Abstract
Cytogenetic and loss of heterozygosity (LOH) studies demonstrated chromosome 3p deletions in transitional cell carcinoma (TCC). We recently cloned the tumor suppressor gene FHIT (fragile histidine triad) at 3p14.2, one of the most frequently deleted chromosomal regions in TCC of the bladder, and showed that it is the target of environmental carcinogens. Abnormalities at the FHIT locus have been found in tumors of the lung, breast, cervix, head and neck, stomach, pancreas, and clear cell carcinoma of the kidney. We examined six TCC derived cell lines (SW780, T24, Hs228T, CRL7930, CRL7833, and HTB9) and 30 primary TCC of the bladder for the integrity of the FHIT transcript, using reverse transcriptase–polymerase chain reaction (RT-PCR) to investigate a potential role of the FHIT gene in TCC of the bladder. In addition, we tested expression of the Fhit protein in the six TCC-derived cell lines by Western blot analysis and in 85 specimens of primary TCCs by immunohistochemistry. Three of the six cell lines (50%) did not show the wild-type FHIT transcript, and Fhit protein was not detected in four of the six cell lines (67%) tested. Fhit expression also was correlated with pathological and clinical status. A significant correlation was observed between reduced Fhit expression and advanced stage of the tumors. Overall, 26 of 30 (87%) primary TCCs showed abnormal transcripts. Fhit protein was absent or greatly reduced in 61% of the TCCs analyzed by immunohistochemistry. These results suggested that loss of Fhit expression may be as important in the development of bladder cancer as it is for other neoplasms caused by environmental carcinogens.
Bladder cancer is the fourth most frequent cause of cancer death in men in the United States. 1 Transitional cell carcinoma (TCC) comprises 90% of all bladder cancers and is the second most frequent malignancy of the genitourinary tract. In 1999 an estimated 54,200 patients were diagnosed with bladder cancer. 2 TCC is classified histopathologically into three types: superficial, confined to the bladder wall, and invasive according to the TNM guideline. 3 Neoplasms of the transitional epithelium represent one of the first tumors to have been associated with environmental risk factors. The single largest etiological factor in the development of bladder cancer in the United States is smoking, although industrial exposure to arylamines also has been implicated as a frequent cause. 4 Numerous recent cytogenetic and molecular genetic investigations have demonstrated many genetic alterations in solid tumors, most of which affect tumor suppressor genes. The RB, TP53, waf1/cip1, and CDKN2 genes have been shown to be mutated and/or deleted in a fraction of the TCC tumors of the bladder. 5-10
Deletions of the short arm of chromosome 3 (3p) have been detected by loss of heterozygosity (LOH) and cytogenetic studies in TCC of the bladder. 11-13 Three discrete regions spanning 3p12–14, 3p21–24, and 3p24–26 often have been shown to be deleted, suggesting that these regions might harbor genes that are important in the development of TCC. 13 Vieten et al 12 have developed an in vitro multistep model of human uroepithelial transformation. In a study of chromosome 3 losses associated specifically with immortalization of five independent human papillomavirus-transformed urothelial cell lines, the smallest common region of deletion was 3p14.1–14.2. These results suggest that loss of a gene(s) in this region may be responsible for immortalization of the uroepithelial cells.
We cloned the tumor suppressor gene FHIT at 3p14.2, 14 one of the most frequently deleted chromosomal regions in TCC of the bladder. 11 FHIT gene lies on a region of over 1 Mb of chromosome 3p14.2 that encompasses the FRA3B fragile region and encodes a protein of 147 amino acids (16.8 kd) with diadenosine triphosphate hydrolase activity. 15 A papillomavirus insertion site, 16 plasmid integration sites, 17 and cancer-specific translocation 18,19 have been mapped within the FRA3B fragile site in the FHIT gene.
The purpose of this report is to show that loss of expression of the Fhit protein occurs in the majority of transitional cell carcinomas of the bladder, suggesting that loss of FHIT function plays a critical role in bladder carcinogenesis.
Materials and Methods
Cell Lines and Tissues
TCC-derived cell lines (SW780, T24, Hs228T, CRL7930, CRL7833, HBT9) and 293 human kidney cells were obtained from the American Type Culture Collection (ATCC) and were maintained in the recommended media. A total of 30 transurethrally resected transitional carcinomas of the bladder were obtained from the Department of Urology, Jefferson Medical College of Thomas Jefferson University (Philadelphia, PA). Samples were taken immediately after resection and snap frozen in liquid nitrogen. In addition, sections from 85 formalin-fixed, paraffin-embedded specimens of TCC of the bladder were obtained from archival blocks of radical cystectomy cases. Forty-one of these specimens came from the Department of Urology, University of Padova (Padua, Italy), and 44 specimens came from the Department of Urology, Jefferson Medical College of Thomas Jefferson University. All samples were obtained from patients who gave informed consent to use excess pathological specimens for research purposes.
