Abstract
Chromosomal imbalances in 113 primary head and neck squamous cell carcinomas (HNSCCs) determined by comparative genomic hybridization were correlated with patients survival using custom-made computer software which enabled the assessment of individual chromosomal loci. The Kaplan-Meier analysis revealed that overrepresentations of 2q12, 3q21-29, 6p21.1, 11q13, 14q23, 14q24, 14q31, 14q32, 15q24, 16q22, and deletions of 8p21-22 and 18q11.2 were significantly associated with both shorter disease-free interval and disease-specific survival in this tumor collective. Multivariate Cox proportional hazards regression models consistently identified the gains of 3q21-29, 11q13, and the loss of 8p21-22 as independent prognostic markers carrying a higher significance than the nodal status as the only clinicopathological parameter with statistical importance. In addition, these three markers allowed a molecular dissection of the patients with low clinical risk (pN0 and pT2 tumors). Thus, the genomic data being derived from the evaluation of primary HNSCC enabled a stratification of the patients into subgroups with different survival highlighting the necessity of a genetically based tumor classification for refining diagnosis and treatment of HNSCC patients.
Histopathological tumor classification is still considered the gold standard for the characterization of solid tumors. However, it is well known that these established parameters do not satisfactorily predict the clinical outcome in individual cases. This is primarily because of the different biological behavior of the tumors that partially escape the conventional histological analysis. In head and neck squamous cell carcinoma (HNSCC) the nodal status is the best prognostic parameter as for many other tumor types. 1-4 It reflects a specific biological behavior being the capacity for lymphangiogenic dissemination. However, for its correct determination an extended operation is necessary. Although surgery is the major option for initial treatment, primary radiochemotherapy has shown promising results in HNSCC. 5 For the improvement of different treatment modalities there is an increased demand for a refined initial characterization which is generally performed by a small biopsy of the primary tumor. So far, these specimens are only used to establish the diagnosis of the disease and a semiquantitive estimate of its malignancy expressed by the tumor grade. In an ideal setting, the pathologist should also answer the question whether or not the tumor carries the potential for metastatic spread or if it is resistant to radiation and/or chemotherapy and he should provide a statement on the patient’s prognosis.
Multiple genetic changes have been reported in HNSCC which include both activation of proto-oncogenes and inactivation of tumor suppressor genes. 6 Until now, the genetic alterations underlying the malignant behavior and progression of these tumors are only partially understood. The only alteration that has been repeatedly associated with worse prognosis in HNSCC is the amplification of chromosome 11q13 harboring the cycD1 gene. 7
To get a more comprehensive picture about which additional chromosomal alterations are associated with prognosis in HNSCC, we correlated our comparative genomic hybridization (CGH) data of 113 primary carcinomas with patients survival using computer software that allowed the assessment of individual loci by Kaplan-Meier analysis. The study highlighted several imbalances with prognostic impact and with each carrying a higher significance than the classical parameters.
Materials and Methods
Patients and Tumor Samples
The study comprised 113 patients with single primary HNSCC. None of the patients had previous malignancies or received treatment before initial tumor biopsy. They were all treated for cure by surgical removal of the primary carcinoma along with a neck dissection being complemented by adjuvant postoperative radiation in advanced stages. All specimens were obtained from surgical resections of the primary tumors that were operated on at the Department of Otorhinolaryngology, Charité Hospital, Humboldt University in Berlin during the period 1994 through 1996. Operation specimens were transferred to the Institute of Pathology of the Charité University Hospital within 1 hour after surgical removal. One aliquot of tumor tissue was frozen in liquid nitrogen and kept at −80°C until DNA extraction. DNA was extracted from several 30-μm cryostat tissue sections by proteinase K digestion and phenol-chloroform extraction and which was verified to consist of a minimum of 70% tumor cells in each case. A second aliquot was submitted for formalin fixation and paraffin embedding. The histopathological diagnosis was established in every case according to the World Health Organization guidelines on hematoxylin and eosin (H&E)-stained tissue sections and the tumors were staged using the tumor-node-metastasis classification forwarded by the International Union Against Cancer (UICC). 8
Follow-up of the patients, performed on an ambulant basis after completed therapy, lasted until November 1, 1999. The median duration of follow-up was 44 months overall (95% confidence interval, 40.14 to 48.76). The causes of death were determined at autopsy or by clinical examination at the Charité University Hospital. The distribution of clinicopathological and survival data of all cases is summarized in Table 1 ▶ . None of the patients had distant metastases (pM = 0) at the time of diagnosis.
