Abstract
Proapoptotic Bcl-2 family members activate cell death by neutralizing their anti-apoptotic relatives, which in turn maintain cell viability by regulating the activation of the cell death effectors, the caspases. Bim belongs to a distinct subgroup of proapoptotic proteins that only resemble other Bcl-2 family members within the short BH3 domain. Gene targeting experiments in mice have shown that Bim is essential for the execution of some but not all apoptotic stimuli, for hematopoietic cell homeostasis, and as a barrier against autoimmunity. There are three Bim isoforms, BimS, BimL, and BimEL, which have different proapoptotic potencies due at least in part to differences in interaction with the dynein motor complex. The expression pattern of Bim was investigated by immunohistochemical staining, immunoprecipitation followed by Western blotting, and in situ hybridization. Bim was found in hematopoietic, epithelial, neuronal, and germ cells. BimL and BimEL were coexpressed at similar levels in many cell types, but BimS was not detected. Microscopic examination revealed a punctate pattern of BimL and BimEL immunostaining, indicating association with cytoplasmic structures. These results are discussed in the context of the phenotype of Bim-deficient mice and the post-translational regulation of Bim’s pro-apoptotic activity.
Apoptotic cell death plays a critical role in tissue molding during embryogenesis, as a regulator of cell turnover, and as a host defense mechanism against pathogens. 1,2 The failure of normally doomed cells to undergo apoptosis can contribute to cellular transformation 3 or autoimmunity, 4,5 whereas the premature demise of long-lived cells has been implicated in the pathology of degenerative disorders. 6
Apoptosis is an evolutionarily conserved process that is characterized morphologically by cell shrinkage, plasma membrane blebbing, and chromatin condensation. 7 Cell death can be induced by a variety of physiological as well as experimentally applied stimuli that activate distinct signaling pathways. These pathways ultimately converge on a common effector machinery that is driven by a family of cysteine proteases (caspases) that cleave substrates after aspartate residues. 8,9 Caspases exist in living cells as zymogens with low enzymatic activity. They need to be cleaved at aspartate residues to generate fragments of ∼20 kd and ∼10 kd that are assembled into the fully active tetrameric (p202p102) enzyme. 9 Adaptor proteins, such as mammalian Apaf-1 or FADD (also called MORT1) and Caenorhabditis elegans CED-4, promote aggregation and self-processing of so-called initiator caspases. 10,11 These in turn proteolytically activate so-called effector caspases, giving rise to the proteolytic avalanche that culminates in the degradation of vital cellular constituents and cell collapse.
The Bcl-2 protein family regulates pathways to apoptosis that are activated by growth factor deprivation or many forms of intracellular damage but play little role in apoptosis induced by tumor necrosis factor receptor family members, at least in lymphocytes. 12,13 Pro-survival Bcl-2 proteins, including mammalian Bcl-2, Bcl-xL, Bcl-w, A1/Bfl1, Mcl-1, Boo/Diva, as well as C. elegans CED-9, inhibit apoptosis by blocking the activity of adaptor proteins, such as mammalian Apaf-1 or C. elegans CED-4. 12 The proapoptotic members of the Bcl-2 family can antagonize Bcl-2 and its homologs. Based on their structure, these proapoptotic proteins can be further subdivided into two groups. One includes mammalian Bax, Bcl-xS, Bak, and Bok/Mtd, which share two or three regions of homology (BH regions) with Bcl-2, whereas the other subgroup, including mammalian Bad, Bik/Nbk, Bid, Harakiri/DP5, Blk, and Bim/Bod, 12 as well as C. elegans EGL-1, 14 only have similarities in the short BH3 domain. The proapoptotic Bcl-2 family members bind via their BH3 domain to Bcl-2 or its functional homologs, and it is believed that this initiates apoptosis by unleashing Apaf-1/CED-4-like adaptors that can then activate certain procaspases (eg, mammalian procaspase-9 or C. elegans CED-3). 12
Bim was originally cloned as a Bcl-2-interacting protein by screening a λ phage expression library constructed from a mouse thymic lymphoma. 15 Alternative splicing generates three Bim isoforms, BimS, BimL, and BimEL, which can all neutralize the activity of pro-survival Bcl-2-like proteins through their BH3 domain. However, the three isoforms vary considerably in their pro-apoptotic activity. This is partly due to sequestration of the less potent forms, BimL and BimEL, but not BimS, to cytoskeletal structures via association with dynein light chain LC8. 16
Gene targeting experiments in mice have revealed the essential roles of Bim. 17 Most bim−/− embryos died before E9.5, demonstrating that Bim plays a critical role in development. The remaining bim−/− mice were live-born and had an outwardly normal appearance. They had, however, two- to fivefold more lymphocytes, macrophages, and granulocytes, probably because of their increased resistance to cytokine withdrawal-induced apoptosis. This indicates that Bim is required for hematopoietic cell homeostasis. Cell survival experiments with lymphocytes showed that Bim is essential for death after some but not all apoptotic stimuli that can be blocked by Bcl-2. By 1 year of age most live-born bim−/− mice accumulated huge excesses (30–200-fold) of immunoglobulin (Ig)-secreting plasma cells and succumbed to fatal autoimmune glomerulonephritis, demonstrating that Bim imposes an important barrier against autoimmunity.
Initial studies demonstrated that bim mRNA is expressed at low levels in several transformed B- and T-lymphoid cell lines. To further investigate the physiological roles of Bim, we have generated a panel of monoclonal antibodies (mAbs) that specifically recognize distinct Bim isoforms. Here we describe an analysis of the expression of Bim protein and bim mRNA in transformed cell lines and normal mouse tissues. BimL and BimEL were associated with cytoplasmic structures in lymphocytes, myeloid cells, epithelial cells, neuronal cells, and germ cells, but BimS could not be detected in any cell type. These results have implications for cell death control in development and tissue homeostasis.
Materials and Methods
Experimental Animals
All experiments with animals were performed according to the guidelines of the Royal Melbourne Hospital Research Foundation Animal Ethics Committee. Wistar rats and C57BL/6 mice were obtained from our institute’s breeding facility at Kew (Victoria, Australia). The Bim-deficient knockout (bim−/−) mice have previously been described. 17
Expression Constructs and Protein Purification
Expression vectors for EE epitope-tagged mouse BimS, BimL, BimEL, BimLΔBH3, Bad, and Bax have previously been described. 15,18 The other expression vectors were constructed by cloning cDNAs for human BimEL, mouse BimEL (lacking the internal splice donor and acceptor sites that allow production of BimL), mouse BimL (aa 2–72 [KO]; gift of P. Bouillet), Bcl-2, Bcl-w, and Bok (gift of Aaron Hsueh) into pEF PGKpuro or pEF PGKhygro vectors incorporating the N-terminal epitope tags EE (EYMPME) 19 or HA (YPYDVPDYA). 20 Full-length mouse bimL or bimS cDNAs were cloned into pGEX (128/128) 21 or pQE-9 (Qiagen), respectively, for the production of BimL (GST FLAG BimL) or BimS (His6BimS) proteins in the bacterial strains BL21 (DE3) pLysS or SG13009/[pREP4]. The proteins were produced according to the manufacturers’ protocols (Amersham, Pharmacia, or Qiagen).
Immunization and Hybridoma Fusion
Wistar rats were initially immunized by subcutaneous (s.c.) injection with 100 μg of purified recombinant protein dissolved in complete Freund’s adjuvant (Difco, Detroit, MI). Two subsequent boosts of the immunogen, resuspended in incomplete Freund’s adjuvant (Difco), were injected s.c. 3 and 6 weeks later. A final boost with protein dissolved in phosphate-buffered saline (PBS) was given i.v. and i.p. 4 weeks later. Three days later, hybridomas were generated by fusing spleen cells from immunized rats with the SP2/0 myeloma cell line as previously described. 22,23 Hybridomas producing monoclonal antibodies to Bim were identified and their isotype determined by a screening strategy that we have previously described. 23 Clones of the interleukin-3 (IL-3)-dependent mouse myeloid cell line FDC-P1 stably expressing Bcl-2 alone, or Bcl-2 plus mouse BimL or BimS, were mixed at a 1:1 ratio, fixed in 1% paraformaldehyde/PBS, permeabilized with 0.3% saponin (Sigma), and stained with hybridoma supernatants. Bound antibodies were revealed with fluorescein isothiocyanate-conjugated goat anti-rat Ig antibodies (Southern Biotechnology) and analyzed in a FACScan analyzer (Becton Dickinson). A single peak of background immunofluorescence indicated that a particular antibody did not recognize Bim, whereas a single peak of high intensity indicated binding to a molecule other than Bim, present in FDC-P1 cells. A double peak histogram, one with background fluorescence intensity and the other with high intensity, identified monoclonal antibodies specific to Bim. 23 Hybridomas producing antibodies to Bim were cloned twice and then adapted for growth in low-serum medium. For production of large amounts of antibodies, hybridomas were cultured for several weeks in the miniPERM classic 12.5-kd production and nutrient module (Heraeus). Antibodies were purified on a protein G-Sepharose column (Pharmacia) according to the manufacturer’s protocols.
