Summary
3′-Uridylylation of RNA is emerging as a phylogenetically widespread phenomenon involved in processing events as diverse as uridine insertion/deletion RNA editing in mitochondria of trypanosomes and small nuclear RNA maturation in humans. This reaction is catalyzed by terminal uridylyltransferases (TUTases), which are template-independent RNA nucleotidyltransferases that specifically recognize UTP and belong to a large enzyme superfamily typified by DNA polymerase β. Multiple TUTases, recently identified in trypanosomes, as well as a U6 snRNA-specific TUTase enzyme in humans, are highly divergent at the protein sequence level. However, they all possess conserved catalytic and UTP recognition domains, often accompanied by various auxiliary modules present at the termini or between conserved domains. Here we report identification, structural and biochemical analyses of a novel trypanosomal TUTase, TbTUT4, which represents a minimal catalytically active RNA uridylyltransferase. The TbTUT4 consists of only two domains that define the catalytic center at the bottom of the nucleoside triphosphate and RNA substrate binding cleft. The 2.0 Å crystal structure reveals two significantly different conformations of this TUTase: one molecule is in a relatively open apo conformation, whereas the other displays a more compact TUTase-UTP complex. A single nucleoside triphosphate is bound in the active site by a complex network of interactions between amino acid residues, a magnesium ion and highly ordered water molecules with the UTP’s base, ribose and phosphate moieties. The structure-guided mutagenesis and cross-linking studies define the amino acids essential for catalysis, uracil base recognition, ribose binding and phosphate coordination by uridylyltransferases. In addition, the cluster of positively charged residues involved in RNA binding is identified. We also report a 2.4 Å crystal structure of TbTUT4 with the bound 2′ deoxyribonucleoside, which provides the structural basis of the enzyme's preference toward ribonucleotides.
Keywords: TUTase, Trypanosoma, nucleotidyltransferase, UTP binding, RNA editing
Introduction
Poly(A) polymerases (PAPs) and terminal uridylyltransferases (TUTases) are the only known homoribonucleotide polymerases, the enzymes that catalyze transfer of a particular nucleoside to an acceptor hydroxyl group. Whereas biological roles of RNA 3′ polyadenylation are extremely diverse, ranging from promoting mRNA stability, nuclear export and translation to RNA surveillance by stimulating the degradation of defective transcripts 1, TUTases have been shown to play key roles in a unique RNA processing pathway - uridine insertion/deletion mRNA editing in the mitochondria of trypanosomes 2. RNA Editing TUTase 1 (RET1) 3 is a processive transferase involved in uridylylation of guide RNAs 4 and, possibly, mitochondrial mRNA turnover 5. RNA Editing TUTase 2 (RET2) is an integral part of the multi-protein RNA editing complex, the “20S editosome”, and is responsible for the major guide-RNA-dependent mRNA U-insertion activity 4, 6. Non-mitochondrial TUTases of unknown functions have been reported in trypanosomes 7, as well as in human 8, 9 and plant 10 cells (reviewed in 11). Purification and identification of the first non-trypanosomal TUTase, the human enzyme specific for the U6 small nuclear RNA, has been reported recently 12.
Poly(A) polymerases and TUTases are members of the distinct nucleotidyltransferase superfamily unified by the presence of the signature helix-turn motif hG[G/S]X9-13Dh[D/E]h (X: any, h: hydrophobic amino acids) 13. Referred to as DNA polymerase β, or Pol β, the superfamily also includes ATP(CTP):tRNA nucleotidyltransferases, terminal deoxy nucleotidyltransferases (TdT), protein nucleotidyltransferases, 2′-5′ oligo(A) synthetases and antibiotic resistance nucleotidyltransferases. In most cases, two conserved acidic residues of the signature motif form a metal-binding triad with a third invariant carboxylate in order to coordinate two metal ions essential for catalysis 14. Although the basic chemistry of a nucleotidyl transfer, a metal-coordinated in-line nucleophilic attack by the 3′ hydroxyl group of the nucleic acid substrate on the α-phosphate of the NTP, is conserved throughout polymerases 15, the substrate specificity is achieved by different mechanisms. For example, the mechanisms of ATP selection by the bovine 16-19 and yeast 20, 21 poly(A) ATP polymerases, albeit still under investigation, appear to be different from the vaccinia virus PAP 22. Based on comparative sequence and structural analyses, the Pol β superfamily has been divided into nine groups represented in bacteria, archaea and eukaryotes 23. Subsequent work identified an apparently monophyletic sub-family 24, which is characterized by conserved C-terminal sequences and includes archaeal CCA-adding enzymes, eukaryotic nuclear mRNA poly(A) polymerases, Trf4/5 nuclear surveillance poly(A) polymerases25-27, 2′-5′ oligo(A) synthetases and TUTases. Crystal structures of DNA polymerase β 28, 29, archaeal CCA-adding enzymes 30, yeast and bovine PAPs 17, 21, vaccinia virus PAP 22, kanamycin nucleotidyl transferase 31, terminal deoxynucleotidyltransferase 32 and trypanosomal RNA editing TUTase 2 (RET2) 33 demonstrate that they all share the conserved N-terminal polymerase domain topology: a five-stranded mixed β sheet flanked by two or three α-helices. Additional modules vary significantly in structure and function.
The recent high-resolution structure of the RET2 TUTase with bound UTP 33 confirmed earlier assessments of the discontinuous N-terminal catalytic domain, which is interrupted by an insertion of approximately 100 amino acids 3, 34, and revealed a three-domain organization not previously observed in nucleotidyltransferases. The N-terminal domain (NTD) interacts extensively with the C-terminal domain (CTD), shaping a bi-domain with a deep cleft that forms the UTP binding site. The “insertion” in the NTD is folded into the compact middle domain (MD) that interacts with the CTD and extends out into solution. Deletion of the MD in RET1 led to complete inactivation of the enzyme, suggesting a role in protein folding or binding of RNA substrate 34. The extensive UTPRET2 contacts are localized mostly to the CTD and the Pol β signature sequence of the N-terminal domain. Superposition of RET2 and yeast nuclear poly(A) polymerases demonstrated that the N-terminal domains of RET2 and PAPs are highly conserved 33. Conversely, no common features have been observed between putative RNA binding domains, the C-terminal part of PAP 19, 20 and the middle domain of RET2 33. The structure of the vaccinia virus poly(A) polymerase/processivity factor heterodimer revealed yet another example of the nucleotide binding site in the deep cleft formed by two domains, one of which is homologous to the Pol β catalytic module 35.
Here we report identification and structure-function analysis of a novel 37.8 kDa TUTase from T. brucei designated TbTUT4. This protein lacks the middle domain present in RNA editing TUTases and therefore represents the minimal enzymatically active bi-domain RNA uridylyltransferase. The 2.0 Å crystal structure contains two molecules in the asymmetric unit, which differ markedly in conformation. One molecule represents the apo form of the enzyme, whereas the second molecule contains a single, tightly bound UTP substrate. TbRET2 and TbTUT4 differ in their biological functions, sub-cellular localization, domain composition, processivity, catalytic efficiency, and RNA substrate specificity. TbRET2 is active on double-stranded and single-stranded RNA whereas TbTUT4 accepts exclusively single-stranded RNA as a primer. Notwithstanding these differences, the bi-domain formed by the catalytic and base recognition modules is remarkably conserved among these TUTases. We also show that the UTP binding site is highly conserved among trypanosomal TUTases making it an attractive target for trypanocide development.
Results and Discussion
Identification of TbTUT4
In an attempt to assess the diversity of RNA uridylyltransferases in trypanosomes, we performed iterative PSI-BLAST searches of a non-redundant database (NCBI) with protein sequences of RNA editing TUTases 1 and 2, and non-mitochondrial TUTase 3 7 from Trypanosoma brucei and Leishmania major. The expectation value cut-off for inclusion of new sequences into a profile was set at 0.001 at each iteration. Searches were performed with full-length sequences as well as with individual N-terminal, middle and C-terminal domains. The 333 amino acid protein from T. brucei, which is annotated as poly(A) polymerase (XP829622) in the GenBank database, attracted our attention because of its high similarity to NTDs and CTDs of known TUTases. The corresponding gene, designated as Terminal Uridylyltransferase 4 from T. brucei, TbTUT4, was PCR-amplified, cloned and sequenced (GenBank accession number DQ923393, Figure 1). Several nucleotide changes, which likely represent strain variations, were detected by alignment with XP829622 (not shown). The protein is conserved among trypanosomatids and shows significant homology to the CID family of poly(A) polymerases from Schizosaccharomyces pombe 36 and animal cytoplasmic poly(A) polymerases 37. Remarkably, the middle domain, which is essential for RET1 activity 34, and is present in RET2 6, 33, 38, 39 and TUT3 7, 11, is apparently replaced by a short linker in TbTUT4 (Figure 1). Therefore, the minimal RNA nucleotidyltransferase may be composed of only N-terminal (Pol β catalytic-like) and C-terminal nucleotide-binding domains. The preliminary characterization (Aphasizhev et al, unpublished) of all trypanosomal TUTases predicted in 11 suggests that the enzymatic properties of TbTUT4 and its intracellular distribution most closely resemble the cytoplasmic TUTase activity described by White and Borst in T. brucei 40, although the precise biological role of TUT4 in trypanosomatids remains to be established.
Figure 1.
Partial multiple sequence alignment of trypanosomal and human TUTases. Multiple sequence alignments were performed with the T-coffee algorithm 55 and refined manually. Protein sequences used in the alignment are: TbRET1 (AAK38334), TbRET2 (AAO63567), TbTUT3 (XP_822966) and TbTUT4 (DQ923393). The signature motif of the Pol β nucleotidyltransferase superfamily is boxed. The N-terminal (NTD), middle (MD) and C-terminal (CTD) domains are shown by red, black and blue bars, respectively. An insertion in the human U6 TUTase NTD is shown by a green bar. Identical amino acids are red on yellow; blocks of similar amino acids are black on cyan background; blocks of weakly similar amino acids are black on green background. Amino acid positions mutated in TbTUT4 are indicated by arrows. Metal-coordinating carboxylate residues are shown by asterisks.
