Abstract
DPC4 is a candidate tumor suppressor gene on chromosome 18q21, a region that shows high frequencies of allelic losses in pancreatic and colorectal adenocarcinomas. Biallelic inactivation of DPC4 has been reported in half of pancreatic cancers, but are relatively infrequent in other tumor types. The role of DPC4 inactivation in colorectal neoplasms has not been fully characterized. An immunohistochemical assay for Dpc4 protein expression has been recently developed and shown to be a sensitive and specific surrogate for alterations in the DPC4 gene. In this study we examined the expression of Dpc4 protein in formalin-fixed archival tissue from 83 colorectal lesions, including 19 adenomas and 64 sporadic adenocarcinomas (11 stage I, 13 stage II, 17 stage III, and 23 stage IV cancers). None of the adenomas or stage I adenocarcinomas showed loss of Dpc4 expression, whereas one of 13 (8%) stage II, one of 17 (6%) stage III, and five of 23 (22%) of stage IV cancers showed loss of Dpc4 expression. There was a borderline significant difference in loss of Dpc4 reactivity in colorectal tumors with distant metastasis at presentation (22%) versus primary tumors without distant metastasis (5%) (Fisher’s exact test, P = 0.05; χ2 = 0.04). Poorly differentiated histology or status of pericolonic lymph nodes did not affect Dpc4 expression. Alterations in DPC4 are involved in the progression of a subset of colorectal carcinomas, especially those that present with advanced disease. In the sequential pathogenesis of colorectal tumors, inactivation of DPC4 is likely to be a late event.
The DPC4 gene (for deleted in pancreatic carcinoma, locus 4) has been identified as a candidate tumor suppressor gene in pancreatic adenocarcinomas. 1,2 The DPC4 gene is located on chromosome 18q21.1, a region that also contains the deleted in colon carcinoma (DCC) gene, 3 and is characterized by high frequency of allelic losses in pancreatic and colorectal carcinomas. 4-6 Inactivation of DPC4 can occur by one of two identified mechanisms: a) intragenic mutation of one allele coupled with loss of the other allele, or b) deletion of both alleles (homozygous deletions). Both mutations and homozygous deletions of the DPC4 gene have been observed in a high proportion of pancreatic carcinomas. 7 Germ-line mutations of DPC4 were recently described in familial juvenile polyposis, 8-10 a condition that predisposes to colorectal cancer, and Dpc4 knockout mice have been reported to develop gastrointestinal polyps resembling juvenile polyps. 11 Moreover, compound mutant mice with both Dpc4 and Apc mutations have demonstrated a significant contribution of loss of Dpc4 function to progression of colorectal cancers. 12 In contrast, the role of DPC4 in human colorectal cancers remains less well defined. Although as many as 60% of colorectal cancers show loss of heterozygosity (LOH) at the 18q21.2 locus, 6 only ∼14% show mutations or homozygous deletions of DPC4 (Table 1) ▶ . It is possible that additional epigenetic mechanisms such as promoter hypermethylation 13 may contribute to inactivating the second DPC4 allele in a subset of cases, but the frequency of this pathway has not been reported so far.
Table 1.
Review of Literature on DPC4 Alterations in Colorectal Neoplasms
| Author (reference) | System | Total tumors examined (n) | DPC4 alteration (%) |
|---|---|---|---|
| Thiagalingam et al 6 | Xenografts | 18 | 6 (33%) |
| MacGrogan et al 29 | Cell lines and primary tumors | 17 | 4 (23%) |
| Takagi et al 30 | Primary tumors | 31 | 5 (16%) |
| Akiyama et al 31 | Primary tumors | 30 | 4 (13%) |
| Koyama et al 32 | Primary tumors | 64 | 7 (11%) |
| Tarafa et al 33 | Xenografts | 13 | 4 (31%) |
| Lei et al 34 | Primary tumors | 10 | 0 (0%) |
| Hoque et al 35 | Primary tumors | 6 | 1 (16%) |
| Miyaki et al 36 | Primary tumors | 176 | 21 (12%) |
| Total | 365 | 52 (14.2%) |
Recently, an immunohistochemical assay has been developed for the determination of Dpc4 expression in archival paraffin-embedded tissues. 14,15 The advantages of an immunohistochemical assay are: 1) down-regulation of protein expression can be directly measured irrespective of mechanism(s) of inactivation; 2) it can be applied to a large number of archived tissue samples in a routine manner; and 3) the specific cell types expressing the protein of interest can be assessed by morphology. In pancreatic adenocarcinomas, immunohistochemical labeling for Dpc4 has been found to be an extremely sensitive (91%) and specific (94%) surrogate for DPC4 genetic alterations. 14 We investigated the expression of Dpc4 protein in 83 colorectal lesions, both adenomas and carcinomas, and correlated loss of Dpc4 expression with the stage of the lesion at presentation. This is the first study to systematically examine Dpc4 expression in a series of colorectal neoplasms that have been stratified by stage.