Cell Lines and Tissue Culture
The cell lines used for analysis of Bim expression are indicated in Table 1 ▶ . Details of those cell lines not cited in previous studies from this laboratory 3,24-29 are available from the authors. These cells were cultured in the high-glucose version of Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum, 50 μmol/L 2-mercaptoethanol, 13 μmol/L folic acid, and 100 μmol/L l-asparagine or were grown in Dulbecco’s modified Eagle’s medium or RPMI medium with 10% fetal calf serum only. FDC-P1 and BAF-3 cells or their derivatives were cultured in the presence of IL-3 (1000 U/ml) produced by X63/0 hybridoma cells stably transfected with an IL-3 expression construct. 30 FDC-P1 clones stably expressing human Bcl-2, human Bcl-2 plus EE-mouse BimL, or human Bcl-2 plus EE-mouse BimS have been described previously. 15,18 Granulocyte differentiation of 34.6Myl cells was enhanced by the addition of 1.5% dimethyl sulfoxide. 25 Liposome-mediated transfection (Lipofectamine; Gibco BRL) of 293T cells was performed as previously described. 31
Table 1.
Summary of IP/Western Analysis of Bim Protein Expression Levels in Cell Lines
| Cell line | Origin | Species | BimL | BimEL |
|---|---|---|---|---|
| ALB 8.1 | Pre-B lymphoma | Mouse | + | + |
| 70Z/3 | B lymphoma | Mouse | + | − |
| CH1 | B lymphoma | Mouse | + | + |
| Sp2/0 | Plasmacytoma | Mouse | ++ | ++ |
| NS-1 | Plasmacytoma | Mouse | + | + |
| K052 DA.20 | T lymphoma | Mouse | + | + |
| WEHI 7.1 | T lymphoma | Mouse | + | + |
| WEHI 703* | T lymphoma | Mouse | + | + |
| EL-4.1 | T lymphoma | Mouse | + | + |
| B6.2.16.BW2† | T lymphoma | Mouse | ++ | + |
| Jurkat | T lymphoma | Human | ++ | ++ |
| P388D1 | Macrophage | Mouse | ++ | ++ |
| RAW 264.7 | Macrophage | Mouse | − | − |
| J774 | Macrophage | Mouse | + | − |
| 34.6Myl | Granulocyte | Mouse | + | + |
| P-815X-2.1 | Mastocytoma | Mouse | + | + |
| 416B MEG‡ | Megakaryocyte | Mouse | + | + |
| 416B | Myeloid | Mouse | + | + |
| BAF-3 | Myeloid | Mouse | + | +/− |
| FDC-P1 | Myeloid | Mouse | + | + |
| F4N | Erythroleukemia | Mouse | − | − |
| TS5 | Erythroleukemia | Mouse | − | − |
| DP16 | Erythroleukemia | Mouse | − | − |
| C2C12 | Muscle (myoblasts) | Mouse | − | ND |
| L6 | Muscle (myoblasts) | Rat | − | ND |
| NIH/3T3 | Fibroblast | Mouse | − | − |
| Rat1 | Fibroblast | Rat | − | − |
| L-929 | Fibroblast | Mouse | − | − |
| WEHI 11 | Fibrosarcoma | Mouse | − | − |
| S17 | Stromal | Mouse | − | − |
| MDCK | Kidney | Dog | − | − |
| MCF-7 | Breast carcinoma (epithelial) | Human | ++ | ++ |
| 293T | Embryonic kidney | Human | + | + |
| HK-2 | Kidney proximal | Human | − | − |
| G-401 | Wilm’s tumor (kidney) | Human | − | − |
| TCMK-1 | Kidney | Mouse | − | − |
| Cosm6 | Kidney (fibroblast) | Monkey | + | − |
| MH134 | Hepatoma | Mouse | + | − |
| HepG2 | Liver (epithelial) | Human | + | + |
| SW480 | Colon carcinoma | Human | − | − |
| EB-3 | Colon carcinoma | Human | − | − |
| HeLa | Cervical carcinoma | Human | − | − |
*Activated N-Ras transgenic mouse thymic T-cell lymphoma.
†Mouse T hybridoma expressing the same receptor as anti-HY TCR transgenic cells.
‡Megakaryocyte differentiation induced by GATA-1 expression.
Immunoprecipitation and Western Blotting
Cell lines or transfected 293T cells were harvested, washed twice in cold PBS, and lysed in lysis buffer (20 mmol/L Tris/HCl (pH 8.0); 125 mmol/L NaCl; 1 mmol/L EGTA; 1% Triton X-100; 10% glycerol; 0.5 μg/ml Pefabloc; 1 μg/ml of each of leupeptin, aprotinin, soybean trypsin inhibitor, and pepstatin; 5 mmol/L NaF; and 2 mmol/L Na3VO4; all reagents from Sigma or Roche Diagnostics). For the preparation of tissue lysates, organs were excised, washed in PBS, and immediately frozen in isopentane on dry ice and were later homogenized at 4°C in lysis buffer as detailed elsewhere. 32
Immunoprecipitation was performed according to previously published protocols. 33 Briefly, cell lysates (from 10 7 cells) or tissue lysates (1.5–3 mg total protein) were precleared by an incubation with a control mAb and protein G-Sepharose before immunoprecipitating with anti-Bim 5E5 or 14A8 mAbs plus protein G-Sepharose for 1.5 hours. After extensive washing (six times in lysis buffer), the immunoprecipitated material was eluted by boiling in sodium dodecyl sulfate-polyacrylamide gel loading buffer, size-fractionated on polyacrylamide gels (Novex), and transferred to nitrocellulose membranes by electroblotting. Nonspecific binding of antibodies to membranes was blocked by incubation overnight in 5% skimmed milk, 1% casein, and 0.05% Tween-20. The membranes were then probed with 5E5 or 14A8 anti-Bim mAbs (1–2 μg/ml), followed by either biotinylated mouse anti-rat IgG2b mAb and horseradish peroxidase (HRP)-conjugated streptavidin (Jackson ImmunoResearch) or HRP-conjugated mouse anti-rat IgG2a mAb (SeroTec). Bound antibodies were visualized by enhanced chemiluminescence (ECL) (Amersham Pharmacia). To control for the concentration and integrity of proteins in the tissue lysates, identical blots were probed with mouse anti-HSP70 mAb N6 (a gift from Dr R. Anderson, Peter MacCallum Cancer Research Institute, Melbourne, Australia), followed by HRP-conjugated sheep anti-mouse Ig antibodies (Silenius) and detection by ECL (Amersham Pharmacia).
Immunohistochemistry
Mouse tissues or cell lines were fixed in Histochoice fixative (Amresco, Solon, OH) and processed for paraffin embedding, cell lines were resuspended in 1% low-melting-point agarose before paraffin embedding. Immunohistochemistry was performed according to a modified protocol detailed elsewhere. 34 Paraffin-embedded sections were deparaffinized and gradually rehydrated. Aldehydes were quenched by incubation in 0.2 mol/L glycine for 30 minutes, and sections were treated with 0.3% hydrogen peroxide and 10% methanol to block endogenous peroxidase activity. Nonspecific staining was prevented by incubation in blocking reagent (TSA indirect signal amplification; NEN, Boston, MA), and cells were then permeabilized by incubation in 0.2% Triton X-100 containing 1% bovine serum albumin. Sections were incubated overnight at 4°C with anti-Bim mAb 4E4, 5E5, 9F5, or 14A8, or with one of the following isotype-matched control antibodies: rat IgG2a/κ (clone B35–95), rat IgG2b/κ (clone A95–1 or Ter119), or rat IgG1/κ (clone R3–34) (Pharmingen). After they were washed in PBS containing 0.2% Triton X-100, tissues were incubated with either biotinylated mouse anti-rat Igκ mAb Mar 18.5 or biotinylated mouse anti-rat IgG isotype-specific antibodies (anti-rat IgG1: RG11/39.4; anti-rat IgG2a: RG7/1.30; or anti-rat IgG2b: RG7/11.1). After washing, sections were incubated with HRP-streptavidin (NEN), and the signal was amplified with biotinyl tyramide according to the manufacturer’s instructions (NEN). This was followed by a second incubation with HRP-streptavidin. Finally, sections were stained with diaminobenzidene (Sigma) in 0.07% hydrogen peroxide, counterstained with hematoxylin, and dehydrated in graded concentrations of alcohol and histolene before mounting in DPX (BDH, Poole, UK).
In Situ Hybridization
The full-length mouse bimL cDNA was subcloned into pBSIISK+ (Stratagene). Antisense RNA probes were prepared by in vitro transcription of the pBSIISK+ BimL fragment, using T3 RNA polymerase with digoxigenin-UTP (Roche Diagnostics). Control sense RNA probes were prepared in a similar manner, using T7 RNA polymerase. Standard in situ hybridization was performed as described 35 on paraffin-embedded mouse tissue sections that had been fixed in 4% paraformaldehyde.