Table 1.
Clinicopathological Data of the Study Cohort
| Total | A | AWT | DBT | DOD | |
|---|---|---|---|---|---|
| No. of patients | 113 | 70 | 10 | 33 | 7 |
| Females | 26 | 14 | 5 | 7 | 1 |
| Males | 87 | 56 | 5 | 26 | 6 |
| Tumor sites | |||||
| Larynx | 41 | 31 | 2 | 8 | 3 |
| Supraglottic | 17 | 13 | 2 | 2 | 1 |
| Glottic | 23 | 17 | — | 6 | 2 |
| Subglottic | 1 | 1 | — | — | — |
| Hypopharynx | 29 | 16 | 3 | 10 | 1 |
| Oropharynx | 31 | 18 | 4 | 9 | 3 |
| Oral cavity | 12 | 5 | 1 | 6 | — |
| Stage | |||||
| pT1 | 6 | 6 | — | — | — |
| pT2 | 29 | 13 | 4 | 12 | 1 |
| pT3 | 45 | 33 | 3 | 9 | 4 |
| pT4 | 33 | 18 | 3 | 12 | 2 |
| pN0 | 41 | 33 | 1 | 7 | 4 |
| pN1 | 22 | 11 | 4 | 7 | 2 |
| pN2 | 48 | 25 | 5 | 18 | 1 |
| pN3 | 2 | 1 | — | 1 | — |
| Grade | |||||
| G1 | 3 | 3 | — | — | — |
| G2 | 86 | 56 | 5 | 25 | 7 |
| G3 | 24 | 11 | 5 | 8 | — |
A, alive without disease; AWT, alive with tumor; DBT, dead because of tumor; DOD, dead because of other disease.
CGH
DNA labeling, hybridization, and detection were performed as previously described. 9,10 The protocols are also available at our web site http://amba.charite.de/cgh.
Digital Image Analysis
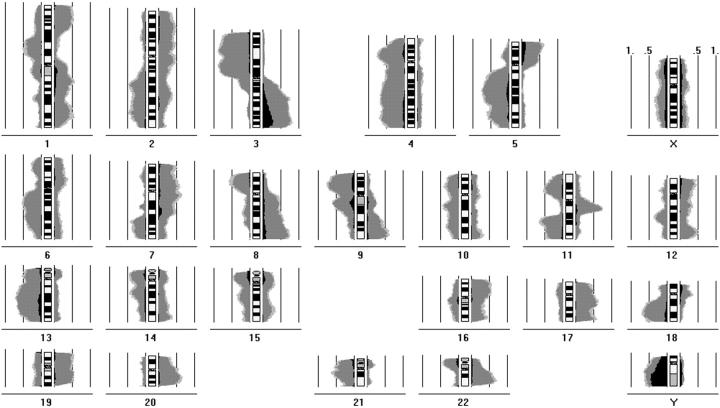
The image acquisition and digital image analysis have also been described in detail. 11,12 At least 10 and generally even 15 metaphases/karyograms were analyzed per case calculating CGH sum karyograms and mean ratio profiles with confidence intervals. The DNA imbalances were determined according to a statistical procedure as described. 10,11 Briefly, the deviations of the mean fluorescein isothiocyanate/tetramethylrhodamine B isothiocyanate profiles from the normal ratio of 1.0 were tested for significance by a two-sided Student’s t-test. Deviations of the ratio profile with at least 99% significance in the Student’s t-test were scored as DNA gains or losses, ie, only those imbalances in which the ratio profile with its 99% confidence interval exceeded the line of the normal ratio 1.0 to the same side were included in the evaluation. This procedure is rather sensitive for scoring chromosomal alterations by CGH. Pronounced DNA gains and losses of HNSCC shown in the histogram of Figure 2 ▶ were defined by those alterations exceeding the ratio values 1.5 and 0.5, respectively. 13 They most likely correspond to high copy amplifications of multicopy deletions. Individual ratio profiles of all tumors are accessible at our CGH Online Tumor Database at http://amba.charite.de/cgh.