RNA Extraction and cDNA Synthesis
RNA was extracted from six TCC-derived cell lines (SW780, T24, Hs228T, CRL7930, CRL7833, HBT9) and from 30 frozen tissue samples with an RNeasy mini Kit (Qiagen, Valencia, CA), according to the manufacturer’s protocol. cDNA was synthesized from 2 μg of total RNA. Reverse transcription (RT) was carried out in a 22-μl volume with 300 units of Superscript II (Life Technologies, Gaithersburg, MD), 500 ng/μl oligo(dT), and 50 ng/μl random hexamers. The reaction was incubated at 42°C for 50 minutes and then was boiled for 5 minutes. The final reaction was diluted with distilled water to 30 μl, and 1 μl of cDNA was used for PCR in consecutive rounds of amplification with FHIT-specific nested primer pairs 5U2/3D2 and 5U1/3D1, as described previously. 14
Amplified DNA fragments corresponding with full-sized and aberrant FHIT transcripts in the cell lines were excised from agarose gel, purified, and sequenced directly. Sequence analysis was performed on 373A and 377 DNA sequencers (Applied Biosystems, Foster City, CA.)
DNA Analysis
DNA from six TCC-derived cell lines (SW780, T24, Hs228T, CRL7930, CRL7833, HBT9) was extracted by standard techniques and amplified by PCR, using oligonucleotide primer pairs for 10 loci within introns 3 (12f/r for 1; 09f/r for 2), 4 (06f/r for 3; 03f/r for 4; Col10kf/r for 5; 0817EF/ER for 6), and 5 (36C3/36C4 for 7; N143kf/r for 8; N100Kf/r for 9; N020Kf/r for 10) and for exons 5, 7, 8, and 9 of the FHIT gene (Figure 1) ▶ . PCR amplification reactions were performed in a 25-μl volume, using 100 μg of template, 1 unit of Taq polymerase (Takara, Pan Vera Corp., Madison, WI), 20 ng of primers, and 0.5 mmol/L of each deoxynucleoside triphosphate. Amplifications were carried out in a Perkin-Elmer thermal cycler for 35 cycles of 94°C for 20 seconds (for denaturation), 57°C for 30 seconds (varied for specific primer pairs), and 72°C for 30 seconds (for extension).
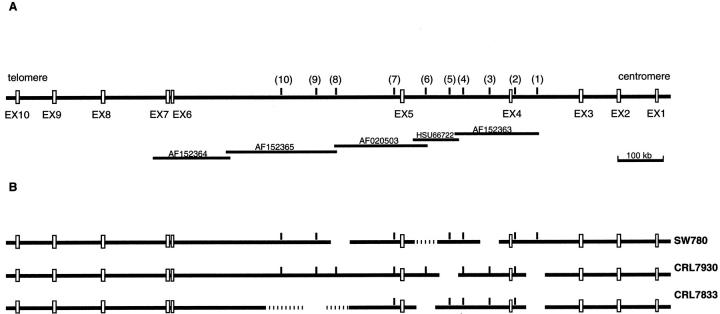
Figure 1.
The FHIT gene. A: FHIT exons 1–10, STSs markers analyzed by PCR (1–10), and the five BACs covering the central portion of the FHIT gene. B: Homozygous deletions of the FHIT gene observed in SW780, CRL7930, and CRL7833 cells.
Amplified products were resolved on a 1.5% agarose gel.
Protein Extraction and Immunoblot Analysis
Cell lines from which protein was extracted were pelleted and rinsed with phosphate-buffered saline (PBS) (Gibco, Grand Island, NY) and were lysed in 400 μl of protein lysis buffer (10 mmol/L Tris, 1% NP-40, 0.1% bovine serum albumin, 1 mmol/L EDTA, PBS) and 50 μl phenylmethylsulfonyl fluoride (PMSF) (125 mmol/L PMSF in ethanol) on ice for 5 minutes. After a 6-minute centrifugation at 8500 rpm at room temperature, the lysate supernatant was removed and was assayed for protein concentration with the Protein Assay reagent (Bio-Rad Laboratories, Melville, NY). Protein was aliquoted and stored at −80°C.