Results
Characterization of Monoclonal Antibodies to Bim
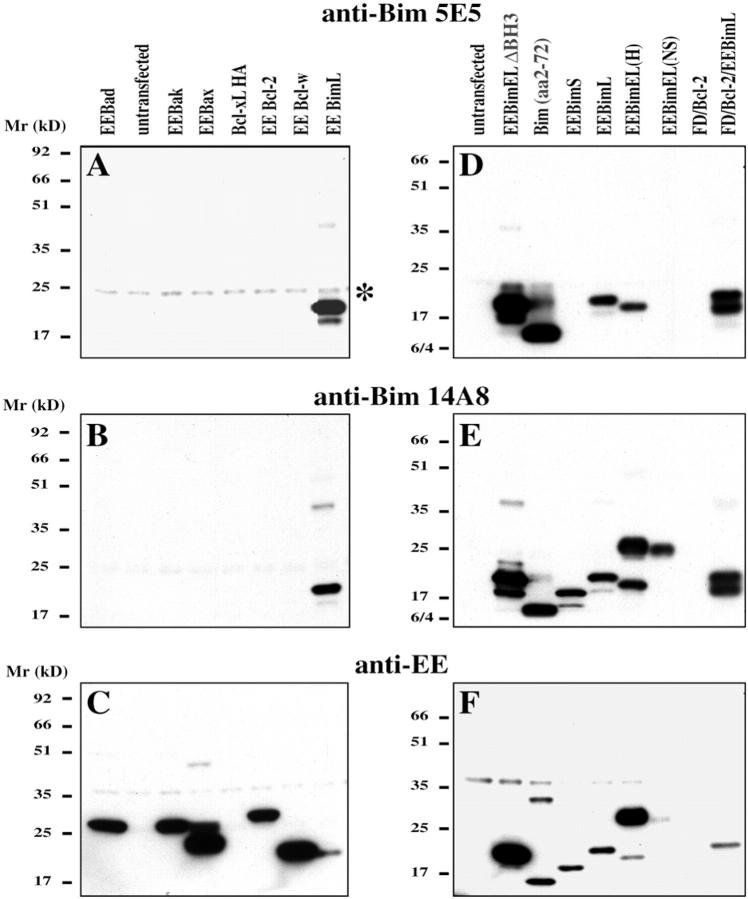
Immunization of rats with mouse BimL or BimS proteins followed by hybridoma fusion yielded several Bim-specific mAbs. 23 Based on our initial studies we selected two mAbs, 5E5 and 14A8, for further experiments. Both recognize mouse and human BimL and were efficacious in many applications, including immunofluorescence staining, 23 Western Blotting (Figure 1, A–F) ▶ , and immunoprecipitation (Figures 2G, 3, and 4) ▶ ▶ ▶ . Immunoblotting of lysates from cells transfected with expression constructs encoding different Bcl-2 family members demonstrated that the two mAbs were highly specific for Bim. The mAbs bound to mouse and human Bim but did not detect Bcl-2, Bcl-w, Bcl-xL, Bak, Bax, or the BH3-only protein Bad (Figure 1, A–C) ▶ . The only cross-reactivity observed was that mAb 5E5 bound to an unknown 26-kd protein present in some kidney cell lines (eg, 293T) but not in other cell types (Figure 1A) ▶ .
Figure 1.
A–C: The specificity of the anti-Bim mAbs 5E5 and 14A8 was determined by immunoblotting of lysates from 293T cells transfected with various EE-tagged Bcl-2 family members (Bad, Bak, Bax, Bcl-2, Bcl-w) or HA-tagged Bcl-xL, using the anti-Bim mAbs 5E5 (A) and 14A8 (B), or as a control anti-EE mAb (C) or anti-HA mAb (not shown). Goat anti-rat Ig or goat anti-mouse Ig antibodies conjugated to HRP were used as secondary reagents followed by ECL detection. *Cross-reacting band in 293T cell detected by 5E5 mAb. D–F: The epitope within Bim recognized by the anti-Bim mAbs was determined by Western blotting of lysates of 293T cells transfected with EE-tagged mutant forms of mouse and human (H) Bim. Blots were probed with anti-Bim mAb 5E5 or 14A8 and as a control with anti-EE mAb. Anti-Bim 5E5 detected EE BimL and anti-Bim 14A8 EE BimS/L/EL; both antibodies detected a truncated form of Bim consisting of the N-terminal 72 aa and a mutant lacking the BH3 region. EE BimEL(NS) is EE BimEL in which the splicing region has been mutated and from which, therefore, no BimL is produced.
Figure 2.
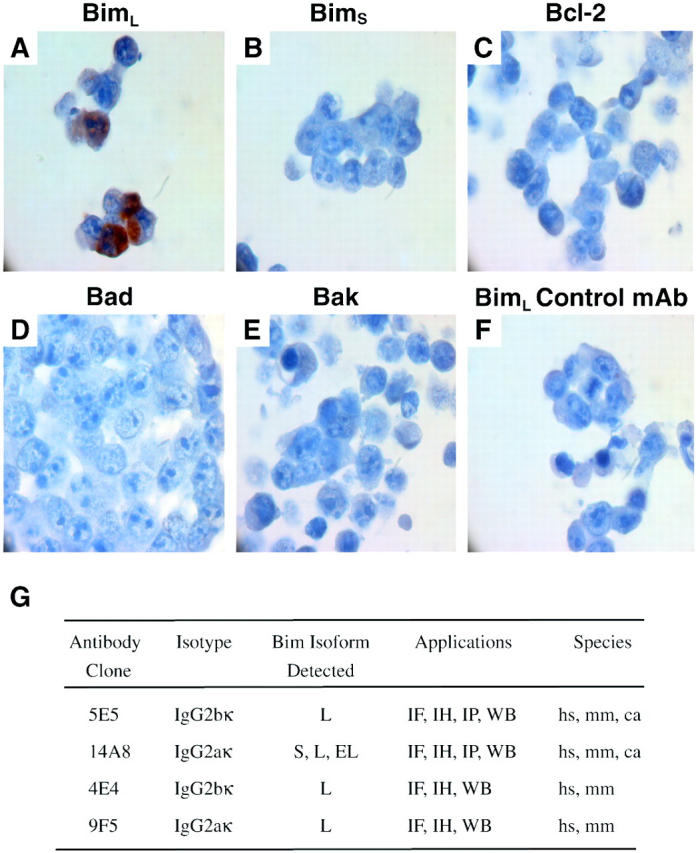
Photomicrographs of representative examples of Bim immunostaining with anti-Bim mAb 5E5 (A–E) or isotype control mAb (F) of sections of 293T cells transfected with various EE-tagged isoforms of Bim, BimL (A and F), BimS (B), or other Bcl-2 family members: Bcl-2 (C), Bad (D), Bak (E). Properties of the anti-Bim mAbs are summarized in G. Most antibodies are suitable for many applications, including immunofluorescence staining (IF), immunohistochemical staining (IH), immunoprecipitation (IP), and Western blotting (WB). 5E5 and 14A8 mAbs recognize human (hs), mouse (mm), and monkey (ca) Bim proteins.
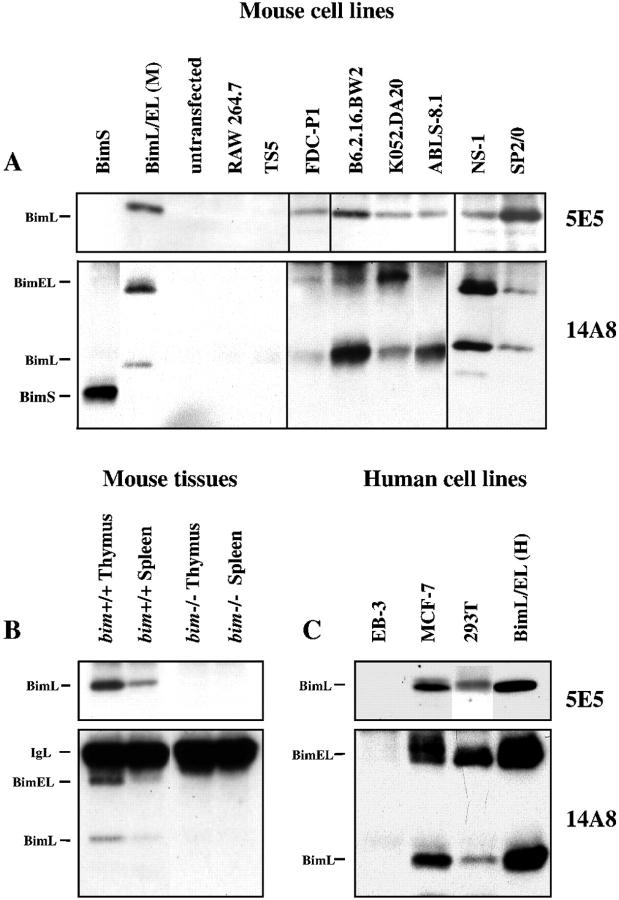
Figure 3.
IP/Western blot analysis of Bim expression in transformed mouse (A) and human (C) cell lines. Bim proteins were immunoprecipitated from lysates of 10 7 cells with anti-Bim mAb 5E5 or 14A8. Western blotting was performed with the same antibodies, followed by biotinylated mouse anti-rat IgG2b antibodies and streptavidin-HRP (5E5) or with HRP-conjugated mouse anti-rat IgG2a antibodies (14A8) and ECL detection. Anti-Bim mAbs 5E5 and 14A8 immunoprecipitate BimL and BimL plus BimEL, respectively, only from lysates of spleen and thymus from normal but not those from Bim-deficient mice (B).