Materials and Methods
Eighty-three cases of colorectal lesions were retrieved from the surgical pathology archives of Parkland Memorial and Zale Lipshy Hospitals, affiliated with the University of Texas Southwestern Medical Center, Dallas. Clinicopathological correlates such as age, sex, site and size of lesions, pathological stage at presentation, histological grade, and presence of lymph node and distant metastases were retrieved from the corresponding surgical pathology reports. The pathological staging designated in the individual cases were jointly verified by two of the authors (AM and GL). The carcinomas were staged using criteria defined by the American Joint Committee on Cancer for staging of colorectal cancers. 16 Multiple hematoxylin and eosin-stained slides from each case were screened by light microscopy for selection of sections with both neoplastic and nonneoplastic colonic tissue. Unstained 5-μm sections were cut from the formalin-fixed paraffin-embedded block selected in each case, and deparaffinized by routine techniques. The slides were treated with sodium citrate buffer (Ventana BioTek Solutions, Tucson, AZ) and steamed at 80°C for 20 minutes. After cooling for 5 minutes, the slides were labeled with monoclonal antibody to Dpc4 (1:100 dilution, clone B8; Santa Cruz Biotechnology, Santa Cruz, CA) using the Bio TekMate 1000 automated immunostainer (Ventana). The detection step was performed by streptavidin-biotin labeling followed by counterstaining with hematoxylin. Positive and negative controls were analyzed in each run, with formalin-fixed normal pancreatic tissue being used as positive control. Normal pancreatic ducts show strongly positive Dpc4 reactivity, whereas moderate expression is present in the acini, islets of Langerhans, stromal fibroblasts, and lymphocytes. 14
The immunohistochemical stains were independently evaluated by two of the authors (AM and GL). When present, Dpc4 is most often seen in the cytoplasmic compartment of cells, with focal nuclear staining only. A two-tier scoring system (positive and negative) was used for analysis, with negative labeling being defined as absence of Dpc4 expression in both the cytoplasmic and nuclear compartments. Positive labeling was defined as either diffuse expression of Dpc4 in the cytoplasm of neoplastic cells, with concomitant focal expression in nuclei or the presence of two distinct populations of cells, those that labeled with the antibody and those that did not. In other words, no distinction was made between diffuse- and focal-positive categories. This was primarily because in our study, the proportion of cases with focal-positive staining were too few to be analyzed separately, a finding also seen in a previous study on pancreatic adenocarcinomas. 14 Nonneoplastic colonic epithelium, stromal fibroblasts, and lymphoid aggregates, all of which expressed Dpc4 with a moderate to strong intensity, served as internal positive controls. The 83 colorectal lesions (including 11 cases in which metastatic lesions were examined simultaneously) were then stratified by stage at presentation and compared with regards to the presence or absence of Dpc4 expression.
Statistical analysis was performed using the Fisher’s exact probability test and chi-square analysis using the SAS statistical software (Cary, NC), to determine whether loss of Dpc4 reactivity in colorectal tumors with distant metastases versus localized primary tumors was significantly different.
Results
Clinicopathological Parameters
The clinical and pathological data in the 83 patients whose colorectal lesions were analyzed in this study are summarized in Table 2 ▶ . The patients were stratified into five subgroups for analysis: 19 patients were diagnosed with benign colorectal adenomas, whereas 64 patients had colorectal cancers (stage I , 11 patients; stage II, 13 patients; stage III, 17 patients; and stage IV, 23 patients). There were 33 males and 50 females (M:F ratio, 1:1.5), with an age range of 41 to 88 years (mean, 65.1 years). The mean ages for the five patient subgroups are summarized in Table 1 ▶ . To the best of our knowledge, none of the 83 patients was diagnosed with familial adenomatous polyposis. There were 44 left-sided, 30 right-sided, and nine transverse colonic lesions (overall ratio, 4.9:3.3:1). The adenomas ranged in size from 1 to 3 cm (mean, 1.3 cm), whereas the carcinomas ranged in size from 1 to 13 cm (mean, 5.0 cm), with an overall mean of 4.2 cm. There were 14 tubular adenomas and five tubulovillous adenomas, two of which had high-grade dysplasia.The majority (55 of 64) of the carcinomas were moderately differentiated adenocarcinomas (G2), whereas nine were poorly differentiated (G3). Of the 40 stage III to IV cancers, six did not have lymph node metastasis in the pericolonic lymph nodes examined (N0), whereas 20 and 14 tumors, respectively, had less than four (N1) and greater than or equal to four (N2) involved lymph nodes in the resected specimen. There were 23 patients with distant metastases (stage IV), 22 of which occurred in the liver. All patients with stage IV cancers had documentation of metastasis by pathological examination at the time of primary resection.