For Western analysis, 100 μg of protein was boiled in loading buffer containing 5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), glycerol, bromophenol blue, and β-mercaptoethanol for denaturing. Samples were electrophoresed on 4–20% precast Tris-glycine gels (Fisher Scientific, Pittsburgh, PA) with the Mini Protean II electrophoresis system (Bio-Rad Laboratories) and subsequently transferred to a nitrocellulose membrane (MSI, Westborough, MA) with the Mini trans-Blot cell apparatus (Bio-Rad Laboratories). Membranes were blocked for 3 hours at room temperature in 5% nonfat dry milk dissolved in TBS-T (Tris-buffered saline Tween20). Primary and secondary antibodies were diluted in 5% nonfat dry milk solution. Western analysis was performed overnight at 4°C with a 1:1000 dilution of the Fhit polyclonal antibody (Zymed Laboratories, San Francisco, CA). Secondary antibody solution (1:2000 dilution) containing anti-rabbit immunoglobulin labeled with horseradish peroxidase (Amersham Life Sciences, Buckinghamshire, England) was applied for 1 hour at room temperature. Membranes were treated with enhanced chemiluminescence detection reagents (Amersham Life Sciences) according to the manufacturer’s recommended protocol. Bands were detected by exposing the membrane to medical X-ray film (Fuji, Stamford, CT) for 1–10 minutes at room temperature.
Blots were stripped in a solution containing 100 mmol/L β-mercaptoethanol, 2% SDS, and 62.5 mmol/L Tris-HCl (pH 6.7) for 30 minutes at 60°C. They were rinsed in TBS-T and reprobed for protein standardization under the previously mentioned conditions, with β-actin (Sigma, St. Louis, MO) as the primary antibody and a 1:2000 dilution of the anti-mouse secondary antibody labeled with horseradish peroxidase (Amersham Life Sciences), and exposed for 1 minute at room temperature before the film was developed.
Immunohistochemistry
Routine deparaffinization of all sections mounted on positive charge slides was carried out according to standard procedures, followed by rehydration through an ethanol series. The slides were immersed in citrate buffer (0.01 mol/L sodium citrate, pH 6.0) and heated in a microwave oven at 600 W (three times for 5 minutes) to enhance antigen retrieval. Endogenous peroxidase was blocked with 0.3% hydrogen peroxidase in methanol for 30 minutes. The sections were then incubated with anti-Fhit antibody, 1:1000 dilution (Zymed Laboratories) overnight at room temperature. The primary antibody was omitted and replaced by PBS in the negative control. After this incubation, sections were treated with biotinylated anti-rabbit antibody and streptavidin-biotin-peroxidase (Histostain-SP Kit; Zymed Laboratories). Antibody localization was detected, with diaminobenzidine as a chromogen substrate. Finally, sections were washed in distilled water and weakly counterstained with Harry’s modified hematoxylin. 20
Statistical Analysis
We used the Fisher exact test with a two-tailed P for analysis of statistical significance of correlation between clinicopathological parameters and Fhit expression.
Results
Homozygous Deletions in TCC-Derived Cell Lines
To determine whether genomic deletions occur in TCC cell lines, we analyzed six TCC-derived cell lines (SW780, T24, Hs228T, HBT9, CRL7833, CRL7930) by PCR amplification, using oligonucleotide primers appropriate for 10 STSs placed in introns 3, 4, and 5 and for exons 5, 7, 8, and 9 of the FHIT gene.
Three cell lines (SW780, CRL7930, CRL7833) exhibit discontinuous homozygous deletions. As shown in Figure 1 ▶ , the fragments that are commonly deleted overlap with regions of the FHIT gene that were deleted in previously reported cancer-derived cell lines. 14,19 Homozygous deletions did not include FHIT exons 5, 7, 8, and 9.
The three cell lines with homozygous deletions within the FHIT gene are the same three cell lines showing both absent or abnormal FHIT transcript and an absence of Fhit protein (see below).
Expression of FHIT Transcripts in TCCs
Using the RT-PCR amplification assay, FHIT cDNAs of altered size were identified in one (CRL7833) of the six cell lines (data not shown). Four cell lines (SW780, T24, Hs228T, HBT9) showed the presence of only a normal-sized FHIT transcript. FHIT cDNA could not be amplified from CRL7930 cells. Sequencing of the amplified DNA products revealed normal FHIT sequences in T24, HS228T, and HBT9 cells. The normal-sized band present in SW780 consisted of a mixture of abnormal FHIT transcripts.