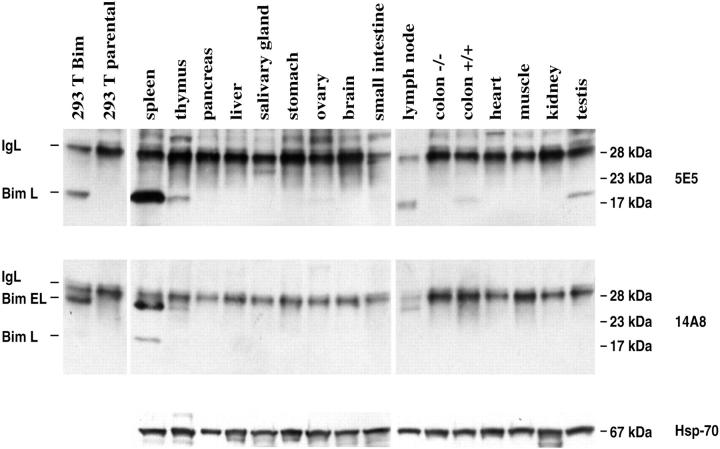
Figure 4.
IP/Western blot analysis of Bim expression in normal mouse tissues. Tissue lysates were prepared and normalized for total protein content. Bim proteins were immunoprecipitated from 1.5–3 mg total tissue protein and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblot analysis as described for Figure 2 ▶ .
To determine more precisely the epitope within Bim recognized by mAbs 5E5 and 14A8, we performed immunoblotting of lysates from cells expressing constructs encoding the different Bim isoforms or mutated versions of Bim. This analysis revealed that the 5E5 mAb detects only the BimL isoform (Figure 1D) ▶ , whereas 14A8 recognizes all three isoforms, BimS, BimL, and BimEL (Figure 1E) ▶ . Importantly, both mAbs bind to an N-terminal fragment (aa 2–72) and not to the BH3 domain of Bim (Figure 1, D–F) ▶ , the only region in Bim that is common in related proteins.
We also tested the specificity of mAbs 5E5 and 14A8 in immunohistochemical staining of cells transfected with expression constructs encoding BimEL, BimL, BimS, BimL lacking the BH3 region (BimLΔBH3), Bcl-2, Bad, or Bak, and processed for paraffin embedding and stained for mAbs 5E5 (Figure 2, A–F) ▶ and 14A8 (not shown). Both mAbs 5E5 and 14A8 showed the same specificity as seen in Western blotting, immunofluorescent staining, and immunoprecipitation.
These results show that mAb 14A8 binds to an N-terminal epitope that is present in all Bim isoforms. In contrast, mAb 5E5 binds to an epitope that is unique to BimL and is generated by amino acids spanning a splice junction unique to BimL. This junction does not exist in BimEL, because of the insertion of an additional coding exon, 15 explaining why mAb 5E5 does not bind this isoform. Because of their high degree of specificity and their ability to work in many applications (Figure 2G) ▶ , mAbs 5E5 and 14A8 were chosen to determine Bim expression and subcellular localization in cultured cell lines and normal mouse tissues.
Expression of Bim in Cultured Cell Lines
Our initial attempts to detect Bim in cultured cell lines by Western blotting of lysates failed because of the low level of expression in these cells. Bim protein could be revealed, however, by immunoprecipitation of lysates from 10 7 cells followed by Western blotting (IP/Western). In many of the cell lines tested, mAb 5E5 detected the ∼19-kd BimL protein, and in some cell lines mAb 14A8 revealed both BimL and the ∼23-kd BimEL protein. BimS was never observed in any of these cell lines, indicating that this isoform is not normally expressed or is present at levels below the limits of detection.
Examples of IP/Western blots are shown in Figure 3 ▶ ; the results are summarized in Table 1 ▶ . The highest levels of BimL and BimEL were found in the human breast cancer-derived epithelial cell line MCF-7 and in several lymphoid lines, such as the mouse T-lymphoid lines B6.2.16.BW2 and K052.DA20, the human T-lymphoma-derived cell line Jurkat and the mouse pre-B-lymphoma line ABLS8.1, and the mouse plasmacytoma lines SP2/0 and NS1. Relatively low levels of BimL and BimEL were detected in the mouse IL-3-dependent myeloid line FDC-P1 and in the human kidney cell line 293T. Some cell lines, such as the mouse macrophage line J774 and the monkey kidney cell line Cos M6 only expressed BimL, and others, such as the fibroblast cell lines NIH3T3, Rat1, and L929; the thymic stromal line S17; and the erythroleukemia-derived lines FN4, DP16, and Ts5 did not express any of the Bim isoforms. These studies demonstrate that BimL and BimEL are expressed in some cell lines, mainly those of hematopoietic and epithelial origins.
IP/Western Analysis of Bim Expression in Mouse Tissues
IP/Western was also used to investigate the expression of Bim in normal mouse tissues. In lymphoid tissues, such as spleen, thymus, and lymph nodes, both mAbs 5E5 and 14A8 detected the BimL protein and mAb 14A8 also recognized the BimEL protein (Figure 4) ▶ . Low levels of BimL were observed in the pancreas, small intestine, and stomach, and low levels of BimEL could be seen in the testes. BimS was not detected in any of the tissues analyzed, consistent with our analysis of its expression in cell lines. The specificity of the mAbs was confirmed by the observation that they immunoprecipitated Bim only from tissues of normal but not Bim-deficient mice (Figure 3B) ▶ . These results demonstrate that BimL and BimEL are expressed in cells from hematopoietic tissues, in the gastrointestinal tract, and in reproductive organs.
Immunohistochemical Analysis of Bim Expression in Mouse Tissues
The expression pattern of the Bim protein in mouse tissues was investigated in detail by immunohistochemical staining, using mAbs 5E5 and 14A8. Because of the low levels of Bim we used the potent tyramide amplification system (see Materials and Methods), which increases the signal up to 1000-fold. Confirmatory results were obtained with two additional, independently derived anti-Bim mAbs (4E4 and 9F5), and as negative controls tissues were stained with isotype-matched rat mAbs. The specificity of the immunostaining was further validated by demonstrating that immunostaining by mAbs 5E5 and 14A8 could be blocked by preadsorption with recombinant GST-BimL. The results of Bim immunostaining were also confirmed by showing that they coincided perfectly with the pattern of in situ hybridization produced with a bim antisense probe.
Bim Expression in Hematopoietic Tissues
Among the hematopoietic cells of the bone marrow, megakaryocytes were intensely stained by both anti-Bim mAbs, and polymorphonuclear cells also had strong BimL/EL immunoreactivity. The intensity of BimL/EL immunoreactivity within the lymphocyte population was variable; some cells were stained strongly but others only weakly. Granulocytes had low but significant levels of Bim, whereas both erythroid precursors and mature erythroid cells were negative.
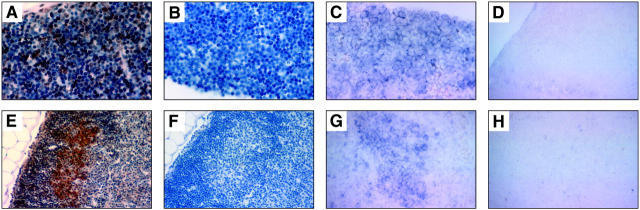
Examination of Bim expression in the thymus showed intense BimL/EL immunoreactivity (Figure 5, A and B) ▶ and bim mRNA expression (Figure 5, C and D) ▶ in the cortical thymocytes, where the immature short-lived CD4+8+ pre-T cells are located. In contrast, expression of Bim protein and bim mRNA was significantly lower (or absent) in the thymic medulla, which contains the mature CD4+8− and CD4−8+ T cells. The only cells in the medulla that stained weakly for Bim protein and bim mRNA were large epithelial and macrophage-like cells.
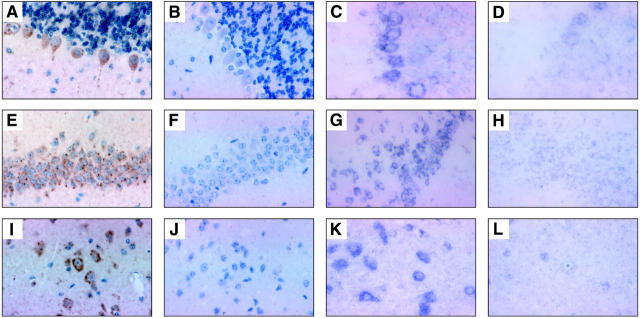
Figure 5.
Photomicrographs of representative examples of Bim immunostaining and bim in situ hybridization in mouse lymphoid tissues. A–D: Thymic cortex. Original magnification, ×400. E–H: Lymph node germinal center. Original magnification, ×200. Paraffin-embedded tissues were immunostained (A, B, E, F), using a diaminobenzidine-based colorimetric detection method with tyramide enhancement (brown). Nuclei were counterstained with hematoxylin (blue). In situ hybridization (C, D, G, H) was performed using the digoxigenin system. Immunostaining with anti-Bim mAb 5E5 (A, E) or with isotype-matched control mAb (B, F). In situ hybridization was conducted with a bim antisense probe (C, G) or with a control bim sense probe (D, H).
