Abstract
The cellular response to hypoxia includes the hypoxia-inducible factor-1 (HIF-1)-induced transcription of genes involved in diverse processes such as glycolysis and angiogenesis. Induction of the HIF-regulated genes, as a consequence of the microenvironment or genetic changes, is known to have an important role in the growth of experimental tumors. Hypoxia-inducible factors 1α and 2α (HIF-1α and HIF-2α) are known to dimerize with the aryl hydrocarbon receptor nuclear translocator in mediating this response. Because regulation of the α chain protein level is a primary determinant of HIF activity, our aim was to investigate the distribution of HIF-1α and HIF-2α by immunohistochemistry in normal and pathological tissues using monoclonal antibodies (mAb). We raised a new mAb to detect HIF-1α, designated 122, and used our previously validated mAb 190b to HIF-2α. In the majority of solid tumors examined, including bladder, brain, breast, colon, ovarian, pancreatic, prostate, and renal carcinomas, nuclear expression of HIF-1α and -2α was observed in varying subsets of the tumor cells. HIF-2α was also strongly expressed by subsets of tumor-associated macrophages, sometimes in the absence of any tumor cell expression. Less frequently staining was observed in other stromal cells within the tumors and in normal tissue adjacent to tumor margins. In contrast, in normal tissue neither molecule was detectable except within subsets of bone marrow macrophages, where HIF-2α was strongly expressed.
The ability of cells to adapt to periods of hypoxia is important for their survival in both physiological and pathophysiological states. 1 Regions of hypoxia are known to exist within many tumors, and the extent of tumor hypoxia correlates with prognosis in a number of tumor types. 2,3
A widespread oxygen sensing system exists in mammalian cells. 4 This regulates expression of a diverse group of genes including erythropoietin, glucose transporters, glycolytic pathway enzymes, vascular endothelial growth factor (VEGF), transferrin, heme oxygenase, and inducible nitric oxide synthetase, 5 many of which are known to be up-regulated in cancer. The response is mediated by a transcriptional complex termed hypoxia-inducible factor-1 (HIF-1), which consists of a heterodimer of HIF-1α and HIF-1β, identical to the previously identified aryl hydrocarbon nuclear translocator (ARNT). 6 Both are members of a family of transcription factors, termed bHLH/PAS proteins, which control a variety of critical embryogenic and physiological events. 7-9 An alternative dimerization partner for ARNT, which also transactivates genes via HIF DNA recognition sites, has been identified and termed endothelial PAS domain protein 1 (EPAS-1), 10 HIF-1α-like factor (HLF), 11 and mouse HIF-related factor (HRF). 12 In keeping with its functional homology with HIF-1α, we have used the term HIF-2α to describe this protein. Recently another family member with sequence homology to HIF-1α and the ability to act as a dimerization partner of ARNT has been identified and termed HIF-3α. 13
Hypoxia in the tumor microenvironment is sufficient to activate HIF-dependent gene expression. 14 A major role for HIF in determining gene expression, tumor angiogenesis, and growth has been demonstrated in xenograft experiments with a cell line deficient in ARNT (HIF-1β). 15 These tumors grow more slowly and have reduced vascularity compared to those from wild-type cells. Studies of HIF-1α knockouts and deficient embryonic stem cells have affirmed its essential role in solid tumor vascular formation and embryonic vascularization. 8,16,17
HIF can also be activated in cancer as a consequence of tumor suppressor gene inactivation. For example, in von Hippel-Lindau syndrome the genetic defect results in constitutive stabilization of HIF α chains, 18 contributing to the highly angiogenic phenotype of tumors in these patients.
We previously reported that HIF-2α is widely expressed in a panel of human cell lines and demonstrated concordance between the HIF-1α and HIF-2α response to a range of experimental conditions. 19 The critical determinant of HIF activity is the level of HIF-1α or HIF-2α protein, since ARNT is constitutively present. 6,20,21 On exposure to hypoxia, both HIF-1α and HIF-2α protein accumulate rapidly in the nucleus and disappear on reoxygenation. The protein levels of HIF-1α and HIF-2α are determined mainly by their rate of proteasomal degradation. 19,22 Although other mechanisms, including positive and negative regulation of co-activator recruitment, phosphorylation, cellular redox state, and intracellular compartmentalization, 23 may also have a role in determining HIF activity, the dominant mode of regulation is HIF α stabilization. 21
Descriptions of the distribution within human tissue of both HIF-1α and HIF-2α have primarily been of mRNA. In situ studies have found HIF-1α RNA to be prevalent in all tissues other than peripheral blood leukocytes, 24 and HIF-2α RNA to be highly expressed in vascular tissues such as lung, heart, placenta, and kidney. 10,12 We have previously reported nuclear localization of these transcription factors in hypoxic cells cultured on slides by immunostaining. 19 Immunostaining using a polyclonal antibody to HIF-1α in frozen lung tissue and retina from animal models of hypoxia has recently been described, showing nuclear localization in a range of cells. 25,26
We now report the use of two monoclonal antibodies that recognize HIF-1α and HIF-2α epitopes and that survive formalin fixation and paraffin embedding. This has allowed us to study the expression and distribution of both HIF-1α and HIF-2α protein in a wide range of normal and pathological human tissues.
Materials and Methods
Cell Culture
Cell lines were cultured as recommended by the European Collection of Animal Cell Cultures. Medium was obtained from Clare Hall (Imperial Cancer Research Fund, London). Cells were plated 24 hours before experiments such that they were approaching confluence at the start of each experiment. Hypoxic exposure was for 4 hours in a NAPCO (Winchester, VA) incubator with 0.1% oxygen, 5% CO2, and the balance nitrogen.
Antibodies
The antibodies used are described in Table 1 ▶ . Immunogens were prepared by cloning the indicated restriction fragments from human HIF-1α or human or mouse HIF-2α in-frame into pGEX-4t-1 (Amersham Pharmacia Biotech, Little Chalfont, UK) with a modified polylinker. Expression of glutathione-S-transferase (GST) fusion protein was induced by exposure of transformed Escherichia coli BL21 or DH5α cells to 0.1 mmol/L isopropyl b-D-thiogalactopyranoside. After ultrasonic bacterial lysis, protein was affinity purified with glutathione Sepharose 4B (Amersham Pharmacia Biotech). Rabbit polyclonal and mouse monoclonal antibodies (mAbs) were generated by standard techniques. Hybridoma supernatants were affinity purified using protein A Sepharose (Pharmacia). The absorbed protein was eluted using 0.1 mol/L citrate buffer, pH 3.0, and neutralized using 10 mmol/L NaOH and desalted into phosphate-buffered saline (PBS).
Table 1.
Details of the Primary Antibodies Used
| Antibody | Immunogen | Isotype | Ref. |
|---|---|---|---|
| 122 | Human HIF-1α 329–530 | Mouse IgG1 | |
| 190b | Human HIF-2α 535–631 | Mouse IgG1 | 19 |
| PG-M1 | Human CD68 | Mouse IgG3 | 41 |
| KB 90 | Human CD11c | Mouse IgG1 | 42 |
| RK5C1 | S. cerevisiae GAL4 1-147 | Mouse IgG2a (Santa Cruz) | |
| PM 8 | Mouse HIF-2α 337–439 | Rabbit polyclonal antiserum |
Antisera and hybridoma supernatants were screened by enzyme-linked immunosorbent assay against the GST fusion protein and GST alone (0.1 ng of protein per well), by immunolabeling of frozen and paraffin-embedded transfected COS-1 cells and by immunoblotting (see below).