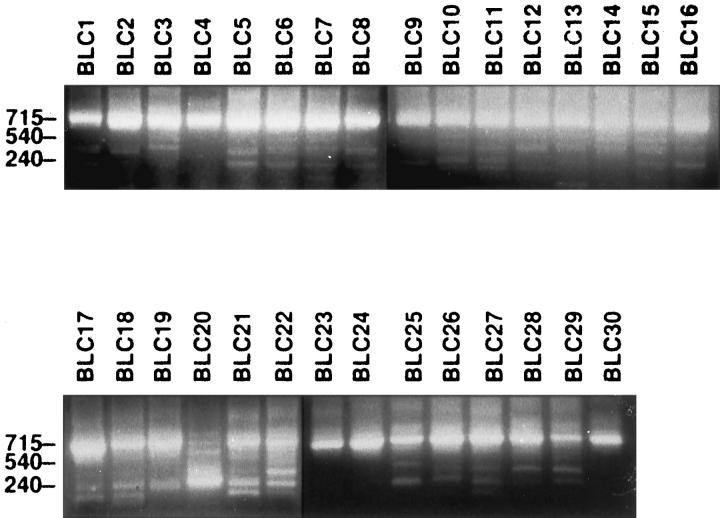
Similar experiments then were performed using RNA extracted from primary TCCs of the bladder. Overall, 26 of 30 (87%) primary TCCs showed a combination of normal and one or more aberrant transcripts (Figure 2) ▶ . Abnormalities of the FHIT transcripts consisted of the absence of one or more exons of the FHIT gene or the absence of exons with insertion of intronic sequences, as described previously. 14,19
Figure 2.
Analysis of expression of the FHIT gene by nested RT-PCR. FHIT RT-PCR products in 30 primary TCCs are shown. Case names are shown at the top; the size of the amplified product is to the left. The 715-bp band represents the size of the wild-type FHIT transcript.
Expression of Fhit Protein in TCCs
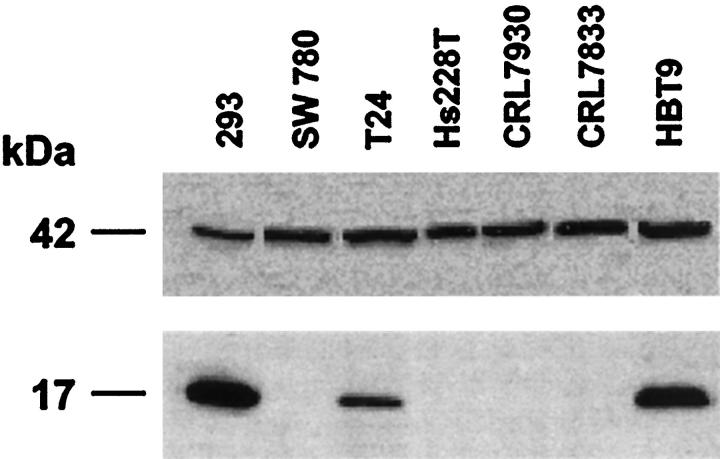
Immunoblot experiments using protein lysate from the six transitional carcinoma-derived cell lines showed an absence of Fhit protein in the three cell lines (SW780, CRL7930, CRL7833) with absent or abnormal FHIT transcripts (Figure 3) ▶ . Conversely, two cell lines, T24 and HBT9, expressed abundant Fhit protein in concordance with the apparent absence of FHIT gene alteration in these cells. The absence of Fhit protein in Hs228T cells in discordance with the presence of a normal FHIT transcript might be because of the expression of a low level of FHIT transcript, as described previously for RCC cell lines. 20
Figure 3.
Expression of the Fhit protein in TCC cell lines. Western blot analysis of protein from TCC-derived cell lines with anti-Fhit antiserum is shown. The size of the protein is shown at the left. Bottom: Absence of the Fhit protein in SW780, Hs228T, CRL7930, and HBT29 protein lysates; the 293 lysate is from a transformed human kidney cell line previously shown to express Fhit. 18 Top: β-Actin expression for the same membranes.
Primary TCCs were tested for expression of Fhit protein by immunohistochemical analysis with the Fhit-specific polyclonal antiserum. The 85 TCC specimens listed in Table 1 ▶ were characterized for tumor grade and stage, and sections were assessed for Fhit expression.
Table 1.
Clinical-Pathological Features and Fhit Expression
| Case | Sex | T | G | Fhit |
|---|---|---|---|---|
| 26 (30.6%) | M 23 | 4TA 5T3b | 1 Cis 2G2/3 | |
| F 3 | 7T2 IT3/4 | 2 G1 | ||
| 1T2a 3T4a | 15 G3 | − | ||
| 5T3 | 1 G1/2 | |||
| 5 G2 | ||||
| 17 (20%) | M 14 | 1TA 5T3b | 1 G1 | |
| F 3 | 6T2 2T4a | 3 G2 | −/+ | |
| 3T3 | 13 G3 | |||
| 9 (10.6%) | M 5 | 5TA | 1 Cis | |
| F 4 | 3T1 | 2 G1 | ||
| 1T2 | 1 G1/2 | +/− | ||
| 3 G2 | ||||
| 2 G2/3 | ||||
| 33 (38.8%) | M 26 | 10TA 1T3b | 1 Cis | |
| F 7 | 7T1 6T4a | 2 G1 | ||
| 9T2 | 4 G1/2 | |||
| 11 G2 | + | |||
| 1 G2/3 | ||||
| 13 G3 | ||||
| 1 G4 |
Sections containing a portion of normal urothelium, representing an internal positive control, were analyzed (Figure 4) ▶ . A four-tier scoring system was used: 100% Fhit-positive cells were scored as +, >50% positive as +/−, <50% positive as −/+, and 98–100% negative as −.