In lymph nodes, BimL/EL immunohistochemical staining and in situ hybridization with a bim antisense probe were most striking in primary follicles as well as cells within germinal centers and the mantle zone of the cortex, which contain mostly activated B cells (Figure 5, E and F) ▶ . Because of their larger cytoplasm, immunohistochemical staining is probably more prominent in activated, compared to quiescent, lymphocytes. The paracortical areas and reticular cells in the medullary cords also showed significant levels of Bim protein expression, although this was more heterogeneous than that seen in primary follicles.
In the spleen, BimL/EL immunoreactivity appeared to be limited to lymphoid cells in the white pulp, where staining was most intense in the germinal centers and the periarteriolar sheath/marginal zone. In the red pulp, scattered megakaryocytes and some macrophage-type cells exhibited moderate BimL/EL immunoreactivity, whereas erythrocytes did not. Bim mRNA expression in the spleen was limited to germinal center cells and a subset of cells in the white pulp (probably the T cell zone).
Bim Expression in the Central Nervous System
In the nervous system, immunostaining and in situ hybridization revealed Bim expression primarily in neurons in the gray matter, with no expression in glial, astrocytes, or oligodendrocytes or various types of supporting cells. Expression of BimEL was dominant in both intensity and distribution compared to BimL.
In the cerebellum, mAb 5E5 (anti-BimL) stained within the granular layer, probably the Purkinje axons that extend into this region, but not the stellate or granule cells. In contrast, mAb 14A8 (anti-BimS,L,EL) and in situ hybridization with a bim antisense probe strikingly labeled the Purkinje neurons and their associated dendrites projecting into the molecular layer (Figure 6, A–D) ▶ .
Figure 6.
Photomicrographs of representative examples of Bim immunostaining and bim in situ hybridization in the mouse CNS. A–D: Staining of the Purkinje cells of the cerebellum. E–H: Hippocampus. I–L: Cerebral cortex (original magnification, ×400). Immunostaining was done with anti-Bim mAb 14A8 (A, E, I) or with an isotype-matched control mAb (B, F, J). In situ hybridization was conducted with a bim antisense probe (C, G, K) or with a control bim sense probe (D, H, L).
Neuronal cells of the hippocampus were strongly labeled by both mAbs (Figure 6, E–H) ▶ . In the cerebral cortex varying numbers of small pyramidal neurons and large motor neurons displayed a punctate cytoplasmic distribution of Bim immunoreactivity (Figure 6, I–L) ▶ . The staining intensity varied according to the layer examined, and in general many more neurons were labeled with mAb 14A8 (anti-BimS,L,EL) than with mAb 5E5 (anti-BimL). Intense BimL/EL immunoreactivity was also observed in the cuboidal cells of the choroid plexus. In situ hybridization analysis of the brain confirmed the immunohistochemical analyses (Figure 6) ▶ .
Bim Expression in the Gastrointestinal and Respiratory Systems
In addition to cells of hematopoietic and neuronal origin, BimL/EL protein and bim mRNA were also detected in several cells of epithelial origin within the digestive tract. Moderate to strong BimL/EL immunoreactivity and prominent signal from in situ hybridization with a bim antisense probe were observed in the serous secretory cells of the submandibular and parotid glands. BimL protein expression extended to the columnar or low cuboidal cells of the interlobular and intercalated ducts of the submandibular, parotid, and sublingual glands (Figure 7, A–D) ▶ .
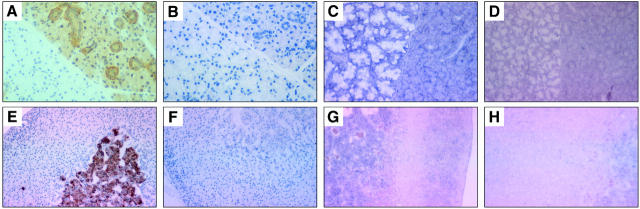
Figure 7.
Photomicrographs of representative examples of Bim immunostaining and bim in situ hybridization in mouse epithelial and endocrine tissues. A–D: Submandibular gland. A: Staining of serous cells and interlobular excretory ducts but not mucus cells (left side of each panel). C: In situ hybridization with a bim antisense probe. E–H: Adrenal gland (original magnification, ×200). Immunostaining was done with anti-Bim mAb 5E5, showing expression within the adrenal medulla (E). B and F: Staining with an isotype-matched control mAb. In situ hybridization was conducted with a bim antisense probe (C, G) or with a control bim sense probe (D, H).
Bim immunoreactivity was observed in many of the complex epithelia lining the digestive and respiratory tracts (data not shown). Moderate BimL immunoreactivity was observed in the basal layer of the stratified squamous epithelia of the esophagus. The ciliated epithelia of the trachea also showed moderate levels of BimL/EL immunoreactivity extending through to the bronchi but not into the alveoli of the lung. This was confirmed by mRNA expression analysis using in situ hybridization.
Moderate BimL/EL immunoreactivity was observed in the stratified squamous epithelia of the tongue (data not shown). In situ hybridization further revealed that the stratum granulosum and, to a lesser extent, the germinativum expressed bim mRNA, but the basal layer of the germinativum of the tongue did not. The serous secreting cells composing the tubuloalveolar glands (Von Ebner’s glands) of the vallate papillae in the tongue also had moderate to intense BimL or BimEL staining and bim mRNA expression.
The smooth muscle, the submucosa, and the serosa of the nonglandular portion of the stomach were marked by the absence of Bim immunoreactivity (with the minor exception of low-level staining of fibrocytes within the connective tissue) (data not shown). In contrast, Bim expression was readily apparent in the glandular portion of the stomach (fundus), where the parietal cells of the crypts, the outer epithelia, and neck cells all showed moderate BimL/EL immunoreactivity. The chief (zymogenic) cells did not express Bim. The simple columnar mucus epithelium of the pyloric region also showed moderate levels of BimL/EL immunoreactivity, but the pyloric glands did not. All of these observations were confirmed by in situ hybridization.
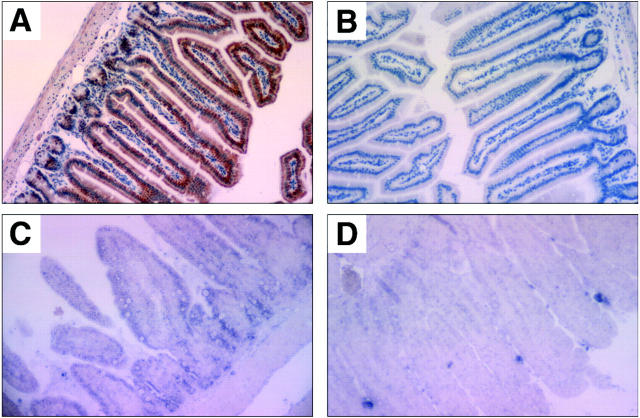
In the small intestine, high levels of BimL/EL immunoreactivity were observed in the simple columnar epithelial cells (enterocytes) of the villi and in crypt cells at the base of the villi (Figure 8, A–D) ▶ . Moderate levels of BimL/EL expression were seen in the epithelial cells of the crypts of the colon. The lymphoid cells of the Peyer’s patches also showed moderate expression of BimL/EL, but the epithelial goblet cells and the lamina propria were negative.
Figure 8.
Photomicrographs of representative examples of Bim immunostaining and bim in situ hybridization in mouse small intestine (original magnification, ×100). Immunostaining was done with anti-Bim mAbs 5E5 (A) within the simple columnar epithelial cells of the villi or with an isotype-matched control mAb (B). In situ hybridization was conducted with a bim antisense probe (C) or with a control bim sense probe (D).
Bim Expression in the Endocrine System
The cuboidal epithelium of the thyroid follicles had moderate levels of BimL immunoreactivity, and the chief (secretory) cells of the parathyroid gland expressed high levels of BimL/EL.
In the pancreas BimL/EL immunoreactivity was moderate in the acinar cells that are responsible for secretion of digestive enzymes, but stronger in the endocrine cells of the islets of Langerhans and the columnar epithelial cells lining the pancreatic ducts. In situ hybridization confirmed that bim expression in the pancreas was restricted to the exocrine acinar cells and that expression of Bim in the islet cells was nonspecific.
The cells of the adrenal gland displayed a heterogeneous pattern of Bim protein and bim mRNA expression. The aldosterone-producing cells of the outer cortical zona glomerulosa and the chromaffin cells of the adrenal medulla both expressed moderate levels of BimL and bim mRNA (Figure 7G) ▶ , whereas BimEL was found only in the adrenal medulla (Figure 7E) ▶ . No Bim immunoreactivity was found in the x-zone in virgin female mice between the cortex and the medulla or in the glucocorticoid-secreting cells of the zona fasiculata.
Bim Expression in Reproductive Organs
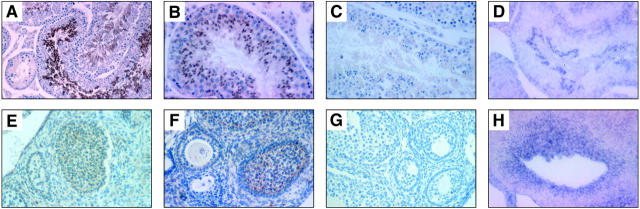
In the testes of adult male mice, intense BimL immunostaining was observed in the cytoplasm of elongating spermatids, becoming concentrated around the midpiece. BimL expression was barely detectable in the spermatogonia and spermatocytes (Figure 9A) ▶ . In contrast, BimEL expression could be demonstrated in elongating spermatids as well as in Sertoli cells, whereas labeling of spermatocytes was much less intense (Figure 9B) ▶ . The ciliated columnar epithelia of the epididymal ducts and the epithelial cells of the ductus deferens displayed moderate BimL staining and high-intensity BimEL immunolabeling. In situ hybridization reproduced the staining seen with anti-Bim mAb 14A8 (Figure 9D) ▶ .