Figure 2.
Summary of chromosomal alterations in 113 HNSCC in a histogram representation. The chromosomal imbalances are shown as incidence curves along each chromosome. Areas on the left side of the chromosome ideogram correspond to loss of genetic material; those on the right side to DNA gains. The frequency of alterations can be determined from 0.5 (50%) and 1.0 (100%) incidence lines depicted parallel to the chromosome ideograms. DNA changes with 99% significance are colored in gray, additional changes with 95% significance are depicted in light gray. The proportion of pronounced DNA gains and losses being defined as imbalances for which the ratio profiles exceeded the thresholds of 1.5 and 0.5, respectively, are visualized in black.
Statistical Methods
Survival analysis was applied to all 113 patients comprising clinicopathological as well as genetic parameters. The clinical parameters included tumor site, tumor size (pT), lymph node metastases (pN), histopathological grade, and UICC stage. The genetic parameters consisted of a comprehensive set of chromosomal alterations determined by CGH corresponding to the 400-band ISCN nomenclature. All noninformative bands, ie, centromeres, heterochromatic regions, satellites, and the sex chromosomes were excluded from the analysis, which in total led to 303 individual bands. A genetic imbalance was scored if at least 75% of the digital chromosome segments were affected by an imbalance.
Survival analysis of the data were performed with SPSS software (SPSS, Munich, Germany) controlled by custom-made software. Values of P < 0.05 were considered to be statistically significant. Deaths from causes other than the index tumor or recurrence/metastases were not considered treatment failures, and these patients were censored in each analysis involving the length of survival. Kaplan-Meier analysis 14 was sequentially performed on each of the 303 bands by using the criteria for a genetic alteration as described above. Because the criterion is binary (alteration/no alteration) it subdivided the set of 113 patients into two subgroups. By this way the relationship between the single chromosomal alterations and the overall survival and disease-free interval was performed. The differences of the survival curves were tested for statistical significance with the log rank test.
Cox proportional hazards regression models 15 were used to examine the relative impact of the variables demonstrated to be statistically significant in univariate analysis. The stepwise backward/forward procedures provided by the SPSS software were used to further reduce the number of variables in the Cox models. For assessing and comparing the Cox models a Wald test with a significance level of 0.05 was used for both inclusion and exclusion of variables.
Results
Survival Analysis—Correlation with Clinicopathological Parameters
The clinicopathological data for all patients are summarized in Table 1 ▶ . Survival was analyzed for the parameters tumor site, stage (pT, pN, UICC), and grade with respect to the occurrence of metastasis/recurrence (disease-free interval) and death from HNSCC (disease-specific survival), respectively. In the univariate Kaplan-Meier analysis statistically significant differences were observed for the pN stage only when comparing nodal-negative cases (pN0) with nodal-positive tumors (pN1, pN2, or pN3), the tumor site comparing pharyngeal versus laryngeal carcinomas and the pT stage.
For the pT stage, however, the best prognosis was associated with the pT1 tumors followed by pT3, pT2, and pT4 carcinomas. Surprisingly, the pT2 carcinomas had a worse outcome than pT3 tumors. However, this can be explained by the composition of both subgroups regarding the location of the primary tumor. The pT2 group consisted mainly of pharyngeal carcinomas (26 pharyngeal versus three laryngeal carcinomas) carrying a worse prognosis than laryngeal carcinomas. For the pT3 group, in contrast, the carcinomas were almost equally distributed (22 pharyngeal versus 23 laryngeal tumors).