Table 2.
Clinical and Pathological Features in 83 Patients with Colorectal Lesions
| n | Sex ratio | Age range (mean) | Location (L:R:T) | Size range, cm (mean) | Histology | LN status | Distant metastasis | |
|---|---|---|---|---|---|---|---|---|
| Overall | 83 | 1:1.5 | 41–88 | 4.9:3.3:1 | 1–13 | 64 | 6n0: 20N1: | 23 metastases |
| (65.1) | (4.2) | carcinomas | 14N2 | (stage IV only) | ||||
| 55G2: 9G3 | (stage III–IV only) | |||||||
| Adenomas | 19 | 50–79 | 3.7:1:1.7 | 1–3 | 14T: 5TV | All N0 | NA | |
| (62.5) | (1.3) | |||||||
| Stage I | 11 | 44–88 | 5:5:1 | 1–9 | 10G2: 1G3 | All N0 | NA | |
| (67.8) | (3.6) | |||||||
| Stage II | 13 | 46–85 | 1:1:0 | 3–12 | 12G2: 1G3 | All N0 | NA | |
| (66.6) | (5.4) | |||||||
| Stage III | 17 | 57–84 | 2.5:4.5:1 | 1–8 | 12G2: 5G3 | 12N1: 5N2 | NA | |
| (68.8) | (4.5) | |||||||
| Stage IV | 23 | 41–85 | 16:6:1 | 3–13 | 21G2: 2G3 | 6N0: 8N1: | 22 Liver; | |
| (62.6) | (6.0) | 9N2 | 1 uterus and ovaries |
G1, well differentiated; G2, moderately differentiated; G3, poorly differentiated; N1, 1 to 3 pericolonic lymph nodes involved; N2, ≥4 pericolonic lymph nodes involved; L:R:T, left:right:transverse colon.
Immunohistochemical Analysis and Statistical Correlation
The results of immunohistochemical analysis are summarized in Table 3 ▶ . Nonneoplastic colonic mucosa in all patients examined showed strong to moderate immunoreactivity for Dpc4.3 Similarly, 19 of 19 (100%) adenomas and 11 of 11 (100%) stage I lesions had positive Dpc4 expression in the neoplastic cells (Figure 1) ▶ . In contrast, one of 13 (8%) stage II, one of 17 (6%) stage III, and five of 23 (22%) stage IV lesions showed loss of Dpc4 immunoreactivity in the tumor cells (Figures 2 and 3) ▶ ▶ . Overall, three lesions had focal Dpc4 reactivity: two stage IV carcinomas had ∼5% weakly positive cells whereas 95% of the tumor cells were nonreactive—these two cases were classified as negative for statistical analysis. In one stage III carcinoma, there were nearly equal proportions of totally negative and strongly positive islands of tumor cells—this case was classified as positive for statistical analysis. In 11 stage IV cancers, the concomitant metastatic tumors in the liver were also analyzed for Dpc4 expression. Ten of 10 (100%) lesions showed persistence of Dpc4 expression in both the primary and metastatic tumors, whereas one lesion showed loss of expression at both sites (Figure 4) ▶ . Moderate Dpc4 expression was seen in the surrounding hepatocytes in all metastatic sections.
Table 3.
Immunohistochemical Expression of Dpc4 in 83 Colorectal Lesions
| Dpc4-positive | Dpc4- negative | |
|---|---|---|
| Overall | 76/83 (92%) | 7/83 (8%) |
| Adenomas | 19/19 (100%) | 0 |
| Stage I | 11/11 (100%) | 0 |
| Stage II | 12/13 (92%) | 1/13 (8%) |
| Stage III | 16/17 (94%) | 1/17 (6%) |
| Stage IV | 18/23 (78%) | 5/23 (22%) |
Figure 1.