Substrate specificity ofTbTUT4
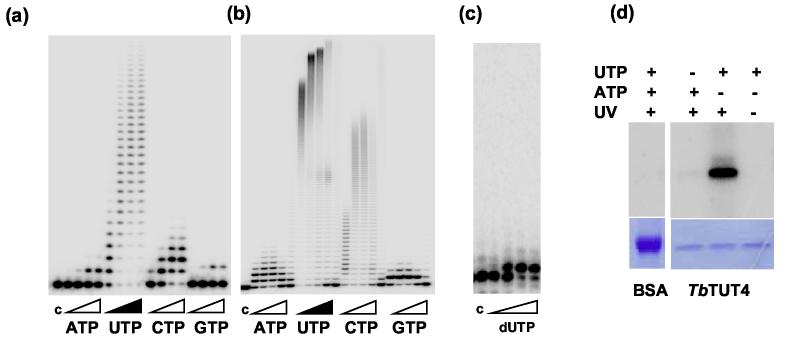
Recombinant TbTUT4 was purified from E. coli to homogeneity and tested for specificity toward nucleoside triphosphate substrates with a synthetic 5′-radiolabeled 24-mer RNA as a primer (Figure 2). In the presence of Mg2+ ions (Figure 2 (a)), TbTUT4 preferentially incorporates uridylyl residues but the reaction with CTP is also detectable, as observed previously for TbRET1 3 and TbTUT3 7. The Mg2+ concentration dependence of the TbTUT4-catalyzed reaction followed an asymmetric bell-shaped curve with maximum rates achieved at approximately 2 mM Mg2+ (not shown). Replacement of magnesium with manganese led to a dramatic stimulation of processivity and partial loss of selectivity for UTP (Figure 2 (b)). Similar effects have been reported for other TUTases 3 and poly(A) polymerases 41. The processivity of nucleoside incorporation by TbTUT4 in the presence of Mn2+ correlated well with NTP preferences observed in the Mg2+-contianing reactions, but was in general reduced at nucleoside triphosphate concentrations above 100 μM (Figure 2(b)). As we have previously demonstrated, the RNA Editing TUTase1 is capable of polymerizing UTP without RNA primer, which implies UTP positioning at the RNA binding site 34. In the case of TbTUT4, no such reaction has been observed in the presence of Mg2+, but it is entirely possible that Mn2+ enhances the UTP/RNA competition for the same binding site. In that case, increasing UTP concentration amounts to primer titration and reduces processivity of the enzyme.
Figure 2.
Nucleoside triphosphate substrate specificity of TbTUT4. Purified TbTUT4 (50 nM), 5∼-labeled 6[U] RNA (0.2 μM) and NTPs were incubated at 27°C for 20 min. c, control – 5∼-labeled RNA primer. (a) Reactions were performed in the presence of 2 mM MgCl>2 and 0.1, 1, 10 or 100 μM NTP. (b) Reactions were performed in the presence of 2.5 mM MnCl>2 and 0.1, 1, 10, 100 or 500 μM NTP. (c) Reactions were carried out as in (a), but in the presence of 0.1, 1, 10 or 100 of μM dUTP. Products were separated on a 15% polyacrylamide/urea gel. (d) Specificity of UV-induced TbTUT4-NTP cross-linking. BSA, bovine serum albumin.
Unlike poly(A) polymerases, however, TbTUT4 incorporates only one deoxynucleotide into RNA (Figure 2 (c)). The enzyme is also inactive in the presence of an RNA primer containing a terminal 2′ deoxy ribose (not shown), indicating the important role of 2′ hydroxyl interactions in positioning of the RNA in the active site.
UV-induced nucleotide-protein cross-linking has been used to elucidate nucleotide binding sites in DNA 42, RNA 43 and poly(A) polymerases 18. Because of short-range action and sensitivity to changes in the immediate vicinity of the heterocyclic base, but not the phosphate moiety 18, we sought to establish an assay that would allow assessing UTP binding by TbTUT4 independently of catalysis. Efficient protein labeling has been observed upon incubation of the TbTUT4 with [α-32P] UTP, but not [α-32P] ATP, followed by UV irradiation at 254 nm (Figure 2 (d)). Interestingly, the presence of divalent cations essential for catalysis had a marginal effect on the efficacy of cross-linking (not shown).
Although the native RNA substrates of TbTUT4 remain to be elucidated, single-stranded RNA with several uridylyl residues at the 3′ end appears to function efficiently as an RNA primer (Figure 2). Overall, these data demonstrate that the NTD-CTD bi-domain represents a minimal functional element of uridylyltransferases and does not require additional sequences for folding, single-stranded RNA binding and enzymatic activity. In addition, the modular organization of TUTases becomes apparent, in that auxiliary domains may be acquired in order to confer a specific function on a conserved catalytic bi-domain module. Such functions may include binding of structured or double-stranded RNA substrates, as single-stranded RNA may be recognized by the catalytic module itself, interaction with other proteins, e.g., RET2 docking into the editosome, or oligomerization, which has been shown for RET1 3. Examples of such auxiliary domains are the middle domain inserted into the NTD of RET1, RET2 and TUT3, the C2H2 zinc finger found at the N-terminus of the processive TUTases (RET1 and TUT3) 7, or the oligomerization domain that has been mapped with low resolution to the C-terminus of RET1 34.
Structure of TbTUT4
As the smallest of the known TUTases, TbTUT4 serves as a minimal model for this class of nucleotidyltransferases. After unsuccessful attempts of heavy atom MIR phasing and selenomethionyl MAD phasing, the structure was solved using the molecular replacement method. With a sequence identity of 30%, portions of the recently published structure of TbRET2 33 (PDB code 2B51) were used to derive search models. TbTUT4 crystals grown in the presence of UTP belong to spacegroup P21 and contain two TbTUT4 molecules in the asymmetric unit (for more details see Materials & Methods).
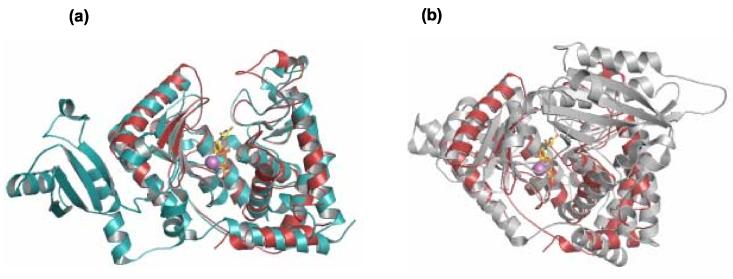
TbTUT4, an RNA-dependent U-specific nucleotidyltransferase, adopts a very compact fold in which the UTP substrate binds in the deep and spacious cleft between N-terminal (NTD) and C-terminal (CTD) domains (Figure 3(a)). The extensive contacts between the NTD and the CTD bury approximately 3,092 Å2 of solvent accessible surface area 44. The enzyme consists of a mixed alpha/beta structure with a two-layer sandwich architecture and the aforementioned beta-polymerase domain 2 topology (CATH), which consists of a twisted mixed beta sheet flanked by two α-helical domains. The β-sheet is comprised of five strands, all of which are anti-parallel with the exception of strand 5 running parallel to strand 2. The middle domain (MD), present in TbRET2 but absent in TbTUT4, would be inserted at the loop connecting strands 4 and 5. The N-terminal α-helix is surprisingly distant from the rest of the NTD and makes hydrophobic contacts with the CTD, including but not limited to the C-terminal α-helix. Analysis of electrostatic surface potential shows that the majority of positive potential resides on the front side of the molecule as an elongated region spanning across the UTP-binding cleft (Figure 3 (b)). This most likely indicates the binding region of the RNA substrate. Lastly, when compared to two of TbTUT4's close nucleotidyltransferase relatives, TbRET2 and ScPAP, the most obvious difference is the lack of RNA binding or any other specialized additional domains (Figure 4).
Figure 3.
Overall structure of TbTUT4. (a) Stereoview ribbon representation of molecule A with the NTD and CTD shown in yellow and blue, respectively. The bound UTP substrate is shown as a stick model and the Mg2+ as a magenta sphere. The dotted yellow line represents the loop region connecting the first two helices of the NTD for which no electron density was observed. A green arrow indicates the position of the middle domain insertion observed in TbRET2. Electrostatic surface potential with positive and negative regions depicted in blue and red, respectively. (b) Front view with UTP binding site. (c) Back view, a 180° rotation around the horizontal axis relative to (b). The majority of positive potential is seen as an elongated region spanning across the UTP-bound cleft. Figure generated using PyMOL (http://www.pymol.org).
Figure 4.
Structural superposition of (a) TbRET2 (teal; PDB code 2B51) with TbTUT4 (red) with the middle domain, which is only present in TbRET2, on the left. (b) Superposition of ScPAP (gray; PDB code 1FA0) with TbTUT4 (red), with an additional domain at the C-terminus, which is only present in ScPAP, on the upper right. UTP/Mg2+ are included for TbTUT4 for visual reference. Figure generated using PyMOL (http://www.pymol.org).
