cycle. 2004;3:1285–1295. doi: 10.4161/cc.3.10.1155. [DOI] [PubMed] [Google Scholar]
- 41.Druker B.J. Inhibition of the Bcr-Abl tyrosine kinase as a therapeutic strategy for CML. Oncogene. 2002;21:8541–8546. doi: 10.1038/sj.onc.1206081. [DOI] [PubMed] [Google Scholar]
- 42.Mantovani F., et al. Pin1 links the activities of c-Abl and p300 in regulating p73 function. Mol. Cell. 2004;14:625–636. doi: 10.1016/j.molcel.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 43.Tsai K.K., Yuan Z.M. c-Abl stabilizes p73 by a phosphorylation-augmented interaction. Cancer Res. 2003;63:3418–3424. [PubMed] [Google Scholar]
- 44.Livasy C.A., et al. Phenotypic evaluation of the basal-like subtype of invasive breast carcinoma. Mod. Pathol. 2005;19:264–271. doi: 10.1038/modpathol.3800528. [DOI] [PubMed] [Google Scholar]
- 45.Strano S., et al. The transcriptional coactivator Yes-associated protein drives p73 gene-target specificity in response to DNA damage. Mol. Cell. 2005;18:447–459. doi: 10.1016/j.molcel.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 46.Basu S., Totty N.F., Irwin M.S., Sudol M., Downward J. Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14-3-3 and attenuation of p73-mediated apoptosis. Mol. Cell. 2003;11:11–23. doi: 10.1016/s1097-2765(02)00776-1. [DOI] [PubMed] [Google Scholar]
- 47.Urist M., Tanaka T., Poyurovsky M.V., Prives C. p73 induction after DNA damage is regulated by checkpoint kinases Chk1 and Chk2. Genes Dev. 2004;18:3041–3054. doi: 10.1101/gad.1221004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Truong T., Sun G., Doorly M., Wang J.Y., Schwartz M.A. Modulation of DNA damage-induced apoptosis by cell adhesion is independently mediated by p53 and c-Abl. Proc. Natl. Acad. Sci. U. S. A. 2003;100:10281–10286. doi: 10.1073/pnas.1635435100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Turner N., Tutt A., Ashworth A. Hallmarks of ‘BRCAness’ in sporadic cancers. Nat. Rev. Cancer. 2004;4:814–819. doi: 10.1038/nrc1457. [DOI] [PubMed] [Google Scholar]
- 50.Decatris M.P., Sundar S., O’Byrne K.J. Platinum-based chemotherapy in metastatic breast cancer: current status. Cancer Treat. Rev. 2004;30:53–81. doi: 10.1016/S0305-7372(03)00139-7. [DOI] [PubMed] [Google Scholar]
- 51.Zangen R., Ratovitski E., Sidransky D. DeltaNp63alpha levels correlate with clinical tumor response to cisplatin. Cell Cycle. 2005;4:1313–1315. doi: 10.4161/cc.4.10.2066. [DOI] [PubMed] [Google Scholar]
- 52.Bradshaw T.D., et al. Elucidation of thioredoxin as a molecular target for antitumor quinols. Cancer Res. 2005;65:3911–3919.. doi: 10.1158/0008-5472.CAN-04-4141. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.