Figure 4.
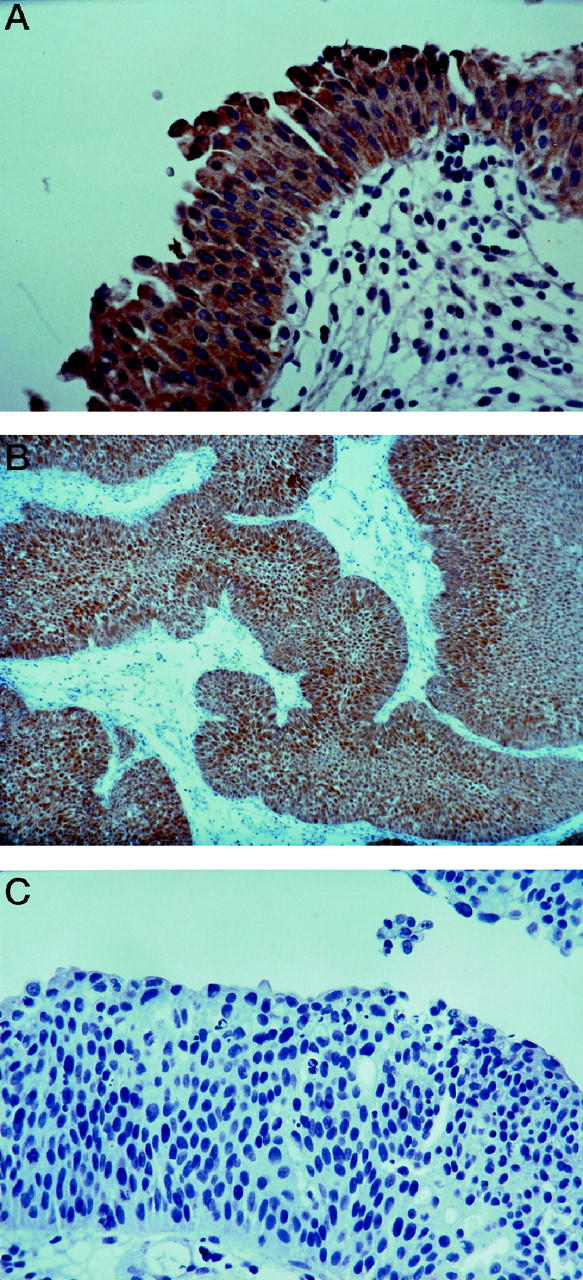
Fhit immunostaining in primary TCCs. A: Uniform strong staining of normal urothelium (×250). B: Papillary TCC showing Fhit expression (×120). C: Complete absence of Fhit staining in a superficial TCC (×250).
Examples of Fhit expression in normal urothelium and TCC sections are shown in Figure 4 ▶ . Fhit protein was undetectable in 26 of 85 (30.6% −) specimens, 17 (20% −/+) specimens showed <50% Fhit-positive cells, and nine specimens (10.6% +/−) showed >50% Fhit-positive cells. Diffuse Fhit immunostaining in all tumor cells was observed in 33 (38.8% +) of the specimens.
Statistical Analysis
Statistical studies showed a significant correlation (P = 0.0013) between absence and/or reduction of Fhit expression and advanced tumor stage. No correlation was found between Fhit expression and tumor differentiation grade.