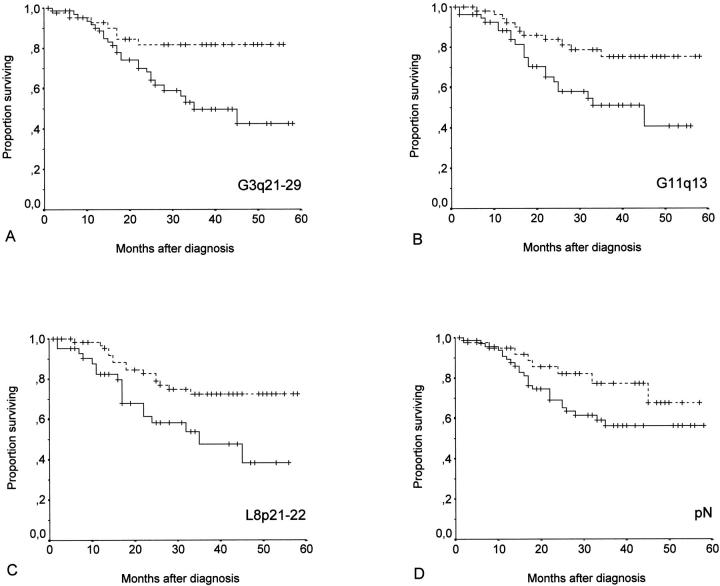
For the nodal stage, the log rank test indicated significance in the correlation with the disease-free interval (P = 0.011) as shown in Figure 1D ▶ whereas there was only a tendency for significance regarding the disease-specific survival (P = 0.078). Similarly, for the tumor site there was a significant P value only for the disease-free interval (P = 0.048) in contrast to the disease-specific survival (P = 0.136).
Figure 1.
A–C: Kaplan-Meier plots comparing disease-specific survival in patients whose tumors showed gains at 3q21-29 (A), gains at 11q13 (B), deletions at 8p21-22 (C) represented by solid lines with that of patients whose tumors did not exhibit the specific chromosomal imbalances (stippled lines), respectively. D: Kaplan-Meier plot comparing disease-specific survival in patients whose tumors had already spread to cervical lymph nodes with that of patients whose tumors did not metastasize at time of diagnosis (stippled line).
Survival Analysis—Correlation with Chromosomal Imbalances
Kaplan-Meier analysis was performed separately for 303 chromosomal imbalances representing the chromosomal bands and subbands of all autosomes. Representative diagrams of chromosomal imbalances showing a statistically significant correlation with patients survival are illustrated in Figure 1, A–C ▶ . All significant alterations are listed in Table 2 ▶ . Overrepresentations of 2q12, 3q21-29, 6p21.1, 11q13, 14q23, 14q24, 14q31, 14q32, 15q24, 16q22, and deletions of 8p21-22 and 18q11.2 had a statistically significant association with both the shorter disease-free interval and the shorter disease-specific survival in this patient population.
Table 2.
Chromosomal Sites Associated with Disease-Free Interval and/or Disease-Specific Survival
| Chromosome | Type | Log rank (p) | Number of cases with/without alteration | Censored cases with/without alteration |
|---|---|---|---|---|
| 1q32 | Gain | 0.049 | 33 /80 | 16/55 |
| 1q42–43 | Gain | 0.034 | 27 /86 | 13/58 |
| 2p24–25 | Gain | 0.013 | 27 /86 | 12/59 |
| 2q12 | Gain | <0.001 (<0.001) | 32 /81 | 14/57 (16/64) |
| 3q21–29 | Gain | 0.007 (0.006) | 66 /47 | 35/36 (40/40) |
| 6p21.1 | Gain | 0.010 (0.008) | 26 /87 | 13/58 (14/47) |
| 6p22–24 | Gain | 0.006 | 20 /93 | 9/62 |
| 7p22 | Gain | 0.045 | 31 /82 | 15/56 |
| 8p21–22 | Loss | 0.014 (0.008) | 43 /70 | 22/49 (25/55) |
| 11q13 | Gain | 0.005 (0.013) | 56 /57 | 29/42 (34/46) |
| 11q23–25 | Loss | 0.042 | 41 /72 | 21/50 |
| 14q23 | Gain | 0.041 (0.021) | 21 /92 | 12/59 (4/85) |
| 14q24 | Gain | 0.003 (0.000) | 21 /92 | 11/60 (11/69) |
| *14q31 | Gain | 0.024 | 21 /92 | 13/67 |
| *14q32 | Gain | 0.030 | 20 /93 | 12/68 |
| 15q24 | Gain | 0.005 (0.005) | 23 /90 | 10/61 (11/69) |
| 16q22 | Gain | 0.015 (0.001) | 24 /89 | 11/60 (11/68) |
| *18q11.2 | Loss | 0.034 | 25 /88 | 14/66 |
| 18q21 | Loss | 0.043 | 64 /49 | 35/36 |
Values for disease-specific survival are indicated in brackets.