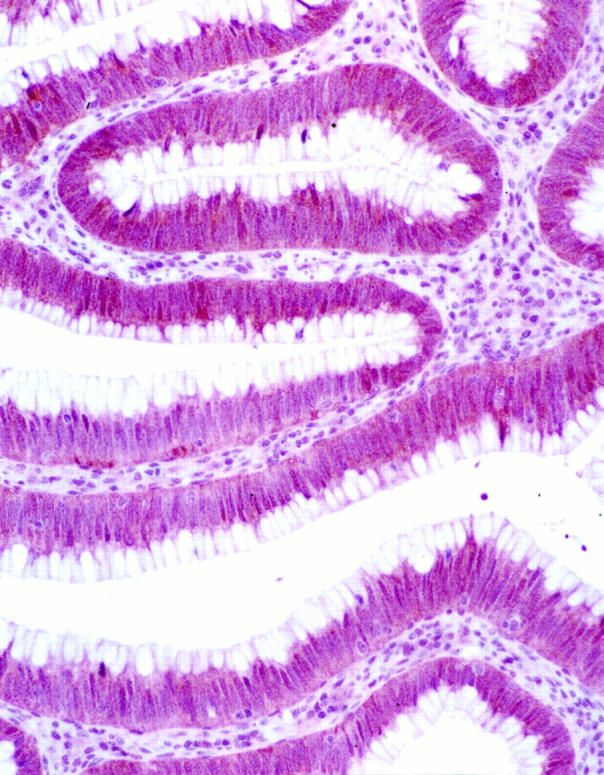
Expression of Dpc4 in tubular adenoma, hematoxylin counterstain. Original magnification, ×40.
Figure 2.

Loss of expression of Dpc4 in primary adenocarcinoma of the colon, hematoxylin counterstain. Original magnification, ×40.
Figure 3.

Loss of expression of Dpc4 in primary adenocarcinoma of the colon, with retention of strong immunoreactivity in the adjacent normal colonic mucosa, hematoxylin counterstain. Original magnification, ×40.
Figure 4.

Loss of expression of Dpc4 in metastatic colon cancer, hematoxylin counterstain. Original magnification, ×40. Adjacent hepatocytes show Dpc4 immunoreactivity.
Table 4 ▶ summarizes the clinicopathological data in the seven cases with loss of Dpc4 expression. The presence of poorly differentiated histology did not affect Dpc4 expression, but this could be because of an overrepresentation of moderately differentiated adenocarcinomas in our series (Table 2) ▶ . Similarly, no trend was seen between loss of Dpc4 expression and the presence or absence of nodal metastasis in the pericolonic lymph nodes, with both N0 and N1/N2 lesions showing loss of Dpc4 expression. There was a borderline statistically significant difference in loss of Dpc4 expression in colorectal carcinomas that presented with distant metastasis (5 of 23 or 22%) versus colorectal cancers without distant metastasis (2 of 41 or 5%) (Fisher’s exact test, P = 0.05, chi-square = 0.04). There was a statistically significant difference in loss of Dpc4 expression in colorectal carcinomas that presented with distant metastasis versus all localized colorectal neoplasms, including adenomas (2 of 60 or 3%) (Fisher’s exact test, P = 0.01, chi-square = 0.007).
Table 4.
Clinicopathological Characteristics of Seven Colorectal Tumors with Loss of Dpc4 Expression
| Stage | Age/Sex | Site | Size | Histology | LN status | Metastasis | |
|---|---|---|---|---|---|---|---|
| Case 1 | IV | 59 /F | Left | 5 | G2 | N1 | Liver |
| Case 2 | IV | 43 /F | Right | 10 | G2 | N0 | Uterus/ovaries |
| Case 3 | IV | 56 /M | Left | 3 | G2 | N2 | Liver |
| Case 4 | IV | 62 /M | Left | 5 | G2 | N1 | Liver |
| Case 5 | IV | 64 /M | Left | 6 | G2 | N0 | Liver |
| Case 6 | III | 75 /F | Transverse | 4 | G2 | N1 | NA |
| Case 7 | II | 85 /M | Left | 4 | G2 | NA | NA |
G1, well differentiated; G2, moderately differentiated; G3, poorly differentiated; N1, 1 to 3 pericolonic lymph nodes involved; N2, ≥4 pericolonic lymph nodes involved; L:R:T, left:right:transverse colon; NA, not applicable.