Transient Transfections of COS-1 Cells
Immunostaining properties of antibodies were tested on frozen and paraffin-embedded pellets of transfected COS cells expressing fusion proteins consisting of HIF-1α or HIF-2α fused to a portion of the yeast transcription factor Gal4 in a subset of the cells. For these experiments, the cells were electroporated with 10 μg of plasmids pGN/HIF-1α 28–826 or pGN/HIF-2α 19–870. 19 After 48 hours, the cells were removed with EDTA, washed with PBS, and either pelleted and fixed in 10% neutral buffered formalin overnight before paraffin-embedding, or snap-frozen in liquid nitrogen. The frozen cell pellets were later embedded in Bright Cryo-M-Bed (Bright Instrument Co. Ltd. Huntingdon, UK), and 8-μm sections were cut onto multiwell slides. A variety of fixatives were tested on these: acetone, methanol, acetone/methanol, and 4% formalin/PBS.
Tissues
Tissues were obtained from the Cellular Pathology Department at the John Radcliffe Hospital (Oxford, UK). Formalin-fixed, paraffin wax-embedded sections were cut at approximately 5 μm and floated on to X-Tra slides (Surgipath Europe Ltd., Peterborough, UK).
Tumor sections were obtained from cases of carcinoma of the bladder, brain, breast, kidney, liver, lung, ovary, pancreas, and prostate. Normal tissue examined included skin, bladder, thymus, spleen, tonsil, lymph node, thyroid, adrenal, pancreas, salivary gland, liver, kidney, heart, esophagus, colon, lung, ovary, testis, uterus, placenta, umbilical cord, brain, prostate, and breast. Sources were diagnostic samples identified from reports as histologically normal, samples originating from healthy individuals (wedge biopsies from living donor kidneys or bone marrow trephines from allogeneic bone marrow donors), and blocks of tissue reported as normal but known to have been located adjacent to resected tumors.
Immunocytochemical Staining
Three-stage peroxidase immunostaining of paraffin sections was performed after dewaxing and rehydrating slides. Endogenous peroxidase was blocked with 0.5% hydrogen peroxide in water for 30 minutes. Antigen retrieval was by pressure cooking in 50 mmol/L Tris and 0.2 mmol/L EDTA buffer, pH 9.0, for 180 seconds or incubating at 60°C for 16 hours in the same buffer (Simon Biddolph, Pathology Department, John Radcliffe Hospital, Oxford, UK, personal communication). To detect HIF-1α and HIF-2α, concentrations of mAbs 122 and 190b were applied at 15 μg/ml to the section and incubated at room temperature for 90 minutes. Substitution of the primary antibody with PBS was used as a negative control. The secondary horseradish peroxidase (HRP)-conjugated goat anti-mouse serum was used at 1/200 (DAKO, Glostrup, Denmark) for 30 minutes and the tertiary HRP-conjugated rabbit anti-goat antiserum 1/100 (DAKO) for 30 minutes. The peroxidase reaction was developed using diaminobenzidine and slides were washed and mounted in aqueous mountant (Aquamount; BDH Laboratory Supplies, Poole, UK). When a rabbit polyclonal serum was used as primary antibody a HRP goat anti-rabbit antiserum 1/100 (DAKO) was used as the secondary. For some of the CD68 immunostaining biotinylated goat anti-mouse serum at 1/400 (DAKO) was used as the secondary followed by streptABComplex/AP (DAKO). The localization of any cellular staining and its intensity were independently assessed by two observers. The intensity of nuclear staining was compared to that seen in parallel stained hypoxic cell pellet sections.
Western Blotting
For whole-cell extracts, adherent cells were washed and removed by scraping and prepared as previously described. 19 Proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in 6% gels under reducing conditions and transferred to Immobilon PVDF membrane (Millipore, Bedford, MA) overnight in 20 mmol/L Tris, 0.1 mol/L glycine, 10% methanol, and 0.05% SDS. The membrane was blocked with PBS, 5% fat-free dried milk, 0.1% Tween 20. For HIF-1α detection, purified mAbs 28b and 122 were used at 4 μg/ml; for HIF-2α, the mAb 190b was used at 4 μg/ml. For detection of the Gal4 DNA binding domain in fusion proteins, RK5C1 (Santa Cruz Biotechnology, Santa Cruz, CA) was used at 0.1 μg/ml. Detection was with HRP-conjugated goat anti-mouse antibody diluted 1/1000 followed by chemiluminescence with enhanced chemiluminescence reagents (Amersham Pharmacia Biotech).
In Situ Hybridization
Sense and antisense probes were labeled with [35S] UTP using cDNA fragments of human HIF-1α 255 bp (nucleotides 764-1018, accession no. U22431), human HIF-2α (nucleotides 2542 to 2762, U81984), and human VEGF 121 (590 nucleotides) 27 as templates.
Eight-micron frozen sections and 4-μm paraffin sections were cut onto Superfrost Plus slides (Surgipath Europe Ltd.). The paraffin sections were dried at 37°C overnight and then dewaxed and fixed in 4% paraformaldehyde in the same manner as the frozen sections. Serial sections were also cut and stored for conventional immunostaining. Subsequent slide preparation and hybridization was by standard techniques as previously described. 15
Results
Selection and Characterization of Antibodies Reactive against HIF-1α and HIF-2α in Immunostaining of Paraffin Sections
Screening our panel of mAbs, we found antibodies 122 and 190b were the best reagents for immunostaining HIF-1α and HIF-2α, respectively, in formalin-fixed, paraffin-embedded material.
Immunoblotting of extracts of COS cells transfected with either pGN/HIF-1α 28–826 or pGN/HIF-2α 19–870 demonstrated that each antibody recognized a single species with no cross-reactivity, which comigrated with the anti-GAL4 signal. Furthermore, in HeLa cell extracts, known to express both HIF-1α and HIF-2α, a single inducible species of the expected mobility was detected by each antibody (Figure 1) ▶ .
Figure 1.