*Values for disease-free interval and disease-specific survival were identical.
Table 2 ▶ indicated only those loci harboring DNA gains or losses in at least 20 of the 113 cases. The overall incidence of imbalances at specific chromosomal sites can be deduced from the histogram of the 113 primary HNSCC shown in Figure 2 ▶ . A frequency of >50% along with significance in the survival analysis was observed for the deletions on 8p21-23, 11q23-25, and 18q as well as the DNA overrepresentations on chromosomes 3q13.3-qter and 11q13. Individual ratio profiles with confidence intervals as well as the histogram are also available at our CGH Online Tumor Database at http://amba.charite.de/cgh where the incidences can be assessed interactively by opening the Infoscreen and pointing at specific chromosomal sites using the computer mouse. Chromosomal imbalances were observed in each case using our methodology for checking the normal cell contamination.
Survival Analysis—Multivariate Regression
Cox proportional hazard models were used to examine the relative impact of the variables identified by the univariate analysis. As a first step, chromosomal loci of Table 2 ▶ were preselected for those imbalances being present in at least 40 tumors and showing ≤55% of censored cases with respect to the disease-free interval. Applying these criteria five chromosomal locations remained, ie, gains of 3q21-29, 11q13, and losses of 8p21-22, 11q23-25, and 18q21. The presence or absence of lymph node metastases (pN0 versus pN1/pN2/pN3) was included as the only clinical parameter in the multivariate analysis resulting totally in six binary variables. For the disease-free interval, the backward and the forward procedure for variable inclusion/exclusion determined the same set of remaining variables in the Cox model which are listed in Table 3 ▶ , ie, nodal status, gains of 3q21-29 and 11q13, as well as deletions of 8p21-22. The Cox model for the disease-specific survival using the backward procedure resulted in the same variables; however, the forward procedure further reduced it to the gains of 3q21-29 and 11q13.
Table 3.
Results of the Cox Multivariate Analysis (Disease-Free Interval)
| Variable | B | P | Exp (B) | 95% CI for Exp (B) | |
|---|---|---|---|---|---|
| Lower | Upper | ||||
| pN | 0.978 | 0.015 | 2.658 | 1.213 | 5.824 |
| 3q21–29 | 1.021 | 0.006 | 2.775 | 1.339 | 5.753 |
| 8p21–22 | 0.642 | 0.043 | 1.901 | 1.022 | 3.538 |
| 11q13 | 0.877 | 0.008 | 2.403 | 1.258 | 4.592 |
B, the estimation for the regression coefficient β; P, probability (rejection of the null hypothesis if it is true); Exp(B), the hazard ratio which measures the relative survival experience of every two compared groups; CI, confidence interval.
Impact of Genetic Markers in Clinicopathological Subgroups
To evaluate the influence of the genetic markers in conventionally defined tumor subgroups we performed Kaplan-Meier analysis for each of the five imbalances that were assessed in the multivariate analysis in patients with pT2, pT3, pT4, pN0, pN1, and pN2 stage, respectively. In particular for the pN0 subgroup, the analysis confirmed the validity of the three independent markers, ie, 3q21-29 gain, 11q13 gain, and 8p21-22 loss, yielding significant P values of the log rank test ranging from <0.001 up to 0.045 both for the disease-specific survival and disease-free interval. Other significant P values (<0.05) were observed for the following combinations: pT2–3q21-29 gain (disease-specific survival and disease-free interval), pT2–11q13 gain (disease-free interval), pT3-8p21-22 loss (disease-specific survival), and pN1-3q21-29 gain (disease-free interval).