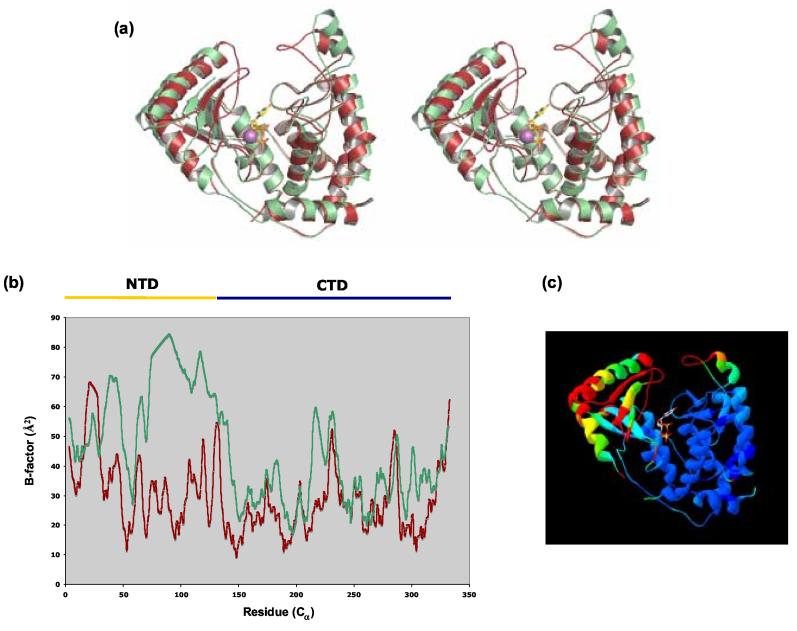
An unusual phenomenon is observed for TbTUT4 in which only one of the molecules in the asymmetric unit contains the UTP substrate, whereas the second molecule shows no trace of UTP in its binding cleft. This is the extreme version of other crystal structures where molecules in the asymmetric unit show varying fractional occupancies for bound ligands 21. The two TbTUT4 molecules exhibit significant structural variations, with the unbound or apo molecule (molecule B) having more regions of disorder and overall higher thermal displacement factors than the molecule with bound UTP (molecule A) (Figure 5). The majority of these deviations occur at the NTD as well as a small portion of the CTD, as shown by structural alignment and B-factor analysis. Upon superposition of the two molecules, it is apparent that molecule A represents a more closed conformation than molecule B, a fact that is particularly evident at the two loop regions at the top of the binding cleft (Figure 5 (a)). Comparison of the r.m.s.d. between the two molecules in the asymmetric unit of TbTUT4 with the r.m.s.d. between molecule A of TbTUT4 and TbRET2 (PDB code 2B51) reveals striking results (Table 2). The two TbTUT4 molecules in the same asymmetric unit are structurally as different from each other as UTP-bound TbTUT4 is from UTP-bound TbRET2 (1.54 Å vs. 1.60 Å), a protein that shares only 30% identity with TbTUT4. Furthermore, apo TbTUT4 shows more structural similarity with ScPAP (PDB code 1FA0) than does UTP-bound TbTUT4 (2.28 Å vs. 3.24 Å). Together, these results suggest that the enzyme undergoes a major conformational change upon UTP binding and that molecule B represents an apo form. The total buried surface area between the two molecules observed in the TbTUT4 asymmetric unit is only 432 Å2 44, suggesting that this “heterodimer” is merely crystallographic in origin. The Stokes' radius of ∼29 Å, as determined by size-fractionation on a Superose 12 column, suggests a native molecular weight of ∼40 kDa, which corresponds to that of monomeric TbTUT4 (not shown). This is further supported by the aforementioned observation that only one of the molecules contains a bound substrate, which is highly unusual for a functional dimer. Interestingly, crystallization attempts in the presence of very high UTP concentrations yield only very small unusable crystals, whereas no crystal growth occurs in the absence of UTP.
Figure 5.
The different conformations of the two molecules in the asymmetric unit. (a) Stereoview representation of TbTUT4 with molecule B (apo, green) superimposed onto molecule A (bound UTP, red). (b) Plot of Cα B-factors for molecule A (red) and molecule B (green). (c) Molecule A colored from blue to red according to its r.m.s.d. from molecule B, with regions with the largest deviations depicted in red. Panels (a) and (b) generated using PyMOL (http://www.pymol.org) and panel (c) using SwissPDBViewer 56.
Table 2.
Comparison of r.m.s.d. values between the structures of TbTUT4 molecules A (UTP-bound) and B (apo), TbRET2 (PDB code 2B51), and ScPAP (PDB code 1FA0). All superpositions and r.m.s.d. calculations were carried out with backbone Cα atoms using the SSM tool58 as implemented in the program Coot 52. Values reported in Å. Comparison of overall structures by r.m.s.d. [Å].
| UTP bound | apo | TbRET2 | ScPAP | |
|---|---|---|---|---|
| TbTUT4 UTP-bound | 0.00 | 1.54 | 1.60 | 3.24 |
| TbTUT4 apo | - | 0.00 | 2.05 | 2.28 |
| TbRET2 | - | - | 0.00 | 3.20 |
| ScPAP | - | - | - | 0.00 |
The UTP binding site
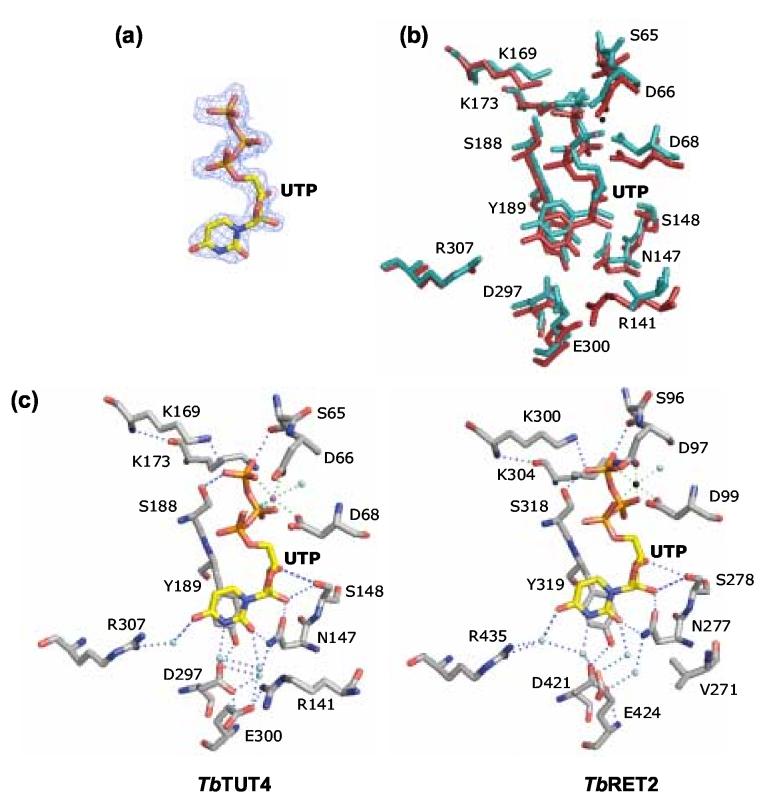
A single UTP is bound to only one of the two crystallographically independent and conformationally divergent TbTUT4 molecules in the asymmetric unit. The UTP binding mode is similar to that of site A described for TbRET2 33 with the notable exception described at the end of this section (Figure 6). The UTP molecule is involved in extensive interactions with the enzyme, primarily with the CTD, including both direct interactions with main-chain and side-chain atoms as well as indirect interactions through ordered water molecules. It is common for members of the nucleotidyltransferase family to exhibit triphosphate coordination via two divalent metal ions that are in turn coordinated by three highly conserved aspartate residues, termed the catalytic triad. However, in the case of TbTUT4, the triphosphate moiety is coordinated by only one metal cation, which participates in one interaction with each phosphate. Additionally, the metal ion is positioned by only two of the three conserved aspartates (D66 and D68), which together with two neighboring water molecules brings about heptacoordination of the cation with distances of 2.7 Å or less (Figure 7(a)). These distances and coordination geometry, in addition to the fact that the crystallization solution contained 100-fold more Ca2+ than Mg2+, suggests that this metal ion may be a Ca2+. However, crystallographic refinement of the structure with this cation as Ca2+ versus Mg2+ (18 vs. 10 electrons) resulted in a 2-fold increase in the B-factor of the ion, coupled with a significant deviation from the B factors of the neighboring atoms, suggesting that Mg2+ is the more likely candidate. To further test these observations, co-crystals of TbTUT4 and UTP were grown in the same conditions as the original crystals while replacing the 200 mM Ca(OAc)2 salt with 200 mM MgCl2, thus removing all sources of calcium from the crystallization buffer. Data collection on one of these crystals (not shown) and generation of electron density maps, including a composite annealed omit map, as well as refinement revealed nearly identical results for metal ion/tri-phosphate coordination, confirming that the metal ion is indeed Mg2+ (not shown). As for the third aspartate in the triad (D136), it seems to have no involvement in ion coordination but instead forms a salt bridge with R126 (Figure 7(b)). However, its presence, like that of the other two aspartates in the triad, has been shown to be absolutely essential for enzymatic activity (Table 3).
Figure 6.
Key protein-UTP contacts in the UTP binding site. (a) UTP observed in molecule A shown with electron density from a composite annealed omit map contoured at 1.0 σ. (b) Superposition of the active sites of TbRET2 (teal) and TbTUT4 (red). Residue labels are for TbTUT4. (c) Comparison of UTP binding via active site residues and their respective hydrogen bond networks for TbTUT4 and TbRET2. Selected water molecules (cyan spheres) were included to illustrate their significance in coordinating the uracil base while others were left out to improve clarity. The metal cations are shown as spheres: for TbTUT4 a Mg2+ (magenta sphere), and for TbRET2 a Mn2+ (black sphere). Figure generated using PyMOL (http://www.pymol.org).
Figure 7.