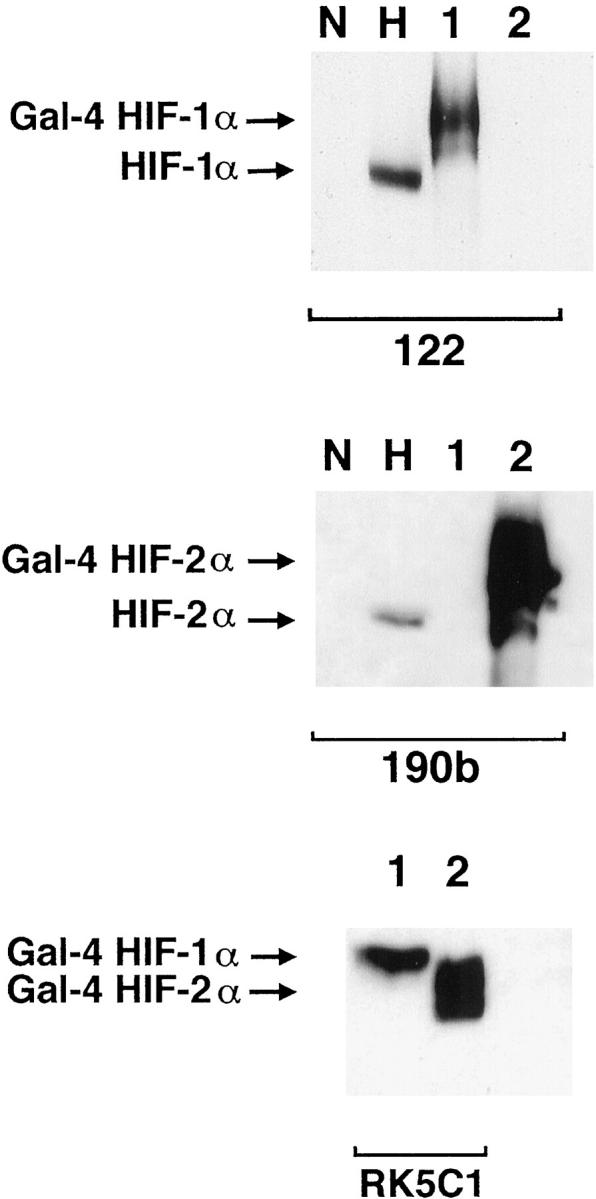
Western blots showing specificity of mAbs used for immunostaining. Whole cell extracts were prepared from HeLa cells cultured in parallel for 4 hours in normoxia (N) and 0.1% hypoxia (H) and COS-1 cells transfected with either pGN/HIF-1α28–826 (lane 1) or pGN/HIF-2α19–870 (lane 2). Transfection resulted in the expression of fusion proteins between the N-terminal Gal4 DNA binding domain and the indicated amino acids of the respective HIF α chains. Preliminary analysis (not shown) using the Gal4 mAb established the amount of each COS extract required to give approximately equal Gal4 signal, indicating similar amounts of HIF-1α and HIF-2α fusion proteins. These were loaded with 50 μg of HeLa extracts and separated by SDS-PAGE, transferred to PVDF membrane and analyzed in parallel using mAb 122 (HIF-1α), 190b (HIF-2α), and RK5C1 (Gal4) as primary antibodies. mAb 122 detects a single band in extracts from COS cells transfected with pGN/HIF-1α28–826 and no bands in extracts from COS cells transfected with pGN/HIF-2α19–870, whereas the opposite pattern is seen with mAb 190b. Detection of comigrating bands with an antibody to the GAL4 DNA binding domain (RK5C1) confirms the identity of these bands as the respective fusion proteins. The antibodies to HIF-1α and HIF-2α recognize hypoxically inducible proteins of 135 kd and 110 kd, respectively, in the HeLa cell extracts, compatible with the known migration of endogenous HIF-1α and HIF-2α on SDS-PAGE.
Transfected COS-1 cells were next used to demonstrate the utility and specificity of these reagents for staining paraffin-embedded material. Cell pellets harvested after transient transfection were processed in a manner identical to that for routine diagnostic histopathological material. Positive nuclear immunostaining was detected in cells expressing GAL4-HIF-1α and GAL4-HIF-2α fusion proteins by mAbs 122 and 190b, respectively (Figure ▶ 2).
The level of endogenous hypoxically induced HIF-1α and HIF-2α protein was manyfold less than that expressed after episomal plasmid amplification in the subset of COS cells that were transfected. Therefore, paired cell pellets were prepared from a number of different human cell lines cultured in parallel in normoxia or 0.1% hypoxia for 4 hours. Although the number of positive cells and the intensity of detectable nuclear staining varied between cell lines, specific staining confined to the nucleus was seen only after hypoxic exposure. Results for HT1080 cells are illustrated in Figure 2 ▶ .
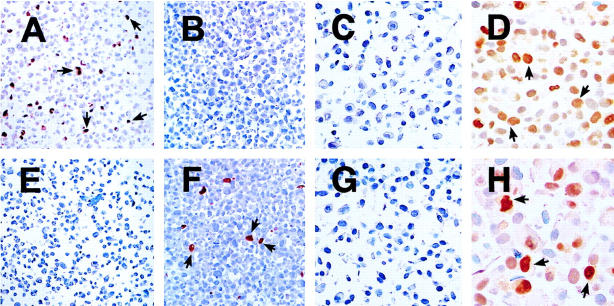
Figure 2.
HIF-1α and HIF-2α protein detection by peroxidase immunohistochemistry on paraffin-embedded material. COS-1 cells were transfected with either pGN/HIF-1α28–826 (A and E) or pGN/HIF-2α19–870 (B and F) resulting in the expression of fusion proteins in a subset of the cells. Cells were then fixed in formalin and processed into paraffin-embedded blocks in a manner analogous to the handling of diagnostic pathological specimens. Likewise HT1080 cells were cultured in parallel for 4 hours in normoxia or 0.1% hypoxia and processed to produce a normoxic cell pellet (C and G) and a hypoxic cell pellet (D and H). Nuclear staining was observed with mAb 122 (A−D) only in the HIF-1α transfectants (A) and the hypoxic cell pellet (D). mAb 190 detected only the HIF-2α fusion protein (F) and HIF-2α was absent in the normoxic cell pellet (G), but hypoxically inducible (H). The intensity of nuclear staining observed within the hypoxic cell pellet was heterogeneous for both antigens. Original magnifications, ×100 (A, B, E, and F) and ×400 (C, D, G, and H).
HIF-1α and HIF-2α Protein Expression in Human Tumors
HIF-1α and HIF-2α protein was detected in most types of human tumors examined, including bladder, breast, colon, glial, hepatocellular, ovarian, pancreatic, prostate, and renal tumors. However, not all tumors stained with both antibodies, and some stained with neither (Table 2) ▶ .
Table 2.
Expression of HIF-1α and HIF-2α within Tumor Cell Nuclei
| Tumor type | No. of cases | HIF-1α and HIF-2α positive | HIF-1α-positive only | HIF-2α-positive only | HIF-1α and HIF-2α negative |
|---|---|---|---|---|---|
| Bladder adenocarcinoma | 5 | 2 | 0 | 0 | 3 |
| Glioblastoma multiforme | 5 | 2 | 0 | 0 | 3 |
| Breast adenocarcinoma | 12 | 10 | 0 | 1 | 1 |
| Colon adenocarcinoma | 5 | 4 | 0 | 0 | 1 |
| Hepatocellular carcinoma | 5 | 2 | 0 | 0 | 3 |
| Hypernephroma | 5 | 2 | 2 | 0 | 1 |
| Lung tumors (squamous cell, adenocarcinoma) | 5 | 0 | 0 | 0 | 5 |
| Ovarian adenocarcinoma | 5 | 5 | 0 | 0 | 0 |
| Pancreatic adenocarcinoma | 3 | 2 | 0 | 1 | 0 |
| Prostatic adenocarcinoma | 5 | 2 | 0 | 1 | 2 |
| Buffy coat chronic myeloid leukemia | 2 | 0 | 0 | 0 | 2 |
| Totals | 57 | 31 | 2 | 3 | 21 |
In addition, HIF-2α expression was detected within subsets of tumor-associated macrophages in all cases.