Discussion
Analyzing primary tumors by CGH we identified several chromosomal imbalances that are associated with bad prognosis of HNSCC patients. Although it is a very powerful tool to screen a tumor genome for chromosomal imbalances most similar CGH studies in other tumor entities gave rather disappointing results often indicating that only a higher overall number of alterations is associated with shorter survival. This discrepancy can be mainly explained by methodological considerations. First, we collected our tumor samples in the context of a prospective study performing hybridizations exclusively with DNA from fresh-frozen tumor samples checking the amount of contaminating normal tissue in each specimen. 9,10 Second, for the calculation of the ratio profiles and confidence intervals at least 10 metaphases were included in the digital image analysis using a statistical evaluation for the determination of chromosomal imbalances which has been reported to be superior to the classical evaluation scheme by fixed ratio thresholds. 16,17 Third, previous studies usually scored the alterations with a resolution that is far behind the power of CGH counting a specific genetic change as an imbalance of the entire chromosome arm. This is understandable because of the complexity of the statistical analysis if each chromosomal subregion is analyzed separately. To overcome this methodological shortcoming we developed computer software enabling the assessment of individual loci and here applied it for the first time on our tumor collective of head and neck carcinomas. The fact that we confirmed several changes that have been previously reported using other methodologies supports the validity of our findings.
Chromosome 11q13 gains have repeatedly been reported to be associated with bad prognosis in HNSCC. 18,19 The main candidate gene is the cycD1 proto-oncogene which drives the cell from the G1 into the S phase of the cell cycle. However, overexpression of this gene seems to be an independent prognostic factor because it is not strictly related to gene amplification. 7 Thus, there must be other genes on chromosome 11q13 which are responsible for poor survival. This is well perceivable because deregulation of the cell cycle alone is probably not sufficient to explain the biological behavior responsible for tumor progression that is mainly characterized by increased invasiveness and the capability of metastasis formation.
Chromosome 3q gain has been defined as the key lesion in the transition from the preinvasive to invasive stage of squamous cell carcinomas of the uterine cervix 20 and was correlated with bad prognosis in our study. Interestingly, losses on the short arm of chromosome 3 did not affect the patients’ outcomes. This is consistent with the observation that 3p deletions along with 9p loss constitute a very early change in tumor development being detectable in precursor lesions. 5
Consistently with our findings, allelic loss on chromosome 8p have been associated with advanced tumor stages and poor survival 20-22 similar to 11qter and 18q deletions. 10,23,24 However, our study also revealed further loci on several other chromosome arms.
For chromosomes 14q and 15q, the histogram in Figure 2 ▶ indicated small regions of deletion that does well correspond with the previously reported minimal regions identified by allelotyping. However, these studies suggested that deletions rather than DNA gains are associated with bad prognosis or tumor progression. 11,25,26 This apparent contradiction can be explained by the consideration that deletions of one allele may be associated with the numerical gain of the second chromosome. Such duplication would result in a shift of the ratio profile, eg, indicating a gain of the telomeric chromosome part instead of a loss of the centromeric region. The histogram curves showing deletions preferentially of the centromeric part as well as overrepresentations in the distal regions are consistent with this hypothesis which however needs to be substantiated by further studies. The findings for chromosomes 14 and 15 also need to be interpreted cautiously because the number of cases carrying these alterations is relatively low compared with other loci. In general, it is important to be aware of the methodological pros and cons of CGH compared to microsatellite analysis being particularly the capacity for the detection of amplifications but the lack of information on the allelic status.
A major result of our study is the fact that the genetic changes, ie, gains of 3q21-29, 11q13, and loss of 8p21-22, were independent markers and carried a higher significance than the classical clinicopathological parameters. In addition, these markers enabled a molecular dissection of conventionally defined subgroups, particularly the patients with pN0 tumors. Importantly, the findings were derived from the investigation of primary tumors. Thus our data clearly point to the feasibility and necessity of a genetic characterization of HNSCC for a refined stratification of patient subgroups which will hopefully pave the way for an individualized treatment finally improving the prognosis of this often lethal disease.