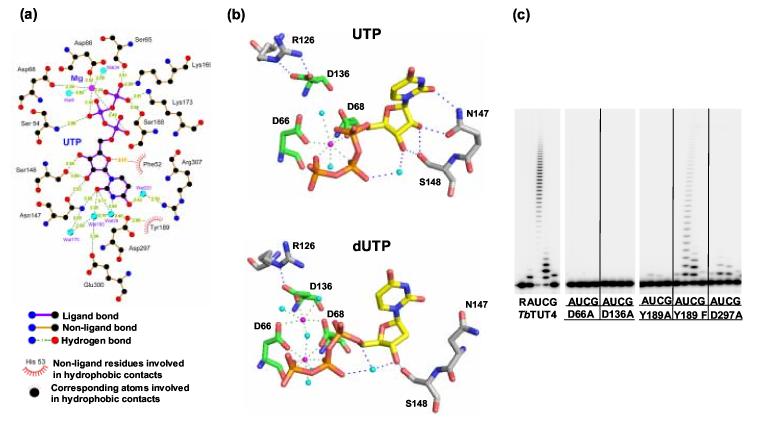
Contribution of base, ribose and phosphate moieties to UTP/dUTP recognition and catalysis. (a) 2D representation of UTP/Mg2+ and active site residues generated using the program LIGPLOT 57. (b) Comparison of UTP and dUTP binding and the respective roles of the catalytic triad aspartates (green). The major differences are observed at positions D136, N147 and for the orientation of the uracil plane. Coordinated Mg2+ ions are shown in magenta and key water molecules in cyan. Panels (a) and (b) generated using PyMOL (http://www.pymol.org). (c) Nucleotidyltransferase activity of TbTUT4 carrying mutations at the metal-coordinating carboxylate (D66), the base-stacking aromatic residue (Y189) and a water-coordinating residue (D297). Reactions were performed as in Figure 1 with 100 nM of enzyme and 100 μM of the respective NTP.
Table 3.
Steady-state kinetic parameters of trypanosomal TbTUT4 mutants with UTP.
| Protein | Km (μM) | ± |
kcat (min−1) |
± |
kcat/Km (min−1 M−1) |
[kcat/Km]mutant/ [kcat/Km]w.t. |
|---|---|---|---|---|---|---|
| TUT4 | 1 | 0.9 | 0.5 | 0.06 | 5 × 105 | 1.0 |
| TUT4/dUTP | 12.2 | 5.4 | 0.2 | 0.02 | 1.6 ×103 | 0.003 |
| F52A | Inactive | Inactive | Inactive | Inactive | ||
| D66A | Inactive | Inactive | Inactive | Inactive | ||
| D68A | Inactive | Inactive | Inactive | Inactive | ||
| R121A | 2.8 | 5.1 | 0.002 | 0.0006 | 8.6 × 102 | 0.002 |
| R121F | Inactive | Inactive | Inactive | Inactive | ||
| R126A | Inactive | Inactive | Inactive | Inactive | ||
| D136A | Inactive | Inactive | Inactive | Inactive | ||
| R141A | 1.0 | 2.1 | 0.007 | 0.0005 | 6.3× 103 | 0.01 |
| N147A | 115.3 | 10.5 | 0.3 | 0.02 | 2.6 × 103 | 0.0005 |
| S148A | 87.3 | 49.2 | 1.3 | 0.5 | 1.4 × 104 | 0.03 |
| S188A | 34.0 | 11.4 | 0.03 | 0.007 | 8.8 × 102 | 0.002 |
| Y189A | Inactive | Inactive | Inactive | Inactive | ||
| Y189F | 21.3 | 3.6 | 0.9 | 0.04 | 4.2 × 104 | 0.09 |
| D297A | 172.8 | 265.5 | 0.02 | 0.003 | 9.1 × 101 | 0.0002 |
| D297N | 13.9 | 25.7 | 0.004 | 0.001 | 2.9 × 102 | 0.0006 |
| E300A | 308.4 | 161.5 | 0.4 | 0.1 | 1.3 × 103 | 0.003 |
| R307A | 0.4 | 1.2 | 0.008 | 0.0004 | 2.0 × 104 | 0.04 |
In addition to metal coordination, the phosphate oxygens serve as hydrogen-bond partners for several active-site serine and lysine residues (Figure 7(a)). Phosphate Oγ1 and Oγ2 are each involved in hydrogen bond interactions with the side chains of one serine and one lysine (S188/K173 and S65/K169, respectively). Phosphate Oγ3 coordinates the Mg2+ cation and receives a hydrogen bond from Wat194 (not shown). Phosphate Oβ1 is also involved in Mg2+ coordination and accepts a hydrogen bond from the backbone NH of S54 and Wat34 while Oβ2 is bridged to the 3′ hydroxyl of the ribose by Wat5. As for the α-phosphate, both Oα1 and Oα2 are stabilized by water molecules (not shown) with Oα1 also contributing to metal coordination. The local environment around the α-phosphate is sterically conducive to in-line nucleophilic attack by the 3′ hydroxyl of the incoming RNA substrate, resulting in nucleoside monophosphate incorporation into the nascent RNA and loss of the pyrophosphate leaving group.
The 3′ hydroxyl group of the ribose sugar donates a hydrogen bond to the oxygen of S148. In fact, the role of S148 may involve preferential binding of ribonucleotides over deoxyribonucleotides as its hydroxyl serves as a hydrogen bond donor to the 2′ O of the ribose. Furthermore, the 2′ hydroxyl donates a hydrogen bond to the carbonyl oxygen of N147, whose amide group in turn donates a hydrogen bond to O2 of the uracil base. Having interactions with both the sugar and the base of UTP, N147 contributes significantly to UTP binding. Furthermore, stacking is observed between one side of the uracil base and the phenol ring of Y189, another interaction that has been shown to be crucial for enzymatic activity (Table 3). In contrast, the region on the side of the uracil not stacking against the Y189 phenol ring appears relatively open and is likely to play a role in RNA substrate binding.
The uracil base is furthermore held in place by numerous neighboring water molecules that are part of an extensive hydrogen-bonded network (Figure 7 (a)). Two waters in particular, Wat28 and Wat220, coordinated by D297 and R307, respectively, may contribute to U-specificity as these waters interact with two significant positions on the pyrimidine base. In the case of UTP binding (standard uracil di-oxo tautomer), the base donates a hydrogen bond from its N3 to Wat28 and receives a hydrogen bond from Wat220 at its carbonyl O4. The hydrogen-bonding roles of these two positions would be reversed in the case of CTP binding (or for the 4-hydroxy tautomer of uracil), where the base N3 becomes a hydrogen-bond acceptor and the NH2, (or 4-OH for the 4-hydroxy tautomer of uracil), would serve as a hydrogen-bond donor. This is the region where the most significant difference in UTP interaction between co-crystals of TbTUT4 and UTP-soaked crystals of cross-linked TbRET2 occurs. In TbRET2, the water interacting with the uracil N3 (Wat26) is donating two hydrogen bonds to the carboxylates of D421 and E424 and it is thus only able to receive a hydrogen bond from the base N3, while it cannot act as a hydrogen bond donor to a unprotonated N3, as would be required for CTP binding33. In contrast, the equivalent water in TbTUT4 (Wat28) only donates one hydrogen bond to the carboxylate of D297 (the equivalent of D421 in TbRET2) while the carboxylate of E300 (the equivalent of E424 in TbRET2) has swung away to form a salt bridge with R141. R141 in TbRET2 is a valine (V271), which is presumably a reason for E424 interacting with the N3-coordinating water instead of forming a salt bridge. This may explain why TbTUT4 is more likely than TbRET2 to accept CTP in its NTP binding pocket.
Mutational analysis of UTP binding site
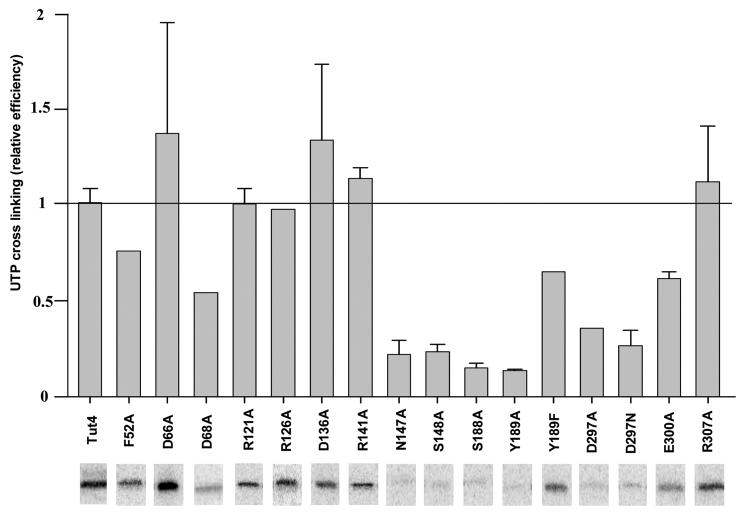
Earlier mutagenesis studies of RNA Editing TUTase1, based on multiple sequence alignments with poly(A) polymerases, established essential roles of three metal coordinating carboxylates (D66, D68 and D136 in TbTUT4 numbers) and positively charged residues of unknown function in positions 121 and 126 (TbTUT4 numbers) 34. A single UTP binding site observed in the TbTUT4-UTP complex (Figures 6(a) and 7(a)) provides an ideal model for the systematic assessment of individual residues contributing to UTP binding and catalysis. The traditional filter-based TUTase activity assay has been found inadequate for measuring kinetic parameters of severely compromised TbTUT4 mutants. Therefore, a new quantitative assay was developed based on high-resolution separation of reaction products by acrylamide/urea gels (Figure 7(c)). The capacity to measure incorporation of a single nucleotide at high UTP concentrations extended the dynamic range of the assay from three to approximately six orders of magnitude. In addition, the UTP-protein cross-linking (Figures 2 (d) and 8) has been applied to evaluate UTP-protein affinity in the absence of RNA substrate.
Figure 8.
UTP binding by TbTUT4 mutants. UTP-protein cross-linking induced by 254 nm UV irradiation was used to assess UTP binding in the absence of RNA primer. Three independent cross-linking reactions have been used to calculate standard deviation. Representative images of [α-32P]UTP-labeled bands are shown below bars for each mutant protein.