Discussion
Many studies have documented genetic aberrations in the development of human transitional cell carcinoma of the urinary bladder, including point mutations of the HA-RAS-1 gene, 21 amplification and/or overexpression of the EGF receptor, 22 mutation and deletion of RB, TP53, waf1/cip1, and CDKN2 genes, 5-10 and as yet unidentified genes on regions of chromosomes 8p and 9p. Allelic losses of chromosome 3p have been reported, and the common regions of allelic loss appear to be 3p12–14, 3p21–24, and 3p24–26. 11-13 The tumor suppressor gene(s) targeted by 3p allelic losses in transitional cell carcinoma of the bladder, however, have not been identified. Because one of the regions of 3p frequently deleted in TCCs contains the FHIT locus (3p14.2), our finding of reduced or absent Fhit protein expression in most TCC-derived cell lines and in primary tumors suggests that FHIT is a tumor suppressor gene involved in the pathogenesis of TCC of the bladder. The FHIT gene also has been shown to be disrupted by translocation 14 in members of a family with heritable renal cell carcinoma that segregates with the t(3;8) (p14.2;q24) chromosome translocation. 23 One family member with the translocation died from bladder cancer and undifferentiated abdominal adenocarcinoma. 23
Western blot analysis shows absence of Fhit protein in four of six TCC-derived cell lines examined. The observation of multiple separate homozygous deletions that did not include FHIT exons in three of the four cell lines showing absence of Fhit protein suggested that in these cells the homozygous deletions are the result of an overlap of independent deletions of the two FHIT alleles, as previously demonstrated in other tumors. 24,25 The immunohistochemical study of 85 primary TCCs of all grades and stages has demonstrated that 30.6% of them were uniformly negative and another 30.6% were a mixture of Fhit-positive and -negative cells. Thus 61.2% of these primary tumors show reduced expression of Fhit. In addition, RT-PCR analysis of the integrity of FHIT transcripts in primary tumors and derived cell lines suggests frequent gene lesions. Similarly, 85% of clear cell renal carcinomas and more than 70% of cervical carcinomas have shown loss or reduction of Fhit expression. 20,26 The inactivation of FHIT in bladder tumors is probably a result of deletions within both alleles, as described for other carcinomas, particularly tumors resulting from exposure to environmental carcinogens. These include cancers of the lung, esophagus, stomach, cervix, and pancreas. 14,26-32 Further studies will be required to define the exact mechanisms leading to loss of Fhit in bladder cancer.
Our study of transitional cell carcinoma of the bladder shows a significant correlation (P = 0.0013) between the absence or reduction of Fhit expression and advanced stage of the primary tumor. The heterogeneous pattern of Fhit loss of expression in TCCs of the bladder is similar to the pattern observed in breast carcinoma. 33 In addition, reduced Fhit expression is correlated with a more aggressive disease in both bladder and breast cancers. Conversely, a uniform pattern of loss of Fhit expression has been observed in tumors such as lung and gastric cancers, in which loss of Fhit has been suggested to be an early event in the tumorigenesis process. 34,35 In conclusion, we show for the first time that FHIT is inactivated in the majority of transitional cell carcinomas of the urinary bladder and suggest that FHIT inactivation is a late event in neoplastic progression.
Footnotes
Address reprint requests to Dr. Raffaele Baffa, Department of Urology, Thomas Jefferson University, 1025 Walnut Street, College Building, Suite 1102A, Philadelphia, PA 19107. E-mail: R_Baffa@lac.jci.tju.edu.
Supported in part by National Institutes of Health grant CA56036 (to C. M. C.) and by the Martin Greitzer Fund.
References
- 1.Thrasher JB, Crawford ED: Current management of invasive and metastatic transitional cell carcinoma of the bladder. J Urol 1993, 149:957-972 [DOI] [PubMed] [Google Scholar]