Discussion
Colorectal cancer accounted for an estimated 129,400 new cases in 1999, including 94,700 of colon cancer and 34,700 of rectal cancer (http://www.cancer.org/). Colorectal cancer is the third most common cancer in men and women. An estimated 56,600 deaths from colorectal cancers occurred in 1999, accounting for ∼10% of cancer deaths (http://www.cancer.org/). Most colorectal carcinomas develop by a pathway of sequential progression from adenomas. 17 Considerable data has accumulated throughout the past decade on the underlying genetic changes that accumulate during the multistep pathogenesis of colorectal neoplasms, primarily based on studies in the two major forms of hereditary colon cancers: familial adenomatous polyposis and hereditary nonpolyposis colorectal cancers. It is now known that mutations of the Ki-ras oncogene, 18 APC (adenomatous polyposis coli gene), 19,20 p53, 5,21 and DCC (located at 18q21) 3,22 are important genetic events in the stepwise progression of colorectal cancers. The 18q21 region is thought to be particularly important locus because as many as 60% of colorectal cancers show LOH at this locus. 6 Despite the high proportion of allelic losses however, there have been few reports of inactivating mutations of DCC in these tumors. In 1996, a second candidate tumor suppressor gene, DPC4, was cloned in the vicinity of DCC raising the possibility that it may be the target of allelic losses at 18q21. 1 The product of DPC4 belongs to the evolutionary conserved family of SMAD proteins which are involved in transforming growth factor-β signal transudation pathways. 23,24 Unlike pancreatic adenocarcinomas, where 50% of tumors show complete loss of DPC4 function (20% mutations and 30% homozygous deletions), 4,7 biallelic inactivation of DPC4 seems to be infrequent in other tumor types. 2,25-28
In the context of colorectal cancers, Thiagalingam et al 6 reported mutations of DPC4 in four of 18 xenografted tumors, and homozygous deletions in two additional tumors, that also spanned the DCC gene. Since that time, several investigators have examined the role of DPC4 in colorectal cancers at the genomic and transcriptional levels (Table 1) ▶ . MacGrogan et al 29 described altered DPC4 sequences in three of 12 colon cancer cell lines and one of five primary tumors, whereas Takagi et al 30 found mutations of DPC4 in five of 31 (16%) primary tumors. Akiyama et al 31 described DPC4 alterations in four of 30 (13%) of early superficial colorectal cancers; interestingly none of their cases harbored either DCC or smad2 alterations, despite a 65% frequency of LOH at 18q21. Similarly, Koyama et al, 32 who found no more than seven of 64 (11%) of tumors with DPC4 mutations despite a 78% LOH at 18q21 in their stage II and III colorectal cancers. Tarafa et al 33 found loss of DPC4 expression by reverse transcriptase-polymerase chain reaction in four of 13 (31%) colorectal xenografts; two tumors contained homozygous deletions and two had coding region mutations. In contrast, Lei et al 34 found no mutations in the coding region of DPC4 in 10 cases of colorectal cancers arising in the background of ulcerative colitis, whereas Hoque et al 35 reported biallelic inactivation of DPC4 in one of six colitis-associated neoplasia. A systematic analysis of DPC4 alterations in progressive stages of colorectal tumors was performed by Miyaki et al 36 in 176 colorectal carcinomas, including 36 metatstatectomy specimens. The authors found DPC4 mutations in six of 17 (35%) primary carcinomas with distant metastases, compared with 0%, 10%, and 7% DPC4 mutations in adenomas, intramucosal carcinomas, and primary carcinomas without distant metastases, respectively. DPC4 mutations were also present in 11 of 36 (31%) distant metastases, including four cases where both the primary and metastasis harbored identical mutations. 36 This was one of the strongest evidences to date that DPC4 is a true target of inactivation in colorectal cancers, and loss of DPC4 function correlated with advanced stages of malignancy, such as distant metastasis. Combining the data from literature on DPC4 inactivation in colorectal neoplasms (including cell lines, xenografts, and primary tumors of all stages) yielded a total of 52 of 365 (14.2%) tumors that have shown genetic alterations of DPC4 at the DNA and/or RNA level (Table 1) ▶ .
We analyzed the expression of Dpc4 protein in a series of 83 colorectal neoplasms (19 adenomas, 11 stage I, 13 stage II, 17 stage III, and 23 stage IV) using a recently developed immunohistochemical assay applicable to paraffin-embedded material. The Dpc4 immunohistochemical assay has been shown to have a high degree of sensitivity and specificity for alterations in DPC4 gene. 14 We found loss of Dpc4 expression in 8% of colorectal neoplasms, all of which were limited to cancers that were stage II or higher. Although this is lower than the proportion of tumors calculated on the basis of literature review (Table 1 ▶ , 14.2%), our series also includes adenomas and low-stage carcinomas, whereas many of the previous studies were performed on xenografts, cell lines, or advanced stage tumors. Both xenografts and cell lines have a potential for selective propagation of aggressive clones or acquisition of additional genetic changes during culture, including those of DPC4. 37-39 When stratified by stage, the highest percentage of loss of Dpc4 reactivity was found in stage IV cancers with distant metastasis (22%), compared with 0% in stage I, 8% in stage II, and 6% in stage III cancers (Fisher’s exact test, P = 0.05, chi-square = 0.04).