Figure 9.
Photomicrographs of representative examples of Bim immunostaining and bim in situ hybridization in mouse reproductive organs (original magnification, ×100). A–D: Testes. A: BimL immunoreactivity within elongating spermatids. B: BimEL immunoreactivity within elongating spermatids, Sertoli cells, and spermatocytes. E–H: Ovary. E: BimL (5E5). F: BimEL (14A8), showing moderate immunoreactivity within the granulosa cells of primordial, primary, secondary, and mature follicles. Immunostaining was done with an isotype-matched control mAb (C, G). In situ hybridization was conducted with a bim antisense probe (D, H).
In adult females, moderate BimL/EL protein and bim mRNA expression was observed within the granulosa cells of primordial, primary, secondary, and mature follicles. Only weak or no staining was seen in the corpus luteum (Figure 9, E–H) ▶ . The simple columnar ciliated epithelium of the oviduct and the epithelial cells of the uterus contained prominent BimL/EL immunoreactivity.
In mammary glands of virgin female mice bim mRNA expression was observed in the ductal and lobular cuboidal epithelial cells (Figure 10A) ▶ . During pregnancy bim mRNA expression extended to the epithelial side branching (Figure 10B) ▶ , and this persisted throughout lactation (Figure 10C) ▶ and involution (Figure 10D) ▶ .
Figure 10.
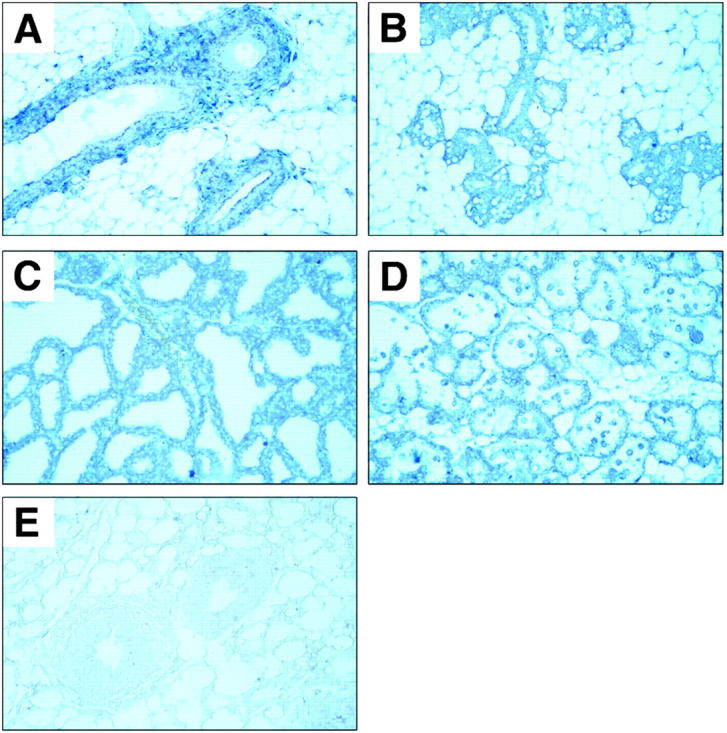
Photomicrographs of representative examples of bim in situ hybridization in the low columnar epithelium of mammary gland lobular tubules. A: Virgin. B: Pregnant. C: Lactating. D: Involution (original magnification, ×400). In situ hybridization was conducted with a bim antisense probe (A–D) or with a control bim sense probe (E).
Bim Expression in the Skin
BimL/EL expression was very striking in the hair follicles, the cylindrical down-growths of the surface epithelium. Intense labeling was seen in the undifferentiated bulb cells within the hair follicle (not shown), the outer epithelial sheath (which may represent a population of keratinocyte stem cells). 36 No staining was observed in the connective tissue root sheath, and only weak labeling was detected in the sebaceous glands. Consistent with these results, high levels of bim mRNA expression were detected by in situ hybridization in hair follicles.
Bim Expression in Cardiovascular and Skeletal Muscles
Consistent with the IP/Western analysis (Figure 4) ▶ , no Bim protein was found in cardiomyocytes or skeletal muscle. Minimal Bim immunolabeling was observed around the outer walls of the cardiac chambers with mAb 5E5, but this was probably nonspecific, because no immunostaining was observed with mAb 14A8 and no bim mRNA could be revealed by in situ hybridization (data not shown).
Bim Expression in the Renal System
In the kidney, BimL/EL expression was strongest in the tubular epithelial cells of the proximal convoluted tubules at the corticomedullary junction. Only weak immunoreactivity was seen in the loops of Henle, the collecting ducts, and the distal convoluted tubules. Prominent bim mRNA expression was observed with certainty only in the ciliated epithelial cells constituting the proximal convoluted tubules within the renal cortex. No Bim immunoreactivity was detected in the glomeruli, indicating that Bim expression in the renal system was restricted to the ductal epithelial cells.
Collectively, these results demonstrate that BimL and BimEL are expressed in lymphocytes, myeloid cells, neurons, many different types of epithelial cells, and neuroendocrine hormone-secreting cells, and in somatic as well as germ cells of the reproductive organs.
Discussion
Bim is a pro-apoptotic member of the Bcl-2 protein family, which shares with its relatives only the short BH3 domain. 15 The three Bim isoforms differ in their pro-apoptotic activity because of differences in their interaction with dynein light chain LC8, which can sequester BimL and BimEL, but not BimS, to the microtubule-associated dynein motor complex. 16 Interestingly, Bim immunoreactivity was present in the cytosol, perhaps suggestive of an association with such intracellular structures (Figures 5–9) ▶ ▶ ▶ . Gene targeting experiments in mice have demonstrated that Bim is required for some but not all apoptotic stimuli that can be inhibited by Bcl-2 or its relatives. Within the whole animal, Bim has a critical role during embryonic development, regulates hematopoietic cell homeostasis, and prevents autoimmunity. 17
Using immunohistochemical staining, IP/Western blotting, and in situ hybridization, we have defined the expression pattern of Bim. The two mAbs 5E5 and 14A8 were used to discriminate between the BimL and BimEL isoforms. No evidence for the expression of the third isoform, BimS, was obtained. This may indicate that BimS is not expressed or is produced only in a very small number of cells that have to be killed very rapidly.
The levels of BimL and BimEL are generally very low because both proteins can only be detected by extremely sensitive techniques. Interestingly, the absence of Bim provides a similar level of protection against cytokine withdrawal-induced apoptosis in lymphocytes 17 as overexpression of Bcl-2. 37,38 Thus Bim (and other BH3-only proteins) may not kill cells simply by inactivating pro-survival Bcl-2 family members in a simple one-to-one interaction. A speculative explanation is that anti-apoptotic Bcl-2 family members exist on intracellular membranes as a loosely aggregated macromolecular structure that regulates the activity of Apaf-1/CED-4-like adaptors. By binding to a small number of antiapoptotic Bcl-2 family members, BH3-only proteins may alter the overall structure of such a complex, thereby allowing caspase activation (see ref 39 ).
Some hematopoietic tissues that normally express BimL and/or BimEL are hyperplastic in Bim-deficient mice, demonstrating that this pro-apoptotic protein is the cause of a nonredundant killing activity in these cells. 17 BimL and, in most instances, BimEL are expressed in B- and T-lymphoid cells, megakaryocytes, and myeloid cells (Figures 3–5 ▶ ▶ ▶ and Table 1 ▶ ). Both proteins are particularly prominent in cells with a high turnover rate, such as cortical CD4+8+ thymocytes, germinal center B cells, and plasma cells. Consistent with this pattern of expression, Bim-deficient mice have an accumulation of B cells, T cells, monocytes, and granulocytes and perturbed T lymphopoiesis. 17 Although numbers of megakaryocytes are normal in Bim-deficient mice, they have a 50% decrease in platelets. 17 Similar observations were made in mice expressing a bcl-2 transgene in megakaryocytes. 55 This may indicate that platelet shedding occurs by an apoptosis-like mechanism that is regulated by Bim and perhaps by other members of the Bcl-2 family. Older bim−/− mice had a 30–200-fold increase in IgG-secreting plasma cells and elevated serum IgM, IgG, and IgA levels, and within a year most succumbed to a systemic lupus erythematosus-like autoimmune disease. 17 Thus, although other BH3-only proteins and Bax-related proapoptotic Bcl-2 family members are expressed in lymphoid and myeloid cells, 40-42 they are unable to compensate for the absence of Bim. It is also possible that other pro-apoptotic Bcl-2 family members can act in conjunction with Bim, and it will therefore be interesting to determine whether the absence of Bad, Blk, Bax, Bak, or related proteins can exacerbate the lymphoid hyperplasia induced by Bim deficiency.