Acknowledgments
The technical assistance of Manuela Pacyna-Gengelbach and Nicole Deutschmann is gratefully acknowledged.
Footnotes
Address reprint requests to Iver Petersen, M.D., Institute of Pathology, Charité Hospital, Humboldt University, Schumannstrasse 20-21, D-10098 Berlin, Germany. E-mail: iver.petersen@charite.de.
Supported by the German Research Foundation (DFG, Pe 602/2).
References
- 1.American Joint Committee on Cancer: Purposes and principles of staging. AJCC Cancer Staging Manual, ed 5. Philadelphia, Lippincott-Raven, 1997, pp 3–9
- 2.Grau JJ, Cuchi A, Traserra J, Firvida L, Arias C, Blanch JL, Estape J: Follow-up study in head and neck cancer. Cure rate according to tumor location and stage. Oncology 1997, 54:38-42 [DOI] [PubMed] [Google Scholar]
- 3.Schantz SP, Harrison LB, Forastiere AA: Tumors of the nasal cavity and paranasal sinuses, nasopharynx, oral cavity, and oropharynx. ed 5 DeVita VT Hellman S Rosenberg SA eds. Cancer: Principles and Practice of Oncology, 1997, :pp 741-801 Lippincott-Raven, Philadelphia [Google Scholar]
- 4.Sessions RB, Harrision LB, Forastiere AA: Tumors of the larynx and hypopharynx. ed 5 DeVita VT Hellman S Rosenberg SA eds. Cancer: Principles and Practice of Oncology, 1997, :pp 802-829 Lippincott-Raven, Philadelphia [Google Scholar]
- 5.Schrader M, Schipper J, Jahnke K, Stuschke M, Sack H, Budach V: Hyperfractionated accelerated simultaneous radiochemotherapy in advanced hypopharyngeal carcinomas. Survival rate, retained function quality of life in a phase II study. HNO 1998, 46:140–145 [DOI] [PubMed]
- 6.Califano J, van der Riet P, Westra W, Nawroz H, Clayman G, Piantadosi S, Corio R, Lee D, Greenberg B, Koch W, Sidransky D: Genetic progression model for head and neck cancer: implications for field cancerization. Cancer Res 1996, 56:2488-2492 [PubMed] [Google Scholar]
- 7.Åkervall JA, Michaelides RJAM, Mineta H, Balm A, Borg A, Dictor MR, Jin Y, Loftus B, Mertens F, Wennerberg JP: Amplification of cyclin D1 in squamous cell carcinoma of the head and neck and the prognostic value of chromosomal abnormalities and cyclin D1 overexpression. Cancer 1997, 79:380-389 [PubMed] [Google Scholar]
- 8.Hermanek P, Sobin LH (Eds): TNM Classification of Malignant Tumors, ed 5. Berlin, Springer, 1997
- 9.Bockmühl U, Schwendel A, Dietel M, Petersen I: Distinct patterns of chromosomal alterations in high and low grade head and neck squamous cell carcinomas. Cancer Res 1996, 56:5325-5329 [PubMed] [Google Scholar]
- 10.Bockmühl U, Petersen S, Schmidt S, Wolf G, Jahnke V, Dietel M, Petersen I: Patterns of chromosomal imbalances in metastasizing and nonmetastasizing head and neck cancer. Cancer Res 1997, 57:5213-5216 [PubMed] [Google Scholar]
- 11.Petersen I, Langreck H, Wolf G, Schwendel A, Psille R, Vogt P, Reichel M, Ried T, Dietel M: Small cell lung cancer is characterized by a high incidence of deletions on chromosomes 3p, 4q, 5q, 10q, 13q and 17p. Br J Cancer 1997, 75:79-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roth K, Wolf G, Dietel M, Petersen I: Image analysis for comparative genomic hybridization (CGH) by a windows-based karyotyping program. Anal Quant Cytol Histol 1997, 19:461-474 [PubMed] [Google Scholar]
- 13.Petersen S, Aninat-Meyer M, Schlüns K, Gellert K, Dietel M, Petersen I: Chromosomal alterations in the clonal evolution to the metastatic stage of squamous cell carcinomas of the lung. Br J Cancer 2000, 82:65-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaplan E, Meier P: Nonparametric estimation from incomplete observations. J Am Stat Assoc 1958, 53:457-481 [Google Scholar]