Substitution of metal-coordinating aspartate residues in the signature sequence (D66A and D68A) predictably inactivated the enzyme (Figure 7(c), Table 3), confirming the critical role of these residues in catalysis. Mutation at position D136, which does not appear to be directly involved in metal coordination or contact with UTP, but rather forms a salt bridge with R126 (Figure 7(b)), also led to a complete loss of activity. Remarkably, none of these mutations, except D68A, resulted in loss of UTP-protein binding as assessed by UV cross-linking (Figure 8). Disruption of the D136-R126 salt bridge by the substitution of R126 to alanine abrogated UMP incorporation into RNA while projecting no effect on UTP binding. Although both D136 and R126 are highly conserved among TUTases, it is unclear whether the salt bridge that locks carboxyl and guanidinium groups is conserved as well. Lack of D136 involvement in metal coordination is consistent with the presence of a single magnesium in the active site (Figure 7(a,b)), which has also been observed in the RET2 structure 33. Another possible salt bridge formed by R141 and E300 in TbTUT4 also does not affect UTP binding - the arginine in position 141 is not universally conserved and the corresponding mutation to alanine did not result in loss of cross-linking or increase in the apparent Km for UTP. The R141A mutation, however, did increase apparent Km for RNA substrate (Table 4). In the TbRET2 structure, E424 (E300 in TbTUT4) forms a salt bridge with a different position R144 (R121 in TbTUT4), and has been proposed to create a structural support for the UTP binding site. However, the alternative role for R121 and three other arginine residues may be envisioned based on the multiple sequence alignment (Figure 1) and superposition of TbRET2, TbTUT4 and ScPAP (Figure 4), which brings into focus a positively charged loop 118 RRTRVPVVRVK 128. This element is highly structurally conserved and contains another invariant arginine in position 126. The residues R121 and R126, together with arginines 141 and 307, lay close in proximity to the non-stacked side (opposite Y189) of the uracil base. As in the cases of R141A and R307A, substitution R121A had no effect on the Km for UTP or cross-linking efficiency but resulted in approximately a 100-fold drop in catalytic rate. Introducing a space-filling phenyl ring at this position (R121F) produced an inactive enzyme. Taken together, these data suggest that the RNA primer approaches the bound UTP from the side of the base that is not involved in stacking with Y189. It is entirely possible that binding of the RNA primer in the active site is facilitated by contacts with a cluster of arginine residues in positions 121, 126, 141 and 307. The lack of conservation at positions 141 and 307 between trypanosomal and human TUTases may reflect the highly specific U6 snRNA substrate requirement of the latter 8, 9.
Table 4.
Steady-state kinetic parameters of trypanosomal TbTUT4 mutants with RNA.
| Protein | Km (μM) | ± |
kcat (min−1) |
± |
kcat/Km (min−1 M−1) |
[kcat/Km]mutant/ [kcat/Km]w.t. |
|---|---|---|---|---|---|---|
| TUT4 | 0.2 | 0.2 | 0.3 | 0.08 | 1.5×106 | 1 |
| R121A | 132.0 | 44.5 | 1.5 | 0.35 | 1.1×104 | 0.007 |
| R141A | 72.6 | 4.5 | 1.3 | 0.05 | 1.8×104 | 0.012 |
| S148A | 0.2 | 0.4 | 1.0 | 0.5 | 5.3 × 106 | 3.5 |
| S188A | 0.3 | 0.3 | 0.1 | 0.05 | 3.0 × 105 | 0.2 |
| R307A | 10.0 | 2.6 | 0.5 | 0.04 | 5×104 | 0.03 |
In order to substantiate the hypothesis of RNA binding by the arginine cluster, the catalytic parameters have been determined for RNA substrate in reactions catalyzed by R121A, R141A and R307A mutants (Table 4). These experiments were possible because the UTP affinity had not been affected by corresponding mutations, which allowed achieving high specific activity of UTP substrate in the reactions. As an internal control, the catalytic parameters for RNA have been measured for the S148A and S188A mutants with compromised UTP binding (Table 3 and Fig. 8). The R126A and R121F mutations rendered proteins inactive under all experimental conditions. The increase in apparent Km ranged from approximately 50 fold in the case of R307 to nearly 600 fold for R121 mutant proteins. Remarkably, the catalytic constant remained virtually unaffected, further confirming the role of these residues in RNA binding. More specifically, clustering of these arginine residues in close proximity to the bound UTP suggest an involvement in coordination of the RNA's terminal nucleoside.
Invariant D297 is directly involved in water molecule-mediated bridging with the N3 amine of the uracil base whereas E300 is not. However, mutation of either position to alanine produced a significant increase in Km for UTP and loss of cross-linking efficiency. Remarkably, the kcat was unaffected by the E300A mutation and dropped approximately 25-fold as a result of the D297A substitution. Restoring one hydrogen bond acceptor relative to the D297A substitution by introducing an asparagine (D297N) partially rescued UTP binding as seen from a 10-fold improvement in the apparent Km. An additional role in UTP binding by TbRET2 has been proposed for another water-mediated contact between R435 33 (R307 in TbTUT4), and O4 of the base. Whereas no effect on Km for UTP or TbTUT4-UTP cross-linking has been observed upon substitution for alanine at this position (R307), a significant drop in catalytic rate (Table 4) and the lack of its conservation between human and trypanosomal TUTases indicates a possible role in RNA binding. Indeed, this suggests that of the two uracil base-specific positions, endocyclic nitrogen N3 and the O4 atom, the former plays a key role in both base recognition and binding whereas contributions of the latter may be deemed insignificant.
The cluster of hydrogen bonds between invariant asparagine and serine residues at positions 147 and 148 as well as the exocyclic oxygen 2 and ribose hydroxyls of the UTP, are crucial for UTP binding, but not catalysis, as can be seen from the approximate 100-fold increase in Km and 80% reduction in UTP cross-linking. Hydrogen bonding to the 2′ hydroxyl group of the ribose is suggestive of a possible role in discrimination of dUTP binding, as reflected by a 10-fold increase in the apparent Km in a single-round dUMP addition (Figure 2(c), Table 3). In agreement with these data, the identical RNA substrate containing a single deoxyuridine at the 3′ end does not act as a UMP acceptor in the TbTUT4-catalyzed reaction (not shown). Apparently, the 2′ hydroxyl group of the RNA substrate is essential for coordination of the nucleophilic attack by the 3′ hydroxyl on the α-phosphate of the UTP.
Although hydrogen bonding is imperative for selective UTP binding, disruption of a stacking interaction between the protein and bound nucleoside triphosphate, which thus far has not been observed in PAPs or other nucleotidyltransferases, had a dramatic effect on the reaction. Replacement of the invariant tyrosine residue at position 189 by alanine completely inactivated the enzyme (Figures 7, 8 and Table 3) and resulted in virtual loss of UTP cross-linking. It should be noted, however, that the stacking interactions of the phenyl ring and uracil base may account for observed photo cross-linking 45. Therefore, in the case of Y189A mutation the lack of cross-linking may not reflect loss of UTP binding. In order to distinguish the effects of phenyl ring removal and loss of hydrogen bonding with D297 (Figures 6 and 7), the Y189F mutation has also been tested. In the case of Y189F, the moderate increase in Km and decrease in UTP cross-linking are consistent, as the hydroxyl group most likely contributes to orientation of the phenyl ring by H-bonding with D297. However, the dramatic effect of the Y189A mutation emphasizes the significance of the stacking interaction between the protein residue and uracil ring as being one of the major UTP binding determinants. The adjacent serine residue 188 points in the opposite direction and, in addition to S65, K73 and K79, recognizes the γ-phosphate. This observation explains the previously reported loss of activity in Leishmania RET1 due to charge reversal at position 79 (TbTUT4 numbers) 34 and a significant decrease of catalytic efficiency in the case of the S188A mutation (Table 3).
Hydroxyl residues of the Pol β superfamily signature sequence hG[G/S]X9-13Dh[D/E]h, which includes the metal-coordinating carboxylates (D66 and D68), provide hydrogen bond donors (S54 and S65) to the non-bridging oxygens of the triphosphate moiety. As expected, none of the residues of this motif are in direct contact with the uracil base. However, the possible clash of the first signature residue (F52 in TbTUT4), which is often large and hydrophobic in Pol β nucleotidyltransferases 23, with the ribose ring oxygen, appears to have a major impact on activity (Table 3), but not on UTP binding (Figure 8). Modeling studies show the lack of substantial alterations of protein structure due to such a mutation, which suggests a role in maintaining ribose conformation, perhaps in conjunction with the invariant Y189 stacked to the uracil base.
Ribonucleotide selection by uridylyltransferases
TbTUT4, an RNA uridylyltransferase, adds a single deoxy uridylyl residue to an RNA primer (Fig. 2 (c)). The overall loss of catalytic efficiency is mostly due to an increase in the apparent Km(Table 3). In order to determine the contribution of the 2′ hydroxyl group in UTP binding, we have also determined a co-crystal structure of TbTUT4 with bound dUTP at 2.4 Å resolution. The structure reveals identical overall protein conformation but significant changes in both nucleotide binding and metal-ion coordination as compared to the TbTUT4-UTP co-crystal structure (Figure 7 (b)). The largest difference observed is the rotation of N147 away from the substrate, resulting not only in the loss of its hydrogen bond with the now non-existent 2′ hydroxyl group but also with the carbonyl oxygen at position 2 of the uracil ring. In addition, an approximately 15° rotation is observed in the plane of the uracil base with respect to its position in the UTP co-crystal structure, resulting in reduced stacking with the aforementioned key residue, Y189. These observations help to explain the enzyme's preference for UTP over dUTP since the substrates only differ by a single hydroxyl group. Another noteworthy difference is the change in positioning of the α-phosphate, resulting in the loss of its contribution to metal-ion coordination. Furthermore, the third catalytic aspartate residue, D136, which has no involvement in metal-ion coordination in the UTP co-crystal structure, is now coordinating a second magnesium ion in the dUTP co-crystal structure. Very weak electron density observed in the active site of the second molecule in the asymmetric unit revealed partial occupancy of a second dUTP molecule (not shown), which was set to 0.5 for refinement, although the overall protein conformation of this molecule still resembles that of the apo molecule of TbTUT4-UTP.