Within positive tumors, the extent, intensity, intracellular location, and distribution of staining seen with both antibodies was heterogeneous. The number of positive tumor nuclei did not correlate with the intensity of staining and ranged from <1% to 95% of tumor cells. In the cases examined, HIF-1α and HIF-2α nuclear staining, respectively, was found in <10% of tumor nuclei in 54% and 61% of the cases, between 10 and 50% of tumor cells, in 9% and 11% of cases, and in >50% in 36% and 28% of tumors. To illustrate these points, examples of immunostaining are shown in Figure 3 ▶ . Sections from a breast adenocarcinoma show predominantly nuclear immunostaining that is of comparable frequency for both HIF-1α and HIF-2α (Figure 3, A and B) ▶ . However, the intensity of nuclear immunoreactivity for each antigen in different tumor cells was variable. A hepatocellular carcinoma is used to demonstrate nuclear and cytoplasmic immunoreactivity for both proteins. Interestingly, in this example the number of nuclei immunostaining for HIF-2α outnumber those staining for HIF-1α (Figure 3, C and D) ▶ . In the renal cell carcinoma, illustrated nuclear and cytoplasmic immunoreactivity for HIF-1α is shown (Figure 3E) ▶ , whereas HIF-2α immunoreactivity was nuclear (Figure 3F) ▶ .
Figure 3.
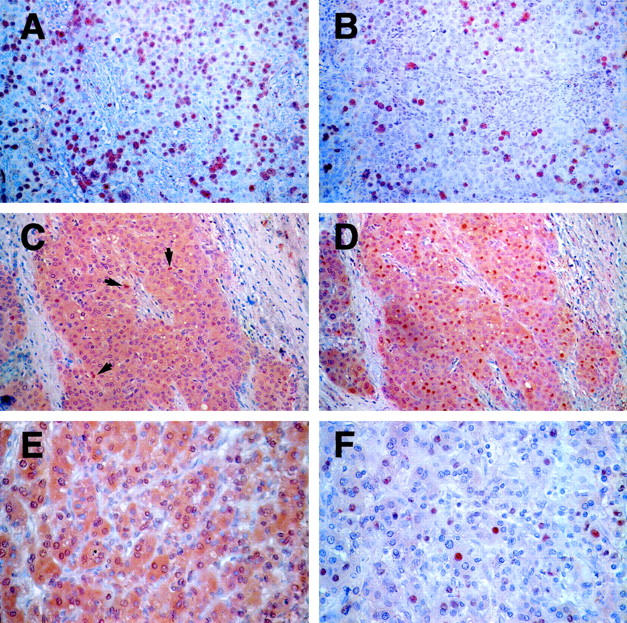
HIF-1α and HIF-2α expression in many common cancers. Immunostaining demonstrated heterogeneous patterns of HIF-1α and HIF-2α expression within subsets of tumor cells in many common cancers. Panels illustrated are from a breast carcinoma (A and B), a hepatocellular carcinoma (C, positive nuclei marked by arrows, and D), and a hypernephroma (E and F). Original magnifications, ×200 (A−D) and ×400 (E and F).
To examine these differences further, we analyzed tissue from a series of breast adenocarcinomas. In cases of breast carcinoma with large areas of intratumoral necrosis, strong expression of both proteins was observed at the hypoxic necrotic/viable tumor margin (Figure 4, A ▶ -D). However, protein was also demonstrable in other areas including the tumor/stromal margin (Figure 4, B ▶ -D). When serial sections of the same case were examined HIF-1α and HIF-2α, expression was generally, but not exclusively, found in overlapping regions of the tumor. HIF-1α protein distribution tended to be more restricted to the perinecrotic regions than HIF-2α protein.
Figure 4.
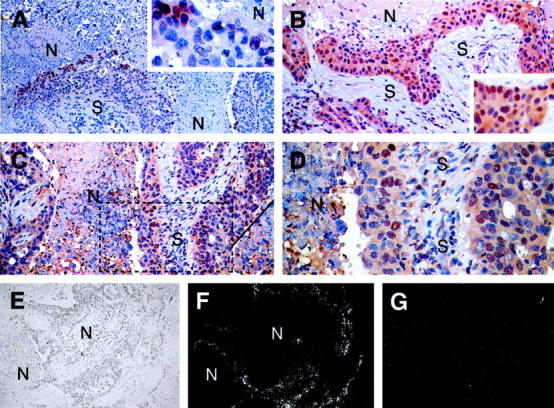
HIF-1α and HIF-2α expression in breast carcinoma. HIF-1α and HIF-2α protein expression was observed in breast tumor cells adjacent to areas of tumor necrosis. In some cases nuclear expression of HIF-1α was found exclusively at the necrotic/viable tumor margin (A; original magnification, ×100, inset; original magnification, ×200; N, necrosis; S, stroma), but in other cases throughout the tumor (B; original magnification, ×200, inset; original magnification, ×300). Nuclear expression of HIF-2α was observed throughout most tumors (C; original magnification, ×200, D; original magnification, ×400 higher power view of the marked section of the field illustrated in C) though like HIF-1α can also be perinecrotic. There were no fundamental differences between the tissue distributions of HIF-1α and HIF-2α. Serial sections of the case of breast carcinoma illustrated in A, C, and D above were hybridized with 35S-labeled VEGF antisense RNA probe (E, bright field illumination; F, dark field view; original magnifications, ×100) and 35S-labeled sense control probe (G, dark field view; original magnification, ×100). VEGF mRNA is strongly expressed around the large area of intratumoral necrosis (N) where HIF-1α and HIF-2α protein was present.
Hypoxia up-regulates expression of a variety of genes important in cancer biology, including VEGF. In situ hybridization experiments were performed to see whether the pattern of HIF α chain expression observed correlated with the distribution of VEGF mRNA. Signal was predominantly seen at necrotic/viable tumor margins (Figure 4F) ▶ , as has been previously reported. 28 These areas of high VEGF mRNA corresponded to the areas where HIF-1α and HIF-2α proteins were localized by immunostaining (Figure 4, A, C, and D) ▶ .
A number of the tumor blocks examined were from the margins of tumors and included adjacent dysplastic and morphologically normal tissue. We observed variable amounts of nuclear expression of HIF-1α and HIF-2α in both the dysplastic and normal cells, perhaps reflecting perturbations of oxygenation (or other influences) emanating from the tumor microenvironment.