- 15.Cox D: Regression models and life tables (with discussion). J R Stat Soc B 1972, 34:187-220 [Google Scholar]
- 16.Kirchhoff M, Gerdes T, Rose H, Maahr J, Ottesen AM, Lundsteen C: Detection of chromosomal gains and losses in comparative genomic hybridization analysis based on standard reference intervals. Cytometry 1998, 31:163-173 [PubMed] [Google Scholar]
- 17.Moore DH 2nd, Pallavicini M, Cher ML, Gray JW: A t-statistic for objective interpretation of comparative genomic hybridization (CGH) profiles. Cytometry 1997, 28:183–190 [DOI] [PubMed]
- 18.Åkervall JA, Jin Y, Wennerberg JP, Zätterström UK, Kjellén E, Mertens F, Willén R, Mandahl N, Heim S, Mitelman F: Chromosomal abnormalities involving 11q13 are associated with poor prognosis in patients with squamous cell carcinoma of the head and neck. Cancer 1995, 76:853-859 [DOI] [PubMed] [Google Scholar]
- 19.Meredith SD, Levine PA, Burns JA, Gaffey MJ, Weiss LM, Erickson NL, Williams ME: Chromosome 11q13 amplification in head and neck squamous cell carcinoma. Association with poor prognosis. Arch Otolaryngol Head Neck Surg 1995, 121:790-794 [DOI] [PubMed] [Google Scholar]
- 20.Heselmeyer K, Schröck E, du Manoir S, Blegen H, Shah K, Steinbeck R, Auer G, Ried T: Gain of chromosome 3q defines the transition from severe dysplasia to invasive carcinoma of the uterine cervix. Proc Natl Acad Sci USA 1996, 93:479-484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scholnick SB, Haughey BH, Sunwoo JB, El-Mofty SK, Baty JD, Piccirillo JF, Zequeira MR: Chromosome 8 allelic loss and the outcome of patients with squamous cell carcinoma of the supraglottic larynx. J Natl Cancer Inst 1996, 88:1676-1682 [DOI] [PubMed] [Google Scholar]
- 22.Wu CL, Roz L, Sloan P, Read AP, Holland S, Porter S, Scully C, Speight PM, Thakker N: Deletion mapping defines three discrete areas of allelic imbalance on chromosome arm 8p in oral and oropharyngeal squamous cell carcinomas. Genes Chromosom Cancer 1997, 20:347-353 [DOI] [PubMed] [Google Scholar]
- 23.Pearlstein RP, Benninger MS, Carey TE, Zarbo RJ, Torres FX, Rybicki BA, Van Dyke DL: Loss of 18q predicts poor survival of patients with squamous cell carcinoma of the head and neck. Genes Chromosom Cancer 1998, 21:333-339 [DOI] [PubMed] [Google Scholar]
- 24.Weber RG, Scheer M, Born IA, Joos S, Cobbers JMJL, Hofele C, Reifenberger G, Zöller JE, Lichter P: Recurrent chromosomal imbalances detected in biopsy material from oral premalignant and malignant lesions by combined tissue microdissection, universal DNA amplification, and comparative genomic hybridization. Am J Pathol 1998, 153:295-303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee DJ, Koch WM, Yoo G, Longo M, Reed A, Califano J, Brennan JA, Westra WH, Zahurak M, Sidransky D: Impact of chromosome 14q loss on survival in primary head and neck squamous cell carcinoma. Clin Cancer Res 1997, 3:501-505 [PubMed] [Google Scholar]
- 26.Wick W, Petersen I, Schmutzler RK, Wolfarth B, Lenartz D, Bierhoff E, Hümmerich J, Müller DJ, Stangl AP, Schramm J, Wiestler OD, von Deimling A: Evidence for a novel tumor suppressor gene on chromosome 15 associated with progression to a metastatic stage in breast cancer. Oncogene 1996, 12:973-978 [PubMed] [Google Scholar]