Conclusions
The crystal structure of a novel trypanosomal terminal uridylyltransferase composed solely of the N-terminal and C-terminal domains demonstrates that a minimal core module of the highly divergent TUTase family is able to form a compact, enzymatically active structure. Multiple sequence alignments of trypanosomal and human TUTase sequences highlight the modular organization of these enzymes. In the more complex members of the family, diverse auxiliary domains are either added to the N-terminus, such as C2H2 zinc fingers (RET1, TUT3 and human U6 TUTase) and RRM RNA binding motifs (human U6 TUTase) 12, or the C-terminus, such as oligomerization domains of RET1 34. Furthermore, an insertion of approximately 100 residues, designated as the middle domain, occurs at a conserved site in the N-terminal domain of trypanosomal RET1, RET2 and TUT3, but not in TUT4. In the case of human U6 TUTase, an approximately 120-amino-acid fragment is inserted into a different position – the loop (amino acids 84-89, Figure 1), which connects the helical turn formed by the Pol β signature motif with an extended conserved α-helix at the C-terminal end of the NTD. Whereas the middle domain in TbRET2 is a tightly folded structure, attempts to detect similarity to any known protein or predict secondary structural elements in the U6 TUTase 120-amino acid insertion have been unsuccessful.
In addition to displaying a minimal active fold for an RNA-dependent nucleotidyltransferase, crystal structure shows two molecules in different conformations, one apo and one with bound UTP, suggesting large conformational changes in TbTUT4 upon substrate binding. Kinetic and UTP cross-linking experiments with TbTUT4 mutants establish a defined set of amino acids that contribute to catalysis and UTP binding. Remarkably, the amino acids providing water-mediated (D297 and E300), or direct (N147) hydrogen bonding with the uracil base, as well as stacking interactions (Y189), are invariant among evolutionarily divergent TUTases and are indispensable for UTP binding. The mutagenesis and cross-linking studies allowed us to distinguish the only essential uracil base-specific contact, the water mediated hydrogen bonding of endocyclic nitrogen N3, from the insignificant contributions of the also U-specific oxygen at position 4. The hydrogen bonds involving ribose hydroxyls are apparently important for recognition of the C3′-endo conformation. In addition, the structure of the TbTUT4-dUTP complex demonstrates the involvement of the 2′ hydroxyl group in coordination of the key residue N147, which also forms a hydrogen bond with the carbonyl oxygen at position 2 of the uracil ring. The latter interaction appears to be important for maintaining co-planarity between the uracil base and essential tyrosine residue 189. Most unexpectedly, the hydrophobic interaction of the sugar pucker with a phenylalanine (F52) preceding the Pol β signature motif is essential for the reaction, likely due to its effect on catalytic rate rather than UTP binding. The observed domain distribution of crucial amino acids allowed us to draw a clear functional distinction between the N-terminal catalytic and C-terminal base recognition domains. The highly conserved NTD bears three universal carboxylates as well as residues involved in the coordination of the triphosphate moiety, the common feature of all NTPs. The specificity for a particular substrate comes from CTD – nucleotide base interactions via hydrogen bonding and stacking contacts.
The positioning of the RNA primer for in-line attack on the UTP α-phosphate remains to be elucidated but some insight has been gained from mutations of four conserved arginine residues (121, 126, 141 and 307), each affecting the catalytic rate of UTP incorporation (Table 3). These arginines are clustered in proximity to the non-stacked face of the uracil base, opposite from conserved Y189. The terminal base of a bound RNA primer could potentially stack against the free face of the UTP's uracil base, resulting in a triple stack of aromatic rings (Y189 phenyl / UTP uracil / RNA 3′ base). Simple modeling shows that there is also ample space for the 3′ ribose of the RNA, and that its 3′ hydroxyl could be situated within 5 Å of the α-phosphate of the UTP, near the three invariant aspartic acids residues D66, D68 and D136. Elucidation of the structure of the ternary complex (TbTUT4 : UTP : RNA), if necessary with the aid of a nonhydrolysable UTP analog, would greatly increase our understanding of how these enzymes interact with the RNA primer.
Trypanosomal TUT4 assumes a bi-domain structure, which most likely originated via recombination between a catalytic domain of the Pol β superfamily13 and the nucleotide-binding ATP-cone domain46, the former providing a conserved catalytic triad of carboxylic residues and the latter the binding site for a nucleotide base. This core platform is shared by 2′-5′ oligoadenylate synthetases, Trf4 and Gld-2-type poly(A) polymerases, archaeal CCA-adding enzymes and uridylyltransferases 24, which implies that different nucleoside triphosphate specificities may be achieved by minor variations in the binding site that do not include key interactions. Indeed, it appears that the conserved positions identified in this work as crucial for UTP binding, such as Y189 and D297, proved to be important for class I archaeal CCA-adding enzyme ATP-incorporating activity 47 and Trf4 poly(A) polymerase function in vivo48, respectively.
Materials and Methods
Protein expression, purification and mutagenesis
The TUT4 genes from T. brucei and L. major were PCR-amplified, cloned and sequenced. Gene sequences were deposited in the GenBank database under accession numbers DQ923393 and DQ923394, respectively. The full-length TbTUT4 gene was inserted into the pET15b vector (Novagen) to generate an N-terminal 6xHis fusion protein. The expression vector was transformed into the BL 21(DE3) RIL (Stratagene) E. coli strain. A bacterial culture was grown in 4 liters of 2YT media with 1% glucose at 37° C to an A600 of ∼0.4. The temperature was then decreased to 10° C and expression was induced with 0.5 mM IPTG (isopropyl-1-thio-β-D-galactopyranoside) for 4 hours. Protein purification was performed at 4° C. The cell pellet (10 g) was washed with PBS, resuspended in 60 ml of lysis buffer (50 mM HEPES, pH 8.0, 50 mM NaCl, 0.2 mg/ml of lysozyme, 1 tablet of Complete™ EDTA-free (Roche) protease inhibitors) and passed through a French press at 12,000 psi. Sodium chloride was adjusted to 300 mM and Triton X-100 to 0.1%. The extract was clarified by centrifugation at 40,000 rpm for 30 minutes in a Ti60 rotor (Beckman) and loaded onto 4 ml of Talon resin (Clontech). The column was washed with 20 column volumes of 300 mM NaCl, 50 mM HEPES, pH 8.0 buffer and with 10 column volumes of the same buffer with 10 mM imidazole. Protein was eluted with 300 mM NaCl, 50 mM HEPES, pH 8.0, 200 mM imidazole buffer, diluted 3-fold with 25 mM HEPES, pH 7.5, loaded onto a 5 ml HiTrap S (GE Healthcare) and eluted in 25 mM HEPES, pH 7.5 using a linear gradient of KCl. The protein solution was adjusted to 10 mg/ml in 10 mM HEPES, pH 7.5, 70 mM KCl, 0.5 mM DTT and used for crystallization immediately. The purity of protein preparation typically exceeded 94%, as determined by SDS-polyacrylamide gel electrophoresis analysis of 1 μg of TbTUT4 followed by staining in Sypro Ruby (Invitrogen).
Point mutations were introduced by a PCR-based method using the QuikChange™ kit (Stratagene) and confirmed by DNA sequencing. Mutant proteins were isolated as above from two liters of bacterial culture.
Size exclusion chromatography
The protein sample (50 μl, 10 mg/ml) was loaded onto a Superose 12 column (GE Healthcare) in 20 mM HEPES, pH 7.5, 150 mM KCl at 0.1 ml/min. Molecular mass standards used to estimate Stokes' radius were aldolase, albumin, ovalbumin and chymotrypsinogen.
Steady-state enzyme kinetics assays
To determine kinetic parameters for UTP substrate, the filter-binding based assays were carried out in 10 μl of 50 mM Tris-HCl, pH 8.0, 10 mM KCl, 1 mM DTT, 2 mM Mg(OAc)2, 1 μM 6[U] RNA and [α-32P]UTP (0.5 μM - 400 μM, ∼10000-500 cmp/pmol). Reactions were started by addition of enzyme to 50-200 nM and incubated at 27°C for 0.25 to 45 min. For RNA substrate parameters, the UTP concentration was kept constant at 10 μM (∼10000 cpm/pmol) and protein was added to 200 nM. Concentrations of RNA varied from 0.01 to 50 μM. Reactions were stopped by adding an equal volume of 0.25 mM NaH2PO4, 0.5% SDS buffer, transferred on DE 81 (Whatman), dried, washed three times per 15 min in 1L of 0.5% sodium pyrophosphate and 0.5 M phosphate buffer pH 7.5 and counted in a liquid scintillation cocktail (Ready Safe, Beckman Coulter) using an LS 6500 counter (Beckman Coulter).
For gel-based assays, reactions were performed in the same buffer with 4 μM of 5′-labeled 6 [U] RNA, 0.1-500 μM of UTP and 100 – 200 nM of the enzyme for 1-30 min. Reactions were stopped by adding 4 volumes of 5 mM EDTA, 95% formamide. The products were separated to single nucleotide-resolution on a 15% acrylamide/8M urea gel and exposed to a phosphor storage screen (Biorad).
Kinetic parameters, the apparent Km, and catalytic rate kcat for UTP incorporation were obtained by fitting the initial velocities as a function of UTP concentration from three experiments into a standard Michaelis kinetics model. The Sigma Plot Enzyme Kinetics software package was used for calculations of Km, Vmax, and standard deviations. The 6[U] RNA (GCUAUGUCUGUCAACUUGUUUUUU) was purchased from Dharmacon Inc. The RNA was purified on a 40 cm 15% acrylamide/urea gel, eluted into 0.1 M sodium acetate, pH 5.0, 0.1% SDS, 1 mM EDTA and precipitated with ethanol.