Expression of HIF-2α in Tumor-Associated Macrophages
An unexpected finding in several tumor types was abundant cytoplasmic 190b immunoreactivity within a subset of cells morphologically identified as tumor-associated macrophages (TAM; Figure 5, A and B ▶ , in breast carcinoma). The cells expressing HIF-2α were confirmed as TAM by immunostaining serial sections with mAb to CD68 and HIF-2α and showing the colocalization of positive cells (Figure 5, C ▶ −F, in breast carcinoma). The assignment of this immunostaining as being due to HIF-2α was confirmed by showing identical patterns when serial sections were immunostained with 190b and the rabbit polyclonal PM8, which were raised against different HIF-2α sequences (Figure 5, G ▶ −I). Furthermore, in situ hybridization confirmed that in cases where there was strong immunostaining of stromal macrophages with 190b, HIF-2α mRNA signal was present over the same areas (Figure 6, A ▶ −C), further supporting our interpretation that macrophage immunoreactivity with 190b represents HIF-2α protein. In contrast, HIF-1α mRNA signal was distributed equally over stromal and tumor areas (Figure 6, D ▶ −F), consistent with previous in situ descriptions. 12
Figure 5.

HIF-2α expression in tumor macrophages. In a variety of tumor types two overlapping distribution patterns were observed with mAb 190b: staining within subsets of the tumor cells and also of stromal cells and macrophages, which was predominantly cytoplasmic. These features are illustrated in sections taken from breast carcinoma. In A, nuclear staining within tumor cells is indicated by arrows and cytoplasmic staining of stromal cells by arrowheads. A further example of stromal staining is shown in B. The immunoreactivity seen in tumor stroma with mAb 190b was investigated further. In serial sections of breast carcinoma colocalization of CD68, using mAb PGM1 (C and D) and mAb 190b (E and F) confirms that the cells staining with 190b in the stroma are macrophages. In serial sections of pancreatic carcinoma, cells shown to be macrophages by their expression of CD68 (detected using mAb PGM1, G) are also labeled by polyclonal antibody PM8 (H) and mAb 190b (I). Because PM8 and 190b recognize different HIF-2α epitopes, this confirms that these macrophages are indeed expressing HIF-2α. Original magnifications, ×200 (A, D, and F−I) and ×100 (B, C, and E)
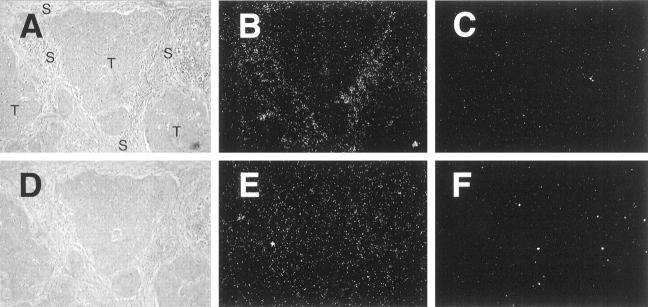
Figure 6.
HIF-2α mRNA expression by tumor stroma. Serial paraffin sections of a case of breast carcinoma demonstrated to have abundant stromal HIF-2α protein by immunostaining (Figure 5B) ▶ were hybridized with 35S-labeled HIF-2α antisense RNA probe (A, bright field view; B, dark field view), 35S-labeled sense control probe (C, dark field illumination). In contrast, HIF-1α mRNA signal was distributed equally over stromal and tumor areas (D bright field view; E dark field view), 35S-labeled sense control probe (F dark field illumination). Original magnifications, ×100.
The intensity of the TAM HIF-2α staining was usually greater than that within tumor cells and in some cases was observed in the absence of any intratumoral HIF-2α expression. For example, no tumor nuclear positivity was observed for either HIF-1α or HIF-2α in the 5 cases of lung carcinoma examined (Table 2) ▶ , but strong HIF-2α expression was seen within TAM in all these cases (not shown).
HIF-1α and HIF-2α Protein Expression in Normal Tissue
Normal tissues examined included skin, bladder, thymus, spleen, tonsil, lymph node, thyroid, adrenal, pancreas, salivary gland, liver, kidney, heart, esophagus, colon, lung, ovary, testis, uterus, placenta, umbilical cord, brain, prostate, and breast. Very little expression of either protein was found in any normal tissue, with the exception of bone marrow, where large numbers of cells morphologically identifiable as macrophages expressed HIF-2α.
The positive cells were confirmed to be macrophages by demonstrating colocalization of CD68- and HIF-2α-positive cells on immunostaining of serial sections (Figure 7, A and B) ▶ . The staining was predominantly within the cytoplasm with additional nuclear positivity in some macrophages. A more extensive examination was undertaken of HIF-2α expression in other populations of normal tissue macrophages; lung, liver, lymph node, spleen and brain. Some normal Kupffer cells were also found to express HIF-2α (not shown).
Figure 7.
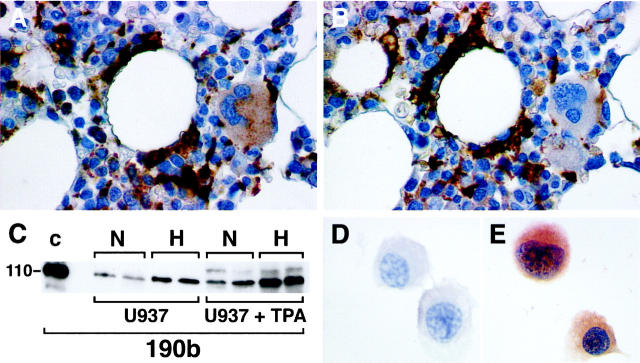
Expression of HIF-2α protein in normal bone marrow macrophages and U937 cell line. Normal bone marrow macrophages show immunoreactivity with mAb 190b. In serial paraffin sections of normal bone marrow trephine staining with CD68 (using mAb PGM1; A), and mAb 190b (B) colocalize in macrophages but not megakaryocytes. U937 cells were cultured with or without PMA (1.6 × 10− 8 M) and incubated in normoxia (N) or 0.1% hypoxia (H) for 4 hours. Whole cell extracts (75 μg) were prepared, separated by SDS-PAGE, transferred onto PVDF membrane, and analyzed with mAb 190b (C). For comparison a hypoxic cell extract of HIF-2α transfected HT1080 cells (50 μg) prepared after culture for 4 hours in 0.1% hypoxia was run in parallel (lane C). The dominant band detected in each case comigrated at the mobility predicted for HIF-2α. In both differentiated and undifferentiated U937 cells the band detected showed hypoxic induction. However, after differentiation the normoxic levels seen were comparable with those seen in hypoxia in undifferentiated cells. U937 cells were cultured with PMA (1.6 × 10− 8 M) to allow differentiation into macrophages, incubated in normoxia or placed in 0.1% hypoxia for 4 hours, and immunostained with mAbs to HIF-2α (190b) and CD11c (KB 90). HIF-2α expression was absent after normoxic culture (D) but detectable after hypoxic culture (E). CD11c expression, confirming macrophage differentiation, was unaffected by normoxic/hypoxic culture (not shown). Original magnifications, ×400 (A and B) and ×300 (D and E)
Expression of HIF-2α in a Macrophage Cell Line
Experiments with untreated and phorbol 12-myristate 13-acetate (PMA) differentiated U937 cells were undertaken to explore further the HIF-2α immunostaining observed in subsets of macrophages. After PMA treatment, cells adopted the morphological phenotype of macrophages and CD11c surface antigen expression. Immunoblots of whole cell extracts show that HIF-2α was expressed and hypoxically inducible in U937 cells. Greater expression was seen after differentiation into macrophages, such that normoxic levels were comparable with those seen in hypoxia in undifferentiated cells. Hypoxic induction was retained after differentiation, resulting in even higher levels of expression (Figure 7C) ▶ .