The expression of Bim is not restricted to hematopoietic cells, but can also be detected by immunohistochemistry and by in situ hybridization in a wide variety of cells of epithelial origin. Epithelia constitute a diverse group of tissues that are involved in a wide range of activities such as absorption, secretion by glandular structures (invaginations of epithelial surfaces), and protection against invading pathogens. As in the case of Bim, the epithelia of many tissues, such as renal tubules, small intestine, breast, prostate, and respiratory tract, also express Bax, 40 Bak, 41 and possibly other BH3-only proteins. Many epithelial cells also express anti-apoptotic proteins, including Bcl-2, 40 Bcl-xL, 43 and Mcl-1. 44 Certain complex epithelia have a striking gradient of expression of pro- versus anti-apoptotic Bcl-2 family members. For example, Bcl-2 was predominantly located in the less differentiated cells lining the epithelial basement membrane, whereas Bax and Mcl-1 were prominent in the more differentiated short-lived cells located in the upper layers of epithelial tissue. Notably, expression of Bim and Bax at the base of the crypts of the small intestinal mucosa is consistent with the reports of high rates of spontaneous and genotoxic damage-inducible apoptosis in this region. 45 Bim expression in the hair follicle bulb is restricted to precursor cells, which, under the inductive influence of the dermal papilla, undergo a highly ordered process of cell death that is critical for hair production. 46 Interestingly, transgenic mice that overexpress Bcl-xL in keratinocytes under the keratin 14 promoter have abnormally short hair, 47 and Bcl-2-deficient mice become prematurely gray. 48 Thus both pro- and anti-apoptotic proteins appear to have a regulatory role in hair production.
So far we have not found any abnormalities in epithelial cells in Bim-deficient mice, 17 and this might be due to the functional overlap between pro-apoptotic Bcl-2 family members. However, the specific function of Bim may only be revealed when epithelia are subjected to mechanical stress or other death-promoting insults.
BimL and, in particular, BimEL are expressed in neuronal cells of the central nervous system (CNS). Indeed, like Bim, Bax has been found in several populations of long-lived neurons, including the Purkinje neurons of the cerebellum and large neurons of the cortex. 40 Expression of such proapoptotic proteins coupled with low Bcl-2 levels may explain why these neurons are very sensitive to induction of apoptosis by neurotrophin withdrawal, hypoxia, and hypoglycemia. 49 It is also possible that BH3-only proteins are involved in the attrition of neurons that occurs normally during development of the mammalian nervous system. 50 Although Bim-deficient mice have no obvious defects in the CNS, 17 under conditions of neuronal stress abnormalities may become apparent. It is also likely that other BH3-only proteins can compensate for Bim in the nervous system. Interestingly, expression of a related protein, Hrk/DP5, is dramatically increased in cultured neurons when they are deprived of neurotrophin. 51 It is therefore possible that this protein is the dominant proapoptotic BH3-only protein in neurons. Alternatively, the function of this subfamily in neurons may only become apparent in mice lacking more than one of these proteins.
BimL and BimEL are both expressed in somatic as well as germ cells of female and male reproductive organs (Figures 4 and 9) ▶ ▶ . Despite this, Bim-deficient mice from both sexes have normal fertility. 17 This may be explained by the expression of many other proapoptotic Bcl-2 family members, such as Bax, Bak, Bok/Mtd, and Bad, in these tissues. 40,41,52 These molecules may have overlapping functions. It is clear, however, that Bcl-2 family members are essential for spermatogenesis, because Bax- 53 and Bcl-w-deficient male mice 32,54 are both infertile because of defective spermatogenesis.
Approximately 65% of Bim-deficient embryos die during development before E9.5. 17 The reason for this premature death remains unknown, but it is interesting to consider this in the context of the sites of Bim expression. Because these bim−/− embryos die so early, this is unlikely to be due to defects in neuronal cells or hematopoietic cells. Abnormalities in epithelial tissues, however, may provide an explanation. In an attempt to identify the essential function of Bim in early development, we are determining Bim expression during embryogenesis and are trying to pinpoint the developmental stage in which Bim plays such an essential role.
Acknowledgments
We thank W. Carter, K. Davern, K. Mackwell, K. Wycherley, and Dr. L. O’Connor for help and advice with immunization and hybridoma fusion; M. Santamaria for expert technical assistance with in situ hybridization; Dr. K. Moriishi for preparation of Bim proteins; Dr. P. Bouillet for the Bim-deficient mice; Dr. K. Loveland and T. Meehan (Monash Institute of Reproduction and Development, Melbourne, Australia) for help with the assessment of Bim expression in murine testes; Dr. F. Battye, D. Kaminaris, J. Parker, and V. Lapatis for help with flow cytometry; and A. Milligan and J. Merryfull for animal care. We are grateful to Prof. S. Cory, Prof. J. Adams, Dr. A. Harris, and Dr. D. Vaux for helpful discussions and critical reading of the manuscript.
Footnotes
Address reprint requests to Dr. Andreas Strasser, Walter and Eliza Hall Institute, Post Office, The Royal Melbourne Hospital, Victoria 3050, Australia. E-mail: strasser@wehi.edu.au.
Supported by grants and fellowships from AMRAD Biotech, the Leukemia and Lymphoma Society of America (New York), the Anti-Cancer Council of Victoria (Melbourne, Australia), the Dr. Josef Steiner Cancer Research Foundation (Bern, Switzerland), the National Health and Medical Research Council (Canberra, Australia), the Cancer Research Institute (New York), and the Victorian Breast Cancer Research Consortium (Melbourne, Australia).
Dr. Print’s present address is Department of Pathology, University of Cambridge, Cambridge, UK.
References
- 1.Vaux DL, Haecker G, Strasser A: An evolutionary perspective on apoptosis. Cell 1994, 76:777-779 [DOI] [PubMed] [Google Scholar]
- 2.Jacobson MD, Weil M, Raff MC: Programmed cell death in animal development. Cell 1997, 88:347-354 [DOI] [PubMed] [Google Scholar]
- 3.Strasser A, Harris AW, Bath ML, Cory S: Novel primitive lymphoid tumours induced in transgenic mice by cooperation between myc and bcl-2. Nature 1990, 348:331-333 [DOI] [PubMed] [Google Scholar]
- 4.Strasser A, Whittingham S, Vaux DL, Bath ML, Adams JM, Cory S, Harris AW: Enforced BCL2 expression in B-lymphoid cells prolongs antibody responses and elicits autoimmune disease. Proc Natl Acad Sci USA 1991, 88:8661-8665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watanabe-Fukunaga R, Brannan CI, Copeland NG, Jenkins NA, Nagata S: Lymphoproliferation disorder in mice explained by defects in Fas antigen that mediates apoptosis. Nature 1992, 356:314-317 [DOI] [PubMed] [Google Scholar]
- 6.Barr PJ, Tomei LD: Apoptosis and its role in human disease. Biotechnology 1994, 12:487-493 [DOI] [PubMed] [Google Scholar]
- 7.Kerr JFR, Wyllie AH, Currie AR: Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer 1972, 26:239-257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vaux DL, Strasser A: The molecular biology of apoptosis. Proc Natl Acad Sci USA 1996, 93:2239-2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thornberry NA, Lazebnik Y: Caspases: enemies within. Science 1998, 281:1312-1316 [DOI] [PubMed] [Google Scholar]
- 10.Srinivasula SM, Ahmad M, Fernandes-Alnemri T, Alnemri ES: Autoactivation of procaspase-9 by Apaf-1-mediated oligomerization. Mol Cell 1998, 1:949-957 [DOI] [PubMed] [Google Scholar]
- 11.Yang X, Chang HY, Baltimore D: Essential role of CED-4 oligomerization in CED-3 activation and apoptosis. Science 1998, 281:1355-1357 [DOI] [PubMed] [Google Scholar]
- 12.Adams JM, Cory S: The Bcl-2 protein family: arbiters of cell survival. Science 1998, 281:1322-1326 [DOI] [PubMed] [Google Scholar]
- 13.Strasser A, Harris AW, Huang DCS, Krammer PH, Cory S: Bcl-2 and Fas/APO-1 regulate distinct pathways to lymphocyte apoptosis. EMBO J 1995, 14:6136-6147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conradt B, Horvitz HR: The C. elegans protein EGL-1 is required for programmed cell death and interacts with the Bcl-2-like protein CED-9. Cell 1998, 93:519-529 [DOI] [PubMed] [Google Scholar]