NTP-protein cross-linking
TbTUT4 (0.1 μg) or bovine serum albumin (10 μg) was incubated with 0.4 pmol of [α-32P] UTP (6,000 Ci/mmol) in 10 μl of buffer containing 50 mM of Tris-HCl, pH 8.0, 10 mM KCl, 1 mm DTT, 2 mM Mg(OAc)2 for 5 min at room temperature and 10 minutes on ice. Cross-linking was performed in an HL 2000 Hybrilinker (UVP) at 100 mJ/cm2. Proteins were separated on an 8-16% SDS/acrylamide gel, transferred to a nitrocellulose membrane and exposed onto a phosphor storage screen (Biorad). The acrylamide gels were stained with Sypro Ruby and digital images were acquired with Gel Doc station (Biorad). The intensity of radioactive signal was normalized vs. intensity of the protein band. The relative cross-linking efficiency in most cases was calculated based on three independent experiments.
Crystallization, data collection, phasing and refinement
Purified recombinant N-terminally His-tagged TbTUT4 at a concentration of 10 mg/mL in 10 mM Hepes pH 7.6, 70 mM KCl, 0.5 mM DTT, and in the presence of 4 mM MgCl2 and 50 μM UTP, was used for initial screening with commercial crystallization screens from Hampton Research and Emerald Biosciences using the vapor diffusion technique at 4 °C. Crystallization trials in the absence of UTP yielded no results. Small plate-like co-crystals of TbTUT4 with UTP grew in conditions of 100 mM sodium cacodylate pH 6.5, 200 mM calcium acetate, 18% PEG-8000 (Crystal Screen solution 46, Hampton Research). For crystallographic analysis, crystals were flash-cooled in liquid nitrogen using the same conditions supplemented with 15% glycerol as a cryo-protectant. Co-crystals of TbTUT4 with dUTP were grown in the same manner, except that the crystallization solution contained 200 mM magnesium chloride in place of 200 mM calcium acetate; concentrations of protein and dUTP were 8 mg/mL and 400 μM, respectively.
X-ray data for the TbTUT4-UTP structure were collected at the Advanced Light Source (Berkeley), beam-line 4.2.2 (Table 2). The data were processed using d*TREK 49 and scaled to a maximum resolution of 2.0 Å using SCALA 44. The packing density, or V>M 50, (4.25 Å3/Da for one molecule and 2.12 Å3/Da for two molecules of TbTUT4 in the asymmetric unit) suggested the presence of two crystallographically independent molecules. However, inspection of the self-rotation function did not reveal any noncrystallographic two-fold axes.
Numerous data sets from heavy metal soaked crystals did not reveal interpretable difference Patterson maps. A three-wavelength data set from a selenomethionyl derivative of TbTUT4 also failed to reveal the location of the selenium sites. Thus, initial phases for TbTUT4 were derived using the program PHASER 51 by searching for two copies each of the NTD and CTD of TbRET2 (PDB code 2B51, sequence identity to target: 30%), respectively. Peaks in an anomalous difference Fourier map computed with phases derived from molecular replacement confirmed that the selenomethionine data collected earlier indeed contained selenium. However, the anomalous signal from eight selenium sites in an asymmetric unit of 75.6 kDa was too weak to allow MAD phasing. In addition, the lack of 2-fold non-crystallographic symmetry increased the difficulty of site identification and reduced the feasibility of map averaging. A large portion of the final model was built by hand using the program Coot 52 and refined using both REFMAC5 44 and CNS 53. Initial topology and parameter files for refinement of the UTP molecule were generated using the program XPLO2D (http://xray.bmc.uu.se/usf/factory_5.html).
X-ray data for the TbTUT4-dUTP structure were collected at the Stanford Linear Accelerator Center, beam-line 9-1 and were processed and scaled using d*TREK 49. The atomic model was obtained by REFMAC5 44 refinement of the TbTUT4-UTP model against the structure factors obtained from the TbTUT4-dUTP dataset, and the dUTP molecules were built in by hand using Coot52. Since the structures of TbTUT4-UTP and TbTUT4-dUTP are essentially identical, the same test set was carried over from the TbTUT4-UTP reflection file to conserve the free reflection set. Final model geometry for both structures was analyzed using PROCHECK 54. The structure factors and atomic coordinates of TbTUT4-UTP and TbTUT4-dUTP have been deposited with the Protein Data Bank under accession codes 2IKF and 2NOM, respectively.
Table 1.
Crystal Data and Refinement Statistics
| Crystal Data | TbTUT4-UTP | TbTUT4-dUTP |
|---|---|---|
| Beamline | 4.2.2* | 9-1** |
| Wavelength (Å) | 0.979 | 0.979 |
| Space Group | P211 | P21 |
| Cell Dimensions | a = 78.8 Å | a = 79.1 Å |
| b = 41.5 Å | b = 41.7 Å | |
| c = 103.8 Å | c = 104.3 Å | |
| β = 94.9° | β = 96.5° | |
| Resolution Range (Å) | 51.71 - 2.00 | 41.19 – 2.40 |
| Total Reflections | 131,965 | 93,588 |
| Unique Reflections | 43,289 | 26,868 |
| Redundancy | 3.0 (2.9) | 3.5 (3.6) |
| Completeness (%) | 94.5 (95.6) | 99.5 (99.8) |
| Mean I/σI | 7.3 (2.6) | 4.9 (2.0) |
| Rsym(%) | 9.8 (57.3) | 17.8 (50.9) |
| Refinement | TbTUT4-UTP | TbTUT4-dUTP |
|---|---|---|
| Resolution range (Å) | 51.71 - 2.00 | 41.20 - 2.40 |
| No. of structure factors | 43,270 | 25,464 |
| Rwork/Rfree* (%) | 22.8 / 27.5 | 23.0 / 30.1 |
| R.m.s.d. bonds (Å) | 0.014 | 0.011 |
| R.m.s.d. angles (°) | 1.78 | 1.42 |
| Number of atoms / avg. B value (Å2) | ||
| Protein | 5,087 / 37.1 | 4,645 / 37.1 |
| Waters | 216 / 35.0 | 125 / 29.1 |
| Metal ion | 1 / 21.9 | 2 / 23.0 |
| Nucleotide | 29 / 27.1 | 56 / 35.8 |
| Ramachandran plot (favored/allowed/generously allowed, %)** | 89.2 / 10.8 / 0 | 88.4 / 11.6 / 0 |
Advanced Light Source, Berkeley, CA
Stanford Linear Accelerator Center, Menlo Park, CA
(Values) are calculated for highest resolution shell.
Rfree based on a test set size of 5% of all structure factors
PROCHECK54
Acknowledgements
We thank Dr. Wim Hol for an early copy of the TbRET2 atomic coordinates (PDB code 2B51) and members of Aphasizhev and Luecke laboratories for discussions. This work was supported by NIH grant AI064653 to RA and NIH grant GM56445 to HL.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome 2. Nat. Rev. Mol. Cell Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- 2.Stuart KD, Schnaufer A, Ernst NL, Panigrahi AK. Complex management: RNA editing in trypanosomes. Trends Biochem. Sci. 2005;30:97–105. doi: 10.1016/j.tibs.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 3.Aphasizhev R, Sbicego S, Peris M, Jang SH, Aphasizheva I, Simpson AM, Rivlin A, Simpson L. Trypanosome Mitochondrial 3′ Terminal Uridylyl Transferase (TUTase): The Key Enzyme in U-insertion/deletion RNA Editing. Cell. 2002;108:637–648. doi: 10.1016/s0092-8674(02)00647-5. [DOI] [PubMed] [Google Scholar]
- 4.Aphasizhev R, Aphasizheva I, Simpson L. A tale of two TUTases. Proc. Natl. Acad. Sci. U. S. A. 2003;100:10617–10622. doi: 10.1073/pnas.1833120100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ryan CM, Read LK. UTP-dependent turnover of Trypanosoma brucei mitochondrial mRNA requires UTP polymerization and involves the RET1 TUTase. RNA. 2005;11:763–773. doi: 10.1261/rna.7248605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ernst NL, Panicucci B, Igo RP, Jr., Panigrahi AK, Salavati R, Stuart K. TbMP57 is a 3′ terminal uridylyl transferase (TUTase) of the Trypanosoma brucei editosome. Mol. Cell. 2003;11:1525–1536. doi: 10.1016/s1097-2765(03)00185-0. [DOI] [PubMed] [Google Scholar]
- 7.Aphasizhev R, Aphasizheva I, Simpson L. Multiple terminal uridylyltransferases of trypanosomes. FEBS Lett. 2004;572:15–18. doi: 10.1016/j.febslet.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 8.Trippe R, Richly H, Benecke BJ. Biochemical characterization of a U6 small nuclear RNA-specific terminal uridylyltransferase. Eur. J. Biochem. 2003;270:971–980. doi: 10.1046/j.1432-1033.2003.03466.x. [DOI] [PubMed] [Google Scholar]
- 9.Trippe R, Sandrock B, Benecke BJ. A highly specific terminal uridylyl transferase modifies the 3′-end of U6 small nuclear RNA. Nucleic Acids Res. 1998;26:3119–3126. doi: 10.1093/nar/26.13.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zabel P, Dorssers L, Wernars K, Van Kammen A. Terminal uridylyl transferase of Vigna unguiculata: purification and characterization of an enzyme catalyzing the addition of a single UMP residue to the 3′-end of an RNA primer. Nucleic Acids Res. 1981;9:2433–2453. doi: 10.1093/nar/9.11.2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aphasizhev R. RNA uridylyltransferases. Cell Mol. Life Sci. 2005;62:2194–2203. doi: 10.1007/s00018-005-5198-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trippe R, Guschina E, Hossbach M, Urlaub H, Luhrmann R, Benecke BJ. Identification, cloning, and functional analysis of the human U6 snRNA-specific terminal uridylyl transferase 1. RNA. 2006 doi: 10.1261/rna.87706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holm L, Sander C. DNA polymerase beta belongs to an ancient nucleotidyltransferase superfamily. Trends Biochem. Sci. 1995;20:345–347. doi: 10.1016/s0968-0004(00)89071-4. [DOI] [PubMed] [Google Scholar]
- 14.Pelletier H, Sawaya MR. Characterization of the metal ion binding helix-hairpin-helix motifs in human DNA polymerase beta by X-ray structural analysis. Biochemistry. 1996;35:12778–12787. doi: 10.1021/bi960790i. [DOI] [PubMed] [Google Scholar]