To examine HIF-2α localization and hypoxic inducibility by immunocytochemistry, PMA differentiated U937 cells were placed in parallel in normoxia or 0.1% hypoxia for 4 hours. HIF-2α was undetectable in normoxia (Figure 7D) ▶ . Hypoxic incubation resulted in both nuclear and strong cytoplasmic staining (Figure 7E) ▶ .
Discussion
A major finding in this survey of malignant and normal tissues was the up-regulation of HIF-α protein levels in cancers. The increase in expression was substantial, such that positive nuclear staining was readily detected in many cancers, whereas with one exception staining was negative in normal tissues. This pattern extended across a broad range of common human malignancies and was observed for both HIF-1α and HIF-2α.
Though initial studies of HIF-2α mRNA in normal fetal and adult tissues indicated a tissue-specific pattern of expression with a strong endothelial bias, 10 our previous study of tissue culture cells demonstrated widespread expression across a range of non-endothelial cell lines. 19 The current study extends these findings in demonstrating up-regulation of HIF-2α as well as HIF-1α in many types of tumor in the native environment.
Since HIF-α subunits are labile in oxygenated cells, we cannot be sure that these findings in fixed tissue will represent the position in vivo in every detail. Nevertheless, tumor and normal tissues were fixed and processed in the same way, providing security that the overall contrast in HIF-α expression represents an important biological difference between the two types of tissue. This finding raises important questions about both the mechanism of HIF-α up-regulation and its consequences for the tumor. Altered patterns of gene expression in cancer could arise both from genetic alterations in the tumor cells and from stimulation by an abnormal microenvironment within the tumor. It is likely that both processes contribute to the observed up-regulation of HIF-α subunits. These proteins are strongly induced by hypoxia, and tumor tissues are commonly hypoxic. Perinecrotic patterns of expression for HIF target genes or HRE-driven reporter genes in experimental studies of tumor xenografts have provided strong indirect evidence for induction of HIF activity by the hypoxic tumor microenvironment. 15
In the current work, we observed expression of both HIF-1α and HIF-2α in perinecrotic regions, providing direct support for microenvironmental mechanisms of HIF activation in diverse types of human tumor. Nevertheless, by no means all positively staining cells were in perinecrotic regions. Measurements of pO2 gradients and blood flow within tumors have shown complex and dynamic patterns. 29 Varying zones of tumor hypoxia could well exist outside the perinecrotic regions, possibly accounting for some of these signals. In the normal tissue at some tumor margins, expression of both HIF-1α and 2α was observed, which would be consistent with the presence of low pH and hypoxia. 30 However, large differences in the staining pattern between apparently similar tumors and incompletely overlapping patterns of expression for each HIF-α subunit suggest that mechanisms other than microenvironmental hypoxia contribute to the up-regulation of these proteins in cancers.
A clear example of genetic up-regulation of this system is seen in the hereditary cancer syndrome von Hippel-Lindau (VHL) disease. 18 The VHL tumor suppressor protein targets HIF-α subunits for ubiquitin-mediated proteolysis. Thus, in cells bearing inactivating mutations of both VHL alleles, HIF-α subunits are stabilized and accumulate at high levels irrespective of cellular hypoxia. VHL mutations are common in both inherited and sporadic renal cell carcinoma and consistent with this the frequency of tumor cell nuclear HIF-1α expression was higher in renal cell carcinoma than in other tumors and positive tumor cells were distributed homogeneously throughout the tumors. In contrast, VHL mutations are uncommon in other sporadic cancers and unlikely to account for HIF up-regulation.
Interestingly, recent studies in tissue culture monolayers have defined a variety of trophic stimuli that amplify the induction of HIF by hypoxia rather than activate the pathway constitutively. In different settings these include transformation with v-src, 31 stimulation with serum, insulin, or insulin-like growth factors, 32,33 and activation of p44/42 MAP kinase. 34,35 Though the precise mechanism of these interfaces with the hypoxia-sensitive pathway is still not clear, the findings suggests a more general influence of growth-promoting stimuli on HIF activity, which could be relevant to the observed up-regulation of HIF in many different types of cancer.
Though HIF-α subunits were commonly up-regulated in cancers, positive staining was not universal. Most tumors stained positively for both α subunits, though some were negative for one, and some were apparently negative for both. At present we have no certain explanation for these findings. Assessment of the effects of duration of fixation on pellets of hypoxic cells indicated that long periods of fixation substantially reduced antigen detection, so that failure to stain some tumors might have been artifactual. In surveys of tissue culture cells by immunoblot analysis, we have found that under maximal hypoxic stimulation, all cells had detectable levels of at least one subunit, though the levels were quite variable. 19 It is possible that relatively low levels of induced expression were still below the detection threshold in this immunohistochemical analysis, or that some tumors were relatively well oxygenated and that HIF was uninduced in the sections examined.
Our findings are, overall, rather similar to those of a survey of HIF-1α expression in human tumor samples that was published while this manuscript was being revised. 36 That analysis concerned expression patterns for HIF-1α rather than for both HIF-α subunits but also showed that up-regulation was a common, though not universal, finding in human tumors.
Despite some negative staining, the overall contrast in HIF-α expression between normal and tumor tissues suggests that the pathway could provide opportunities for diagnostic or therapeutic exploitation. Studies in wild-type and HIF-deficient cells have indicated a major role for HIF activation in the promotion of tumor angiogenesis, 15-17 so that assessment of HIF activity in human tumors might provide a guide to angiogenic potential, or to other phenotypes which have been associated with hypoxia, such as resistance to radiotherapy or chemotherapy. The differential activity between tumor and normal tissues also suggests that antagonism of the HIF pathway could provide a new approach to treatment. 14
An unexpected finding in this study was dense staining of a population of stromal cells both within and close to the tumors with the anti-HIF-2α mAb 190b. Morphological identification of these cells as macrophages was confirmed by CD68 expression. The specificity of the HIF-2α immunoreactivity was confirmed by similar positive staining with a polyclonal antiserum raised against a non-overlapping portion of the HIF-2α immunogen. In situ hybridization studies demonstrated high levels of HIF-2α mRNA in the regions of strong HIF-2α immunoreactivity suggesting that, in part, the mechanism of up-regulation might be at the mRNA level. The role played by hypoxia in this expression pattern is unclear, though some tumor-associated property is presumably responsible for the localization.