The figures in our study correlated fairly well with what has been reported by Miyaki et al 36 in the stage-wise progression of colorectal cancers, although they had a slightly higher frequency of DPC4 mutations in stage IV cancers. Taken in conjunction, these findings suggest that inactivation of DPC4 1) is a late event in colorectal carcinogenesis, and 2) might be permissive for acquisition of a metastatic phenotype in these tumors. Interestingly, we failed to find loss of Dpc4 expression in distant metastatic deposits from 10 lesions in which the primary tumor also expressed Dpc4. In one case, both the primary and metastasis showed loss of Dpc4 expression. Miyaki et al 36 also did not find significant differences in the rate of DPC4 mutations in stage IV cancers versus distant metastases (36% versus 31%). This suggests that DPC4 inactivation may be permissive for acquisition of a metastatic phenotype, but additional genetic changes at DPC4 rarely occur during dissemination. In the current study, tumors with a poorly differentiated histology or presence of pericolonic nodal metastasis did not show any trend toward loss of Dpc4 expression.
In conclusion, increasing evidence from the literature suggests an important role for DPC4 inactivation in a subset of colorectal cancers, based on genetic analyses. This is the first study to perform a systematic immunohistochemical analysis of Dpc4 expression in a series of sporadic colorectal tumors stratified by stage, and show that the loss of Dpc4 expression is a late event in colorectal tumors, that correlates with the presence of metastatic disease at presentation. The overall low frequency of Dpc4 inactivation in this study contrasts with the high frequency of LOH at 18q21, suggesting the existence of another putative tumor suppressor gene at this locus in addition to DCC and DPC4.
Footnotes
Address reprint requests to Anirban Maitra, M.D., Department of Pathology, University of Texas Southwestern Medical Center, 5323 Harry Hines Blvd., Dallas, TX 75390-9073. E-mail: maitra.anirban@pathology.swmed.edu.
References
- 1.Hahn SA, Schutte M, Hoque AT, Moskaluk CA, da Costa LT, Rozenblum E, Weinstein CL, Fischer A, Yeo CJ, Hruban RH, Kern SE: DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science 1996, 271:350-353 [DOI] [PubMed] [Google Scholar]
- 2.Riggins GJ, Kinzler KW, Vogelstein B, Thiagalingam S: Frequency of Smad gene mutations in human cancers. Cancer Res 1997, 57:2578-2580 [PubMed] [Google Scholar]
- 3.Fearon ER, Cho KR, Nigro JM, Kern SE, Simons JW, Ruppert JM, Hamilton SR, Preisinger AC, Thomas G, Kinzler KW, Vogelstein B: Identification of a chromosome 18q gene that is altered in colorectal cancers. Science 1990, 247:49-56 [DOI] [PubMed] [Google Scholar]
- 4.Hahn SA, Hoque AT, Moskaluk CA, da Costa LT, Schutte M, Rozenblum E, Seymour AB, Weinstein CL, Yeo CJ, Hruban RH, Kern SE: Homozygous deletion map at 18q21.1 in pancreatic cancer. Cancer Res 1996, 56:490-494 [PubMed] [Google Scholar]
- 5.Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM, Bos JL: Genetic alterations during colorectal-tumor development. N Engl J Med 1988, 319:525-532 [DOI] [PubMed] [Google Scholar]
- 6.Thiagalingam S, Lengauer C, Leach FS, Schutte M, Hahn SA, Overhauser J, Willson JK, Markowitz S, Hamilton SR, Kern SE, Kinzler KW, Vogelstein B: Evaluation of candidate tumour suppressor genes on chromosome 18 in colorectal cancers. Nat Genet 1996, 13:343-346 [DOI] [PubMed] [Google Scholar]
- 7.Hruban RH, Offerhaus GJA, Kern SE, Goggins M, Wilentz RE, Yeo CJ: Tumor-suppressor genes in pancreatic cancer. J Hepatobiliary Pancreat Surg 1998, 5:383-391 [DOI] [PubMed] [Google Scholar]
- 8.Howe JR, Roth S, Ringold JC, Summers RW, Jarvinen HJ, Sistonen P, Tomlinson IP, Houlston RS, Bevan S, Mitros FA, Stone EM, Aaltonen LA: Mutations in the SMAD4/DPC4 gene in juvenile polyposis. Science 1998, 280:1086-1088 [DOI] [PubMed] [Google Scholar]