- 2.Landis SM, Murray T, Bolden S, Wingo PA: Cancer statistics. CA Cancer J Clin 1999, 49:8-31 [DOI] [PubMed] [Google Scholar]
- 3.Sobin LH, Fleming ID: TNM Classification of Malignant Tumors, 5th ed. Union Internationale Contre le Cancer and American Joint Commitee on Cancer. Cancer 1997, 80:1803–1804 [DOI] [PubMed]
- 4.Ross RK, Jones PA, Yu MC: Bladder cancer epidemiology and pathogenesis. Semin Oncol 1996, 23:536-545 [PubMed] [Google Scholar]
- 5.Olumi AF, Tsai TC, Nichols PW, Skinner DG, Cain DR, Bender LI, Jones PA: Allelic loss of chromosome 17p distinguishes high grade from low grade transitional cell carcinoma of the bladder. Cancer Res 1990, 50:7081-7083 [PubMed] [Google Scholar]
- 6.Presti JC, Jr, Reuter VE, Galan T, Fair WR, Cordon-Cardo C: Molecular genetic alteration in superficial and locally advanced human bladder cancer. Cancer Res 1991, 51:5405-5409 [PubMed] [Google Scholar]
- 7.Miyao N, Tsai YC, Lerner SP, Olumi AF, Spruck CH, III, Gonzalez-Zulueta M, Nichols PW, Skinner DG, Jones PA: Role of chromosome 9 in human bladder cancer. Cancer Res 1993, 53:1230-1232 [PubMed] [Google Scholar]
- 8.Ruppert JM, Tokino K, Sidransky D: Evidence for two bladder cancer suppressor loci on chromosome 9 in human bladder cancer. Cancer Res 1993, 53:5093-5095 [PubMed] [Google Scholar]
- 9.Cairns P, Proctor AJ, Knowles MA: Loss of heterozygosity at the RB locus is frequent and correlates with muscle invasion in bladder carcinomas. Oncogene 1991, 6:2305-2309 [PubMed] [Google Scholar]
- 10.Xu H-J, Cairns P, Hu S-X, Knowles MA, Benedict WF: Loss of RB protein expression in primary bladder cancer correlated with loss of heterozygosity at RB locus and tumor progression. Int J Cancer 1993, 53:781-784 [DOI] [PubMed] [Google Scholar]
- 11.Li M, Zhang ZF, Reuter VE, Cordon-Cardo C: Chromosome 3 allelic losses, and microsatellite alteration in transitional cell carcinoma of the urinary bladder. Am J Pathol 1996, 149:229-235 [PMC free article] [PubMed] [Google Scholar]
- 12.Vieten L, Belair CD, Savelieva L, Julicher K, Brocker F, Bardenheur W, Schutte J, Opalka B, Reznikoff CA: Minimal deletion of 3p13–14.2 associated with immortalization of human uroepithelial cells. Genes Chromosom Cancer 1998, 21:39-48 [DOI] [PubMed] [Google Scholar]
- 13.Voorter C, Joos S, Bringuier PP, Vallinga M, Poddighe P, Schalken J, Du Manoir S, Ramaekers F, Litchter P, Hopman A: Deletion of chromosomal imbalance in transitional cell carcinoma of the bladder by comparative genomic hybridization. Am J Pathol 1995, 146:1341-1354 [PMC free article] [PubMed] [Google Scholar]
- 14.Ohta M, Inoue H, Cotticelli MG, Kastury K, Baffa R, Palazzo JP, Siprashvili Z, Mori M, McCue P, Druck T, Croce CM, Huebner K: The FHIT gene spanning the chromosome 3p14.2 fragile site, and renal carcinoma-associated t(3; 8) breakpoint, is abnormal in digestive tract cancers. Cell 1996, 84:587-597 [DOI] [PubMed] [Google Scholar]
- 15.Barnes LD, Garrison PN, Siprashvili Z, Guaranowsky A, Robinson AK, Ingram SW, Croce CM, Ohta M, Huebner K: FHIT a putative tumor suppressor in humans, is a dinucleoside 5′,5″-P1, P3-triphosphate hydrolase. Biochemistry 1996, 35:11529-11535 [DOI] [PubMed] [Google Scholar]
- 16.Wilke CM, Hall BK, Hoge A, Paradee W, Smith DI, Glover TW: FRA3B extends over a broad region, and contains a spontaneous HPV 16 integration site: direct evidence for the coincidence of viral integration site and fragile sites. Hum Mol Genet 1996, 5:187-195 [DOI] [PubMed] [Google Scholar]
- 17.Rassool F, Le Beau MM, Shen ML, Neilly ME, Espinosa R, III, Ong ST, Boldog F, Drabkin H, McCarroll R, Mckeithan TW: Direct cloning of DNA sequences from the common fragile site region at chromosome band 3p14.2. Genomics 1996, 35:109-117 [DOI] [PubMed] [Google Scholar]
- 18.Kastury K, Baffa R, Druck T, Ohta M, Cotticelli MG, Inoue H, Negrini M, Rugge M, Huang D, Croce CM, Palazzo JP, Huebner K: Potential gastrointestinal tumor suppressor locus at the 3p14.2 FRA3B site identified by homozygous deletions in tumor cell lines. Cancer Res 1996, 56:978-983 [PubMed] [Google Scholar]