The interplay between tumor and stromal cells has been highlighted in a recent study of transgenic mice expressing green fluorescent protein (GFP) under the control of the human VEGF promoter. 37 Implanted tumors were able to induce the transgene, strongly indicating the potential for the tumor environment to induce this HIF-responsive promoter in stromal cells of host origin. The potential importance of gene expression patterns in tumor-associated stromal cells is supported by the association of high levels of macrophage infiltration with high vascular grade and reduced relapse-free survival in breast cancer. 38
Though positive cells were much less frequent outside the tumor environment similar HIF-2α expression was observed in some Kupffer cells and some normal bone marrow macrophages, suggesting that it is a feature of some stages of differentiation, or activation, in this cell lineage. Within normal bone marrow areas of hypoxia may have a role in determining stem cell differentiation. 39 Comparison of the levels of HIF-2α protein detected in U937 cells before and after treatment with PMA supports this concept. Interestingly, HIF-2α staining patterns in TAM, bone marrow macrophages, and PMA-treated U937 cells were unusual in being distributed more uniformly throughout the cell, as has been reported in cultured cells lines treated with proteasomal inhibitors. A recent report has described the blocking of proteasomal degradation of HIF-1α by a macrophage-derived peptide. 40 Whether a similar mechanism could account for the current findings is unclear.
In conclusion, we have demonstrated that HIF-1α and HIF-2α can be detected at the protein level in routinely processed material from a subset of most, if not all, common types of cancers. Tumor cells showed nuclear expression of one or both molecules, with or without cytoplasmic staining. In addition, expression of HIF-2α has been shown in tumor-associated macrophages. In future studies it will be of interest to examine the relationship of these expression patterns to tumor prognosis and response to therapy.
Footnotes
Address reprint requests to Prof. A .L. Harris, ICRF Molecular Oncology Laboratory and Angiogenesis Group, Institute of Molecular Medicine, John Radcliffe Hospital, Oxford, OX3 9DU, UK. E-mail: aharris.lab@icrf.icnet.uk.
Supported by Imperial Cancer Research Fund, the Wellcome Trust, and the Medical Research Council (United Kingdom).
References
- 1.Bunn HF, Poyton RO: Oxygen sensing and molecular adaptation to hypoxia. Physiol Rev 1996, 76:839-885 [DOI] [PubMed] [Google Scholar]
- 2.Hockel M, Schlenger K, Hockel S, Aral B, Schaffer U, Vaupel P: Tumor hypoxia in pelvic recurrences of cervical cancer. Int J Cancer 1998, 79:365-369 [DOI] [PubMed] [Google Scholar]
- 3.Stadler P, Becker A, Feldmann HJ, Hansgen G, Dunst J, Wurschmidt F, Molls M: Influence of the hypoxic subvolume on the survival of patients with head and neck cancer. Int J Radiat Oncol Biol Phys 1999, 44:749-754 [DOI] [PubMed] [Google Scholar]
- 4.Maxwell PH, Pugh CW, Ratcliffe PJ: Inducible operation of the erythropoietin-3′ enhancer in multiple cell-lines: evidence for a widespread oxygen-sensing mechanism. Proc Natl Acad Sci USA 1993, 90:2423-2427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ratcliffe PJ, Ebert BL, Firth JD, Gleadle JM, Maxwell PH, Nagao M, Orourke JF, Pugh CW, Wood SM: Oxygen regulated gene expression: erythropoietin as a model system. Kidney Int 1997, 51:514-526 [DOI] [PubMed] [Google Scholar]
- 6.Wang GL, Jiang BH, Rue EA, Semenza GL: Hypoxia-inducible factor-1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O-2 tension. Proc Natl Acad Sci USA 1995, 92:5510-5514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crews ST: Control of cell lineage-specific development and transcription by bHLH-PAS proteins. Genes Dev 1998, 12:607-620 [DOI] [PubMed] [Google Scholar]
- 8.Iyer NV, Kotch LE, Agani F, Leung SW, Laughner E, Wenger RH, Gassmann M, Gearhart JD, Lawler AM, Yu AY, Semenza GL: Cellular and developmental control of O-2 homeostasis by hypoxia-inducible factor 1 alpha. Genes Dev 1998, 12:149-162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tian H, Hammer RE, Matsumoto AM, Russell DW, McKnight SL: The hypoxia-responsive transcription factor EPAS1 is essential for catecholamine homeostasis and protection against heart failure during embryonic development. Genes Dev 1998, 12:3320-3324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tian H, McKnight SL, Russell DW: Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev 1997, 11:72-82 [DOI] [PubMed] [Google Scholar]
- 11.Ema M, Taya S, Yokotani N, Sogawa K, Matsuda Y, FujiiKuriyama Y: A novel bHLH-PAS factor with close sequence similarity to hypoxia-inducible factor 1 alpha regulates the VEGF expression and is potentially involved in lung and vascular development. Proc Natl Acad Sci USA 1997, 94:4273-4278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flamme I, Frohlich T, vonReutern M, Kappel A, Damert A, Risau W: HRF, a putative basic helix-loop-helix-PAS-domain transcription factor is closely related to hypoxia-inducible factor-1 alpha and developmentally expressed in blood vessels. Mech Dev 1997, 63:51–60 [DOI] [PubMed]
- 13.Gu YZ, Moran SM, Hogenesch JB, Wartman L, Bradfield CA: Molecular characterization and chromosomal localization of a third alpha-class hypoxia inducible factor subunit, HIF3 alpha. Gene Expr 1998, 7:205-213 [PMC free article] [PubMed] [Google Scholar]
- 14.Dachs GU, Patterson AV, Firth JD, Ratcliffe PJ, Townsend KMS, Stratford IJ, Harris AL: Targeting gene expression to hypoxic tumor cells. Nat Med 1997, 3:515-520 [DOI] [PubMed] [Google Scholar]
- 15.Maxwell PH, Dachs GU, Gleadle JM, Nicholls LG, Harris AL, Stratford IJ, Hankinson O, Pugh CW, Ratcliffe PJ: Hypoxia-inducible factor-1 modulates gene expression in solid tumors and influences both angiogenesis and tumor growth. Proc Natl Acad Sci USA 1997, 94:8104-8109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ryan HE, Lo J, Johnson RS: HIF-1 alpha is required for solid tumor formation and embryonic vascularization. EMBO J 1998, 17:3005-3015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carmeliet P, Dor Y, Herbert JM, Fukumura D, Brusselmans K, Dewerchin M, Neeman M, Bono F, Abramovitch R, Maxwell P, Koch CJ, Ratcliffe P, Moons L, Jain RK, Collen D, Keshert E: Role of HIF-1 alpha in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature 1998, 394:485-490 [DOI] [PubMed] [Google Scholar]