- 9.Houlston R, Bevan S, Williams A, Young J, Dunlop M, Rozen P, Eng C, Markie D, Woodford-Richens K, Rodriguez-Bigas MA, Leggett B, Neale K, Phillips R, Sheridan E, Hodgson S, Iwama T, Eccles D, Bodmer W, Tomlinson I: Mutations in DPC4 (SMAD4) cause juvenile polyposis syndrome, but only account for a minority of cases. Hum Mol Genet 1998, 7:1907-1912 [DOI] [PubMed] [Google Scholar]
- 10.Friedl W, Kruse R, Uhlhaas S, Stolte M, Schartmann B, Keller KM, Jungck M, Stern M, Loff S, Back W, Propping P, Jenne DE: Frequent 4-bp deletion in exon 9 of the SMAD4/MADH4 gene in familial juvenile polyposis patients. Genes Chromosom Cancer 1999, 25:403-406 [PubMed] [Google Scholar]
- 11.Wambach J, Barnard J: SMAD4 germline mutations in juvenile polyposis coli. J Pediatr Gastroenterol Nutr 1999, 28:538-539 [DOI] [PubMed] [Google Scholar]
- 12.Takaku K, Oshima M, Miyoshi H, Matsui M, Seldin MF, Taketo MM: Intestinal tumorigenesis in compound mutant mice of both Dpc4(smad4) and Apc genes. Cell 1998, 92:645-656 [DOI] [PubMed] [Google Scholar]
- 13.Belinsky SA, Nikula KJ, Palmisano WA, Michels R, Saccomanno G, Gabrielson E, Baylin SB, Herman JG: Aberrant methylation of p16(INK4a) is an early event in lung cancer and a potential biomarker for early diagnosis. Proc Natl Acad Sci USA 1998, 95:11891-11896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilentz RE, Su GH, Dai JL, Sparks AB, Argani P, Sohn TA, Yeo CJ, Kern SE, Hruban RH: Immunohistochemical labeling for dpc4 mirrors genetic status in pancreatic adenocarcinomas: a new marker of DPC4 inactivation. Am J Pathol 2000, 156:37-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilentz RE, Iacobuzio-Donahue CA, Argani P, McCarthy DM, Parsons JL, Yeo CJ, Kern SE, Hruban RH: Loss of expression of Dpc4 in pancreatic intraepithelial neoplasia: evidence that DPC4 inactivation occurs late in neoplastic progression. Cancer Res 2000, 60:2002-2006 [PubMed] [Google Scholar]
- 16.Cohen AM, Tremiterra S, Candela F, Thaler HT, Sigurdson ER: Prognosis of node-positive colon cancer. Cancer 1991, 67:1859-1861 [DOI] [PubMed] [Google Scholar]
- 17.Fearon ER, Vogelstein B: A genetic model for colorectal tumorigenesis. Cell 1990, 61:759-767 [DOI] [PubMed] [Google Scholar]
- 18.Bos JL, Fearon ER, Hamilton SR, Verlaan-de Vries M, van Boom JH, van der Eb AJ, Vogelstein B: Prevalence of ras gene mutations in human colorectal cancers. Nature 1987, 327:293-297 [DOI] [PubMed] [Google Scholar]
- 19.Bodmer WF, Bailey CJ, Bodmer J, Bussey HJ, Ellis A, Gorman P, Lucibello FC, Murday VA, Rider SH, Scambler P, Sheer D, Solomon E, Spurr NK: Localization of the gene for familial adenomatous polyposis on chromosome 5. Nature 1987, 328:614-616 [DOI] [PubMed] [Google Scholar]
- 20.Leppert M, Dobbs M, Scambler P, O’Connell P, Nakamura Y, Stauffer D, Woodward S, Burt R, Hughes J, Gardner E, Lathrop M, Nasmuth J, Lalouell J-M, White R: The gene for familial polyposis coli maps to the long arm of chromosome 5. Science 1987, 238:1411-1413 [DOI] [PubMed] [Google Scholar]
- 21.Baker SJ, Fearon ER, Nigro JM, Hamilton SR, Preisinger AC, Jessup JM, vanTuinen P, Ledbetter DH, Barker DF, Nakamura Y, White R, Vogelstein B: Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science 1989, 244:217-221 [DOI] [PubMed] [Google Scholar]
- 22.Kikuchi-Yanoshita R, Konishi M, Fukunari H, Tanaka K, Miyaki M: Loss of expression of the DCC gene during progression of colorectal carcinomas in familial adenomatous polyposis and non-familial adenomatous polyposis patients. Cancer Res 1992, 52:3801-3803 [PubMed] [Google Scholar]
- 23.Kawabata M, Imamura T, Inoue H, Hanai J, Nishihara A, Hanyu A, Takase M, Ishidou Y, Udagawa Y, Oeda E, Goto D, Yagi K, Kato M, Miyazono K: Intracellular signaling of the TGF-beta superfamily by Smad proteins. Ann NY Acad Sci 1999, 886:73-82 [DOI] [PubMed] [Google Scholar]