- 15.O’Connor L, Strasser A, O’Reilly LA, Hausmann G, Adams JM, Cory S, Huang DCS: Bim—a novel member of the Bcl-2 family that promotes apoptosis. EMBO J 1998, 17:384-395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Puthalakath H, Huang DCS, O’Reilly LA, King SM, Strasser A: The pro-apoptotic activity of the Bcl-2 family member Bim is regulated by interaction with the dynein motor complex. Mol Cell 1999, 3:287-296 [DOI] [PubMed] [Google Scholar]
- 17.Bouillet P, Metcalf D, Huang DCS, Tarlinton DM, Kay TWH, Kšntgen F, Adams JM, Strasser A: Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science 1999, 286:1735-1738 [DOI] [PubMed] [Google Scholar]
- 18.Huang DCS, Cory S, Strasser A: Bcl-2, Bcl-XL and adenovirus protein E1B19kD are functionally equivalent in their ability to inhibit cell death. Oncogene 1997, 14:405-414 [DOI] [PubMed] [Google Scholar]
- 19.Grussenmeyer T, Scheidtmann KH, Hutchinson MA, Eckhart W, Walter G: Complexes of polyoma virus medium T antigen and cellular proteins. Proc Natl Acad Sci USA 1985, 82:7952-7954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson IA, Niman HL, Houghten RA, Cherenson AR, Connolly ML, Lerner RA: The structure of an antigenic determinant in a protein. Cell 1984, 37:767-778 [DOI] [PubMed] [Google Scholar]
- 21.Blanar M, Rutter W: Interaction cloning: identification of a helix-loop-helix zipper protein that interacts with c-Fos. Science 1992, 256:1014-1018 [DOI] [PubMed] [Google Scholar]
- 22.Galfre G, Howe SC, Milstein C, Butcher GW, Howard JC: Antibodies to major histocompatibility antigens produced by hybrid cell lines. Nature 1977, 266:550-552 [DOI] [PubMed] [Google Scholar]
- 23.O’Reilly LA, Cullen L, Moriishi K, O’Connor L, Huang DCS, Strasser A: Rapid hybridoma screening method for the identification of monoclonal antibodies to low abundance cytoplasmic proteins. BioTechniques 1998, 25:824-830 [DOI] [PubMed] [Google Scholar]
- 24.Culvenor JG, Harris AW, Mandel TE, Whitelaw A, Ferber E: Alkaline phosphatase in hematopoietic tumor cell lines of the mouse: high activity in cells of the B lymphoid lineage. J Immunol 1981, 126:1974-1977 [PubMed] [Google Scholar]
- 25.Elefanty AG, Cory S: bcr-abl-induced cell lines can switch from mast cell to erythroid or myeloid differentiation in vitro. Blood 1992, 79:1271-1281 [PubMed] [Google Scholar]
- 26.Kongsuwan K, Webb E, Housiaux P, Adams JM: Expression of multiple homeobox genes within diverse mammalian haemopoietic lineages. EMBO J 1988, 7:2131-2138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strasser A, Harris AW, Jacks T, Cory S: DNA damage can induce apoptosis in proliferating lymphoid cells via p53-independent mechanisms inhibitable by Bcl-2. Cell 1994, 79:329-339 [DOI] [PubMed] [Google Scholar]
- 28.Visvader J, Begley CG, Adams JM: Differential expression of the LYL, SCL and E2A helix-loop-helix genes within the hemopoietic system. Oncogene 1991, 5:187-194 [PubMed] [Google Scholar]
- 29.Visvader J, Adams JM: Megakaryocytic differentiation induced in 416B myeloid cells by GATA-2 and GATA-3 transgenes or 5-azacytidine is tightly coupled to GATA-1 expression. Blood 1993, 82:1493-1501 [PubMed] [Google Scholar]
- 30.Karasuyama H, Melchers F: Establishment of mouse cell lines, which constitutively secrete large quantities of interleukin 2, 3, 4 or 5, using modified cDNA expression vectors. Eur J Immunol 1988, 18:97-104 [DOI] [PubMed] [Google Scholar]
- 31.Huang DCS, O’Reilly LA, Strasser A, Cory S: The anti-apoptosis function of Bcl-2 can be genetically separated from its inhibitory effect on cell cycle entry. EMBO J 1997, 16:4628-4638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Print CG, Loveland KL, Gibson L, Meehan T, Stylianou A, Wreford N, de Kretser D, Metcalf D, Köntgen F, Adams JM, Cory S: Apoptosis regulator Bcl-w is essential for spermatogenesis but appears otherwise redundant. Proc Natl Acad Sci USA 1998, 95:12424-12431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moriishi K, Huang DCS, Cory S, Adams JM: Bcl-2 family members do not inhibit apoptosis by binding the caspase-activator Apaf-1. Proc Natl Acad Sci USA 1999, 96:9683-9688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Reilly LA, Gu D, Sarvetnick N, Edlund H, Phillips JM, Fulford T, Cooke A: α-Cell neogenesis in an animal model of IDDM. Diabetes 1997, 46:599-606 [DOI] [PubMed] [Google Scholar]
- 35.Wilkinson DG: In Situ Hybridization. Oxford, New York, IRL Press, 1992
- 36.Rochat A, Kobayashi K, Barrandon Y: Location of stem cells of human hair follicles by clonal analysis. Cell 1994, 76:1063-1073 [DOI] [PubMed] [Google Scholar]
- 37.Sentman CL, Shutter JR, Hockenbery D, Kanagawa O, Korsmeyer SJ: bcl-2 inhibits multiple forms of apoptosis but not negative selection in thymocytes. Cell 1991, 67:879-888 [DOI] [PubMed] [Google Scholar]
- 38.Strasser A, Harris AW, Cory S: Bcl-2 transgene inhibits T cell death and perturbs thymic self-censorship. Cell 1991, 67:889-899 [DOI] [PubMed] [Google Scholar]
- 39.Straker A, O’Connor L, Dixit UM: Apoptosis signaling. Annu Rev Biochem 2000, 69:217-245 [DOI] [PubMed] [Google Scholar]
- 40.Krajewski S, Krajewska M, Shabaik A, Miyashita T, Wang HG, Reed JC: Immunohistochemical determination of in vivo distribution of Bax, a dominant inhibitor of Bcl-2. Am J Pathol 1994, 145:1323-1336 [PMC free article] [PubMed] [Google Scholar]
- 41.Krajewski S, Krajewska M, Reed JC: Immunohistochemical analysis of in vivo patterns of bak expression, a proapoptotic member of the Bcl-2 protein family. Cancer Res 1996, 56:2849-2855 [PubMed] [Google Scholar]
- 42.Mok C-L, Gil-Gómez G, Williams O, Coles M, Taga S, Tolaini M, Norton T, Kioussis D, Brady HJM: Bad can act as a key regulator of T cell apoptosis and T cell development. J Exp Med 1999, 189:575-586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krajewski S, Krajewska M, Shabaik A, Wang HG, Irie S, Fong L, Reed JC: Immunohistochemical analysis of in vivo patterns of Bcl-X expression. Cancer Res 1994, 54:5501-5507 [PubMed] [Google Scholar]
- 44.Krajewski S, Bodrug S, Krajewska M, Shabaik A, Gascoyne R, Berean K, Reed JC: Immunohistochemical analysis of Mcl-1 protein in human tissues. Am J Pathol 1995, 146:1309-1319 [PMC free article] [PubMed] [Google Scholar]
- 45.Merritt AJ, Potten CS, Kemp CJ, Hickman JA, Balmain A, Lane DP, Hall PA: The role of p53 in spontaneous and radiation-induced apoptosis in the gastrointestinal tract of normal and p53-deficient mice. Cancer Res 1994, 54:614-617 [PubMed] [Google Scholar]
- 46.Sperling LC: Hair anatomy for the clinician. J Am Acad Dermatol 1991, 25:1-17 [DOI] [PubMed] [Google Scholar]
- 47.Pena JC, Kelekar A, Fuchs EV, Thompson CB: Manipulation of outer root sheath cell survival perturbs the hair-growth cycle. EMBO J 1999, 18:3596-3603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Veis DJ, Sorenson CM, Shutter JR, Korsmeyer SJ: Bcl-2-deficient mice demonstrate fulminant lymphoid apoptosis, polycystic kidneys, and hypopigmented hair. Cell 1993, 75:229-240 [DOI] [PubMed] [Google Scholar]
- 49.Merry DE, Korsmeyer SJ: Bcl-2 gene family in the nervous system. Annu Rev Neurosci 1997, 20:245-267 [DOI] [PubMed] [Google Scholar]
- 50.Oppenheim RW: Cell death during development of the nervous system. Annu Rev Neurosci 1991, 14:453-501 [DOI] [PubMed] [Google Scholar]
- 51.Imaizumi K, Morihara T, Mori Y, Katayama T, Tsuda M, Furuyama T, Wanaka A, Takeda M, Tohyama M: The cell death-promoting gene DP5, which interacts with the BCL2 family, is induced during neuronal apoptosis following exposure to amyloid β protein. J Biol Chem 1999, 274:7975-7981 [DOI] [PubMed] [Google Scholar]
- 52.Hsu SY, Kaipia A, McGee E, Lomeli M, Hsueh AJW: Bok is a pro-apoptotic Bcl-2 protein with restricted expression in reproductive tissues and heterodimerizes with selective anti-apoptotic Bcl-2 family members. Proc Natl Acad Sci USA 1997, 94:12401-12406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Knudson CM, Tung KSK, Tourtellotte WG, Brown GAJ, Korsmeyer SJ: Bax-deficient mice with lymphoid hyperplasia and male germ cell death. Science 1995, 270:96-99 [DOI] [PubMed] [Google Scholar]
- 54.Ross AJ, Waymire KG, Moss JE, Parlow AF, Skinner MK, Russell LD, MacGregor GR: Testicular degeneration in Bclw-deficient mice. Nat Genet 1998, 18:251-256 [DOI] [PubMed] [Google Scholar]
- 55.Ogilvy S, Metcartf D, Print CG, Bath ML, Harris AW, Adams JM: Constitutive Bcl-2 expression throughout the hematopoietic compartment affects multiple lineages and enhances progenitor cell survival. Proc Natl Acad Sci USA 1999, 96:14943-14948 [DOI] [PMC free article] [PubMed] [Google Scholar]