- 19.Druck T, Hadaczek P, Ohta M, Siprashvili Z, Baffa R, Negrini M, Kastury K, Veronese ML, Rosen D, Rothstein J, McCue P, Cotticelli MG, Inoue H, Croce CM, Huebner K: Structure and expression of the human FHIT gene in normal and tumor cells. Cancer Res 1997, 57:504-512 [PubMed] [Google Scholar]
- 20.Hadczek P, Siprashvili Z, Markiewsky M, Domagala W, Druck T, McCue PA, Pekarsky Y, Otha M, Huebner K, Lubinski J: Absence or reduction of FHIT expression in most clear cell renal carcinoma. Cancer Res 1998, 58:2946-2951 [PubMed] [Google Scholar]
- 21.Fujita J, Yoshida O, Yuasa Y, Rhim JS, Hatanaka M, Aaronson SA: Ha-ras oncogenes are activated by somatic alterations in human urinary tract tumors. Nature 1984, 309:464-466 [DOI] [PubMed] [Google Scholar]
- 22.Wright C, Mellon K, Johnston P, Lane DP, Harris AL, Horne CH, Neal DE: Expression of mutant p53, c-erb-2 and the epidermal growth factor receptor in transitional cell carcinoma of the human urinary bladder. Br J Cancer 1991, 63:967-970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cohen AJ, Berg S, Marchetto DJ, Tsai S, Jacobs SC, Brown RS: Hereditary renal-cell carcinoma associated with chromosomal translocation. N Engl J Med 1979, 301:592-595 [DOI] [PubMed] [Google Scholar]
- 24.Inoue H, Ishii H, Alder H, Snyder E, Druck T, Huebner K, Croce CM: Sequence of the FRA3B common fragile region: implications for the mechanism of FHIT deletion. Proc Natl Acad Sci USA 1997, 94:14584-14589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mimori K, Druck T, Inoue H, Alder H, Berk L, Mori M, Huebner K, Croce CM: Cancer-specific chromosome alterations in the constitutive fragile region FRA3B. Proc Natl Acad Sci USA 1999, 96:7456-7461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greenspan DL, Connolly DC, Wu R, Lei RY, Vogelstein JTC, Kim YT, Mok JE, Munoz N, Bosch FX, Shah K, Cho KR: Loss of FHIT expression in cervical carcinoma cell lines and primary tumors. Cancer Res 1997, 57:4692-4698 [PubMed] [Google Scholar]
- 27.Sozzi G, Veronese ML, Negrini M, Baffa R, Cotticelli MG, Inoue H, Tornielli S, Pilotti S, Degregorio L, Pastorino U, Pierotti MA, Ohta M, Huebner K, Croce CM: The FHIT gene at 3p14.2 is abnormal in lung cancer. Cell 1996, 85:17-26 [DOI] [PubMed] [Google Scholar]
- 28.Fong KM, Biesterveld EJ, Virmiani A, Wistuba I, Sekido Y, Bader SA, Ahmadian M, Ong ST, Rasool FV, Zimmerman PV, Giaccone G, Gazdar AF, Minna JD: FHIT and FRA3B 3p14.2 allele loss are common in lung cancer and preneoplastic bronchial lesions and are associated with cancer-related FHIT cDNA splicing aberrations. Cancer Res 1997, 57:2256-2267 [PubMed] [Google Scholar]
- 29.Yanagisawa K, Kondo M, Osada H, Uchida K, Tkagi K, Masuda A, Takahashi T: Molecular analysis of the FHIT gene at 3p14.2 in lung cancer cell lines. Cancer Res 1996, 56:5579-5582 [PubMed] [Google Scholar]
- 30.Baffa R, Veronese ML, Santoro R, Mandes B, Palazzo JP, Rugge M, Santoro E, Croce CM, Huebner K: Loss of FHIT expression in gastric carcinoma. Cancer Res 1998, 58:4708-4714 [PubMed] [Google Scholar]
- 31.Shridhar R, Shridhar V, Wang X, Paradee W, Dugan M, Sarkar F, Wilke C, Glover TW, Vaitkevicius VK, Smith DI: Frequent breakpoints in the 3p14.2 fragile site FRA3B in pancreatic tumors. Cancer Res 1996, 56:4347-4350 [PubMed] [Google Scholar]
- 32.Sorio C, Baron A, Orlandini S, Zamboni G, Pederzoli P, Huebner K, Scarpa A: The FHIT gene is expressed in pancreatic ductular cells and is altered in pancreatic cancers. Cancer Res 1999, 59:1308-1314 [PubMed] [Google Scholar]
- 33.Campiglio M, Pekarsky Y, Menard S, Tagliabue E, Pilotti S, Croce CM: FHIT loss of function in human primary breast cancer correlates with advanced stage of the disease. Cancer Res 1999, 59:3866-3869 [PubMed] [Google Scholar]
- 34.Sozzi G, Pastorino U, Moiraghi L, Tagliabue E, Pezzella F, Ghirelli C, Tornielli S, Sard L, Huebner K, Pierotti MA, Croce CM, Pilotti S: Loss of FHIT function in lung cancer and preinvasive bronchial lesions. Cancer Res 1998, 58:5032-5037 [PubMed] [Google Scholar]
- 35.Capuzzi D, Santoro E, Hauck WW, Kovatich AJ, Rosato FE, Baffa R, Huebner K, McCue PA: Fhit expression in gastric adenocarcinoma: correlation with disease stage and survival. Cancer 2000, 88:24-34 [PubMed] [Google Scholar]