- 24.Moskaluk CA, Kern SE: Cancer gets Mad: DPC4 and other TGFbeta pathway genes in human cancer. Biochim Biophys Acta 1996, 1288:M31-M33 [DOI] [PubMed] [Google Scholar]
- 25.Powell SM, Harper JC, Hamilton SR, Robinson CR, Cummings OW: Inactivation of Smad4 in gastric carcinomas. Cancer Res 1997, 57:4221-4224 [PubMed] [Google Scholar]
- 26.Zhou Y, Kato H, Shan D, Minami R, Kitazawa S, Matsuda T, Arima T, Barrett JC, Wake N: Involvement of mutations in the DPC4 promoter in endometrial carcinoma development. Mol Carcinog 1999, 25:64-72 [DOI] [PubMed] [Google Scholar]
- 27.Yakicier MC, Irmak MB, Romano A, Kew M, Ozturk M: Smad2 and Smad4 gene mutations in hepatocellular carcinoma. Oncogene 1999, 18:4879-4883 [DOI] [PubMed] [Google Scholar]
- 28.Maesawa C, Tamura G, Nishizuka S, Iwaya T, Ogasawara S, Ishida K, Sakata K, Sato N, Ikeda K, Kimura Y, Saito K, Satodate R: MAD-related genes on 18q21.1, Smad2 and Smad4, are altered infrequently in esophageal squamous cell carcinoma. Jpn J Cancer Res 1997, 88:340-343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacGrogan D, Pegram M, Slamon D, Bookstein R: Comparative mutational analysis of DPC4 (Smad4) in prostatic and colorectal carcinomas. Oncogene 1997, 15:1111-1114 [DOI] [PubMed] [Google Scholar]
- 30.Takagi Y, Kohmura H, Futamura M, Kida H, Tanemura H, Shimokawa K, Saji S: Somatic alterations of the DPC4 gene in human colorectal cancers in vivo. Gastroenterology 1996, 111:1369-1372 [DOI] [PubMed] [Google Scholar]
- 31.Akiyama Y, Arai T, Nagasaki H, Yagi OK, Nakahata A, Nakajima T, Ohkura Y, Iwai T, Saitoh K, Yuasa Y: Frequent allelic imbalance on chromosome 18q21 in early superficial colorectal cancers. Jpn J Cancer Res 1999, 90:1329-1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koyama M, Ito M, Nagai H, Emi M, Moriyama Y: Inactivation of both alleles of the DPC4/SMAD4 gene in advanced colorectal cancers: identification of seven novel somatic mutations in tumors from Japanese patients. Mutat Res 1999, 406:71-77 [DOI] [PubMed] [Google Scholar]
- 33.Tarafa G, Villanueva A, Farre L, Rodriguez J, Musulen E, Reyes G, Seminago R, Olmedo E, Paules AB, Peinado MA, Bachs O, Capella G: DCC and SMAD4 alterations in human colorectal and pancreatic tumor dissemination. Oncogene 2000, 19:546-555 [DOI] [PubMed] [Google Scholar]
- 34.Lei J, Zou TT, Shi YQ, Zhou X, Smolinski KN, Yin J, Souza RF, Appel R, Wang S, Cymes K, Chan O, Abraham JM, Harpaz N, Meltzer SJ: Infrequent DPC4 gene mutation in esophageal cancer, gastric cancer and ulcerative colitis-associated neoplasms. Oncogene 1996, 13:2459-2462 [PubMed] [Google Scholar]
- 35.Hoque AT, Hahn SA, Schutte M, Kern SE: DPC4 gene mutation in colitis associated neoplasia. Gut 1997, 40:120-122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miyaki M, Iijima T, Konishi M, Sakai K, Ishii A, Yasuno M, Hishima T, Koike M, Shitara N, Iwama T, Utsunomiya J, Kuroki T, Mori T: Higher frequency of Smad4 gene mutation in human colorectal cancer with distant metastasis. Oncogene 1999, 18:3098-3103 [DOI] [PubMed] [Google Scholar]
- 37.Reyes G, Villanueva A, Garcia C, Sancho FJ, Piulats J, Lluis F, Capella G: Orthotopic xenografts of human pancreatic carcinomas acquire genetic aberrations during dissemination in nude mice. Cancer Res 1996, 56:5713-5719 [PubMed] [Google Scholar]
- 38.Bartsch D, Barth P, Bastian D, Ramaswamy A, Gerdes B, Chaloupka B, Deiss Y, Simon B, Schudy A: Higher frequency of DPC4/Smad4 alterations in pancreatic cancer cell lines than in primary pancreatic adenocarcinomas. Cancer Lett 1999, 139:43-49 [DOI] [PubMed] [Google Scholar]
- 39.Capella G, Farre L, Villanueva A, Reyes G, Garcia C, Tarafa G, Lluis F: Orthotopic models of human pancreatic cancer. Ann NY Acad Sci 1999, 880:103-109 [DOI] [PubMed] [Google Scholar]