- 18.Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, Wykoff CC, Pugh CW, Maher ER, Ratcliffe PJ: The tumour suppressor protein VHL targets hypoxia inducible factors for oxygen dependent proteolysis. Nature 1999, 399:271-275 [DOI] [PubMed] [Google Scholar]
- 19.Wiesener MS, Turley H, Allen WE, Willam C, Eckardt KU, Talks KL, Wood SM, Gatter KC, Harris AL, Pugh CW, Ratcliffe PJ, Maxwell PH: Induction of endothelial PAS domain protein-1 by hypoxia: characterization and comparison with hypoxia-inducible factor-1 alpha. Blood 1998, 92:2260-2268 [PubMed] [Google Scholar]
- 20.Huang LE, Arany Z, Livingston DM, Bunn HF: Activation of hypoxia-inducible transcription factor depends primarily upon redox-sensitive stabilization of its alpha subunit. J Biol Chem 1996, 271:32253-32259 [DOI] [PubMed] [Google Scholar]
- 21.Huang LE, Gu J, Schau M, Bunn HF: Regulation of hypoxia-inducible factor 1 alpha is mediated by an O-2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc Natl Acad Sci USA 1998, 95:7987-7992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salceda S, Caro J: Hypoxia-inducible factor 1 alpha (HIF-1 alpha) protein is rapidly degraded by the ubiquitin-proteasome system under normoxic conditions - its stabilization by hypoxia depends on redox-induced changes. J Biol Chem 1997, 272:22642-22647 [DOI] [PubMed] [Google Scholar]
- 23.Wenger RH, Gassmann M: Oxygen(es) and the hypoxia-inducible factor-1. Biol Chem 1997, 378:609-616 [PubMed] [Google Scholar]
- 24.Wiener CM, Booth G, Semenza GL: In vivo expression of mRNAs encoding hypoxia-inducible factor 1. Biochem Biophys Res Commun 1996, 225:485-488 [DOI] [PubMed] [Google Scholar]
- 25.Yu AY, Frid MG, Shimoda LA, Wiener CM, Stenmark K, Semenza GL: Temporal, spatial, and oxygen-regulated expression of hypoxia- inducible factor-1 in the lung. Am J Physiol 1998, 275:L818-L826 [DOI] [PubMed] [Google Scholar]
- 26.Ozaki H, Yu AY, Della N, Ozaki K, Luna JD, Yamada H, Hackett SF, Okamoto N, Zack DJ, Semenza GL, Campochiaro PA: Hypoxia inducible factor-1 alpha is increased in ischemic retina: temporal and spatial correlation with VEGF expression. Invest Ophthalmol Vis Sci 1999, 40:182-189 [PubMed] [Google Scholar]
- 27.Zhang HT, Craft P, Scott PAE, Ziche M, Weich HA, Harris AL, Bicknell R: Enhancement of tumor-growth and vascular density by transfection of vascular endothelial-cell growth-factor into Mcf-7 human breast-carcinoma cells. J Natl Cancer Inst 1995, 87:213-219 [DOI] [PubMed] [Google Scholar]
- 28.Brown LF, Berse B, Jackman RW, Tognazzi K, Guidi AJ, Dvorak HF, Senger DR, Connolly JL, Schnitt SJ: Expression of vascular permeability factor (vascular endothelial growth factor) and its receptors in breast cancer. Hum Pathol 1995, 26:86-91 [DOI] [PubMed] [Google Scholar]
- 29.Helmlinger G, Yuan F, Dellian M, Jain RK: Interstitial pH and pO(2) gradients in solid tumors in vivo: high-resolution measurements reveal a lack of correlation. Nat Med 1997, 3:177-182 [DOI] [PubMed] [Google Scholar]
- 30.Gatenby RA, Gawlinski ET: A reaction-diffusion model of cancer invasion. Cancer Res 1996, 56:5745-5753 [PubMed] [Google Scholar]
- 31.Jiang BH, Agani F, Passaniti A, Semenza GL: V-SRC induces expression of hypoxia-inducible factor 1 (HIF-1) and transcription of genes encoding vascular endothelial growth factor and enolase 1: involvement of HIF-1 in tumor progression. Cancer Res 1997, 57:5328-5335 [PubMed] [Google Scholar]
- 32.Punglia RS, Lu M, Hsu J, Kuroki M, Tolentino MJ, Keough K, Levy AP, Levy NS, Goldberg MA, Damato RJ, Adamis AP: Regulation of vascular endothelial growth factor expression by insulin-like growth factor I. Diabetes 1997, 46:1619-1626 [DOI] [PubMed] [Google Scholar]
- 33.Feldser D, Agani F, Iyer NV, Pak B, Ferreira G, Semenza GL: Reciprocal positive regulation of hypoxia-inducible factor 1 alpha and insulin-like growth factor 2. Cancer Res 1999, 59:3915-3918 [PubMed] [Google Scholar]
- 34.Conrad PW, Freeman TL, BeitnerJohnson D, Millhorn DE: EPAS1 trans-activation during hypoxia requires p42/p44 MAPK. J Biol Chem 1999, 274:33709-33713 [DOI] [PubMed] [Google Scholar]
- 35.Richard DE, Berra E, Gothie E, Roux D, Pouyssegur J: p42/p44 mitogen-activated protein kinases phosphorylate hypoxia-inducible factor 1 alpha (HIF-1 alpha) and enhance the transcriptional activity of HIF-1. J Biol Chem 1999, 274:32631-32637 [DOI] [PubMed] [Google Scholar]
- 36.Zhong H, De Marzo AM, Laughner E, Lim M, Hilton DA, Zagzag D, Byechler P, Isaacs WB, Semenza GL, Simons JW: Overexpression of hypoxia-inducible factor 1 alpha in common human cancers and their metastases. Cancer Res 1999, 59:5830-5835 [PubMed] [Google Scholar]
- 37.Fukumura D, Xavier R, Sugiura T, Chen Y, Park EC, Lu NF, Selig M, Nielsen G, Taksir T, Jain RK, Seed B: Tumor induction of VEGF promoter activity in stromal cells. Cell 1998, 94:715-725 [DOI] [PubMed] [Google Scholar]
- 38.Leek RD, Lewis CE, Whitehouse R, Greenall M, Clarke J, Harris AL: Association of macrophage infiltration with angiogenesis and prognosis in invasive breast carcinoma. Cancer Res 1996, 56:4625-4629 [PubMed] [Google Scholar]
- 39.Cipolleschi MG, Dellosbarba P, Olivotto M: The role of hypoxia in the maintenance of hematopoietic stem-cells. Blood 1993, 82:2031-2037 [PubMed] [Google Scholar]
- 40.Li J, Post M, Volk R, Youhe G, Li M, Metais C, Sato K, Tsai J, Aird W, Rosenberg RD, Hampton TG, Li J, Sellke F, Carmeliet P, Simons M: PR39, a peptide regulator of angiogenesis. Nat Med 2000, 6:49-55 [DOI] [PubMed] [Google Scholar]
- 41.Falini B, Flenghi L, Pileri S, Gambacorta M, Bigerna B, Durkop H, Eitelbach F, Thiele J, Pacini R, Cavaliere A, Martelli M, Cardarelli N, Sabattini E, Poggi S, Stein H: Pg-M1: a new monoclonal-antibody directed against a fixative-resistant epitope on the macrophage-restricted form of the Cd68 molecule. Am J Pathol 1993, 142:1359-1372 [PMC free article] [PubMed] [Google Scholar]
- 42.Master PS, Richards SJ, Kendall J, Roberts BE, Scott CS: Diagnostic application of monoclonal-antibody Kb90 (Cd11c) in acute myeloid-leukemia. Blut 1989, 59:221-225 [DOI] [PubMed] [Google Scholar]