- 15.Steitz TA. DNA and RNA polymerases: structural diversity and common mechanisms. Harvey Lect. 1997;93:75–93. [PubMed] [Google Scholar]
- 16.Martin G, Moglich A, Keller W, Doublie S. Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) polymerase. J. Mol. Biol. 2004;341:911–925. doi: 10.1016/j.jmb.2004.06.047. [DOI] [PubMed] [Google Scholar]
- 17.Martin G, Keller W, Doublie S. Crystal structure of mammalian poly(A) polymerase in complex with an analog of ATP. EMBO J. 2000;19:4193–4203. doi: 10.1093/emboj/19.16.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin G, Jeno P, Keller W. Mapping of ATP binding regions in poly(A) polymerases by photoaffinity labeling and by mutational analysis identifies a domain conserved in many nucleotidyltransferases. Protein Sci. 1999;8:2380–2391. doi: 10.1110/ps.8.11.2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin G, Keller W. Mutational analysis of mammalian poly(A) polymerase identifies a region for primer binding and catalytic domain, homologous to the family X polymerases, and to other nucleotidyltransferases. EMBO J. 1996;15:2593–2603. [PMC free article] [PubMed] [Google Scholar]
- 20.Zhelkovsky A, Helmling S, Bohm A, Moore C. Mutations in the middle domain of yeast poly(A) polymerase affect interactions with RNA but not ATP. RNA. 2004;10:558–564. doi: 10.1261/rna.5238704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bard J, Zhelkovsky AM, Helmling S, Earnest TN, Moore CL, Bohm A. Structure of yeast poly(A) polymerase alone and in complex with 3′-dATP. Science. 2000;289:1346–1349. doi: 10.1126/science.289.5483.1346. [DOI] [PubMed] [Google Scholar]
- 22.Moure CM, Bowman BR, Gershon PD, Quiocho FA. Crystal structures of the vaccinia virus polyadenylate polymerase heterodimer: insights into ATP selectivity and processivity 1. Mol. Cell. 2006;22:339–349. doi: 10.1016/j.molcel.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 23.Aravind L, Koonin EV. DNA polymerase beta-like nucleotidyltransferase superfamily: identification of three new families, classification and evolutionary history. Nucleic Acids Res. 1999;27:1609–1618. doi: 10.1093/nar/27.7.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rogozin IB, Aravind L, Koonin EV. Differential action of natural selection on the N and C-terminal domains of 2′-5′ oligoadenylate synthetases and the potential nuclease function of the C-terminal domain. J. Mol. Biol. 2003;326:1449–1461. doi: 10.1016/s0022-2836(03)00055-x. [DOI] [PubMed] [Google Scholar]
- 25.LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 26.Vanacova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS. Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Regnault B, Devaux F, Namane A, Seraphin B, Libri D, Jacquier A. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 28.Pelletier H, Sawaya MR, Wolfle W, Wilson SH, Kraut J. Crystal structures of human DNA polymerase beta complexed with DNA: implications for catalytic mechanism, processivity, and fidelity. Biochemistry. 1996;35:12742–12761. doi: 10.1021/bi952955d. [DOI] [PubMed] [Google Scholar]
- 29.Sawaya MR, Pelletier H, Kumar A, Wilson SH, Kraut J. Crystal structure of rat DNA polymerase beta: evidence for a common polymerase mechanism. Science. 1994;264:1930–1935. doi: 10.1126/science.7516581. [DOI] [PubMed] [Google Scholar]
- 30.Li F, Xiong Y, Wang J, Cho HD, Tomita K, Weiner AM, Steitz TA. Crystal structures of the Bacillus stearothermophilus CCA-adding enzyme and its complexes with ATP or CTP. Cell. 2002;111:815–824. doi: 10.1016/s0092-8674(02)01115-7. [DOI] [PubMed] [Google Scholar]
- 31.Sakon J, Liao HH, Kanikula AM, Benning MM, Rayment I, Holden HM. Molecular structure of kanamycin nucleotidyltransferase determined to 3.0-A resolution 2. Biochemistry. 1993;32:11977–11984. doi: 10.1021/bi00096a006. [DOI] [PubMed] [Google Scholar]
- 32.Delarue M, Boule JB, Lescar J, Expert-Bezancon N, Jourdan N, Sukumar N, Rougeon F, Papanicolaou C. Crystal structures of a template-independent DNA polymerase: murine terminal deoxynucleotidyltransferase 1. EMBO J. 2002;21:427–439. doi: 10.1093/emboj/21.3.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deng J, Ernst NL, Turley S, Stuart KD, Hol WG. Structural basis for UTP specificity of RNA editing TUTases from Trypanosoma brucei. EMBO J. 2005;24:4007–4017. doi: 10.1038/sj.emboj.7600861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aphasizheva I, Aphasizhev R, Simpson L. RNA-editing terminal uridylyl transferase 1: identification of functional domains by mutational analysis. J. Biol. Chem. 2004;279:24123–24130. doi: 10.1074/jbc.M401234200. [DOI] [PubMed] [Google Scholar]
- 35.Moure CM, Bowman BR, Gershon PD, Quiocho FA. Crystal structures of the vaccinia virus polyadenylate polymerase heterodimer: insights into ATP selectivity and processivity. Mol. Cell. 2006;22:339–349. doi: 10.1016/j.molcel.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 36.Read RL, Martinho RG, Wang SW, Carr AM, Norbury CJ. Cytoplasmic poly(A) polymerases mediate cellular responses to S phase arrest. Proc. Natl. Acad. Sci. U. S. A. 2002;99:12079–12084. doi: 10.1073/pnas.192467799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang L, Eckmann CR, Kadyk LC, Wickens M, Kimble J. A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature. 2002;419:312–316. doi: 10.1038/nature01039. [DOI] [PubMed] [Google Scholar]
- 38.Aphasizhev R, Aphasizheva I, Nelson RE, Gao G, Simpson AM, Kang X, Falick AM, Sbicego S, Simpson L. Isolation of a U-insertion/deletion editing complex from Leishmania tarentolae mitochondria. EMBO J. 2003;22:913–924. doi: 10.1093/emboj/cdg083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Worthey EA, Schnaufer A, Mian IS, Stuart K, Salavati R. Comparative analysis of editosome proteins in trypanosomatids. Nucleic Acids Res. 2003;31:6392–6408. doi: 10.1093/nar/gkg870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.White T, Borst P. RNA end-labeling and RNA ligase activities can produce a circular ribosomal RNA in whole cell extracts from trypanosomes. Nucleic Acids Res. 1987;15:3275–3290. doi: 10.1093/nar/15.8.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin G, Keller W. Tailing and 3′-end labeling of RNA with yeast poly(A) polymerase and various nucleotides. RNA. 1998;4:226–230. [PMC free article] [PubMed] [Google Scholar]
- 42.Rush J, Konigsberg WH. Photoaffinity labeling of the Klenow fragment with 8-azido-dATP. J. Biol. Chem. 1990;265:4821–4827. [PubMed] [Google Scholar]
- 43.Mustaev A, Zaychikov E, Grachev M, Kozlov M, Severinov K, Epshtein V, Korzheva N, Bereshchenko O, Markovtsov V, Lukhtanov E, Tsarev I, Maximova T, Kashlev M, Bass I, Nikiforov V, Goldfarb A. Strategies and methods of cross-linking of RNA polymerase active center. Methods Enzymol. 2003;371:191–206. doi: 10.1016/S0076-6879(03)71014-3. [DOI] [PubMed] [Google Scholar]
- 44.Bailey S. The Ccp4 Suite - Programs for Protein Crystallography 1. Acta Crystallographica Section D-Biological Crystallography. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 45.Meisenheimer KM, Koch TH. Photocross-linking of nucleic acids to associated proteins 2. Crit Rev. Biochem. Mol. Biol. 1997;32:101–140. doi: 10.3109/10409239709108550. [DOI] [PubMed] [Google Scholar]
- 46.Aravind L, Wolf YI, Koonin EV. The ATP-cone: an evolutionarily mobile, ATP-binding regulatory domain. J. Mol. Microbiol. Biotechnol. 2000;2:191–194. [PubMed] [Google Scholar]
- 47.Tomita K, Fukai S, Ishitani R, Ueda T, Takeuchi N, Vassylyev DG, Nureki O. Structural basis for template-independent RNA polymerization. Nature. 2004;430:700–704. doi: 10.1038/nature02712. [DOI] [PubMed] [Google Scholar]
- 48.Wang Z, Castano IB, Adams C, Vu C, Fitzhugh D, Christman MF. Structure/function analysis of the Saccharomyces cerevisiae Trf4/Pol sigma DNA polymerase. Genetics. 2002;160:381–391. doi: 10.1093/genetics/160.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pflugrath JW. The finer things in X-ray diffraction data collection 1. Acta Crystallogr. D Biol. Crystallogr. 1999;55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 50.Matthews BW. Solvent Content of Protein Crystals. J. Mol. Biol. 1968;33:491. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 51.Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions 4. Acta Crystallogr. D Biol. Crystallogr. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 52.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics 3. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 53.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination 13. Acta Crystallogr. D. Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 54.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck - A Program to Check the Stereochemical Quality of Protein Structures 15. Journal of Applied Crystallography. 1993;26:283–291. [Google Scholar]
- 55.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 56.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling 6. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 57.Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions 4. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 58.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions 1. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]