Abstract
Gastrointestinal stromal tumors (GISTs), the most common mesenchymal tumors of the gastrointestinal tract, typically express the KIT protein. Activating mutations in the juxtamembrane domain (exon 11) of the c-kit gene have been shown in a subset of GISTs. These mutations lead into ligand-independent activation of the tyrosine kinase of c-kit, and have a transforming effect in vitro. Several groups have studied the clinical implication of the c-kit mutation status of exon 11 in GISTs and a possible relationship between c-kit mutations and malignant behavior has been established. Recently, a 1530ins6 mutation in exon 9 and missense mutations, 1945A>G in exon 13 of the c-kit gene were reported. The frequency and clinical importance of these findings are unknown. In this study we evaluated 200 GISTs for the presence of mutations in exons 9 and 13 of c-kit. Six cases revealed 1530ins6 mutation in exon 9 and two cases 1945A>G mutation in exon 13. All tumors with mutations in exon 9 and 13 lacked mutations in exon 11 of c-kit. None of the analyzed tumors had more than one type of c-kit mutation. All but one of the eight tumors with mutations in exon 9 or 13 of the c-kit gene were histologically and clinically malignant. All four of six cases with exon 9 mutation of which location of primary tumor was known, were small intestinal, suggesting that this type of mutation could preferentially occur in small intestinal tumors. Exon 9 and 13 mutations seem to be rare, and they cover only a small portion (8%) of the balance of GISTs that do not have mutations in exon 11 of c-kit. This finding indicates that other genetic alterations may activate c-kit in GISTs, or that KIT is not activated by mutations in all cases.
Gastrointestinal stromal tumors (GISTs) are the most common mesenchymal tumors of the gastrointestinal tract. GISTs differ from other gastrointestinal mesenchymal tumors histologically, immunohistochemically, and genetically. Immunohistochemically they are typically positive for CD117 (KIT protein) and CD34 but negative for S100-protein and desmin and express smooth muscle actin in 20 to 40% of cases. 1
The c-kit gene encodes for a receptor for a growth factor termed stem cell factor. The KIT protein (stem-cell factor receptor) contains an internal tyrosine kinase component and regulates cell growth and survival. 2-5 Constitutional KIT expression has been shown in germ cells, melanocytes, hematopoietic stem cells, mast cells, and the interstitial cells of Cajal. 6-8
Specific mutations in the c-kit gene have been found in myeloproliferative disorders, 9 mast cell neoplasms, 10 seminoma, 11 and in acute myeloid leukemia. 12 Mutations in the juxtamembrane domain (exon 11) of the c-kit gene have been shown in GISTs, 13,14 and in a mast-cell leukemia cell line. 15 The juxtamembrane domain mutations have been shown to lead into ligand-independent activation (phosphorylation) of the tyrosine kinase of c-kit and have a transforming effect in vitro. 13 The c-kit mutation status of exon 11 has been studied in GISTs by several groups 16-20 and the relationship between presence of mutations and malignant behavior has been demonstrated. 16,17,19
Recently, 1530ins6 mutation in exon 9 (extracellular domain) and 1945A>G mutation in exon 13 (kinase domain) of the c-kit gene were reported in a small group of GISTs, that lacked mutations in exon 11 (juxtamembrane domain) of c-kit. 21 In this study, we evaluated the frequency and clinicopathological importance of these mutations in a large series of benign and malignant GISTs.
Materials and Methods
Tissue Material
Two hundred GISTs were obtained from the files of the Armed Forces Institute of Pathology, Washington, DC; the Haartman Institute of the University of Helsinki, Helsinki, Finland; the Maria Sklodowska-Curie Memorial Institute, Krakow, Poland; and the Medical University, Lodz, Poland.
Immunohistochemistry
The tumors were immunohistochemically analyzed for CD34, CD117 (the c-kit proto-oncogene protein product), α-smooth muscle actin, desmin, and S100-protein. Immunohistochemistry was performed by using the avidin-biotin peroxidase complex system and diaminobenizidine as the chromogen, as previously described. 22 Negative and positive controls were included in each run.
Molecular Studies
Exons 9, 11, and 13 of the c-kit gene was evaluated for the mutations by polymerase chain reaction amplification and direct sequencing of the amplification products. Formalin-fixed tumor and normal tissue were microdissected from paraffin blocks. DNA was extracted as previously described. 17 Primer sequences are shown in Table 1 ▶ . Annealing temperatures for all sets of primers was 56°C. Cycling condition and the reaction mix were standard as recommended by Perkin-Elmer-Cetus (Foster City, Ca). Previously described precautions 23 were followed to avoid and monitor possible cross-contamination. The polymerase chain reaction assays amplified fragments containing the entire sequences of the exons 9, 11, and 13 of the c-kit gene. The amplification products were size-fractionated on 2% agarose gels, purified from the gel, and sequenced directly using forward and reverse primers. The sequences were analyzed using the Lasergene software (DNASTAR, Madison, WI) in connection with the data of the GenBank 110/EMBL55 database (January 99 edition).
Table 1.
Oligonucleotide Primers Used in this Study
| Primer | Sequence | PCR products |
|---|---|---|
| CK9.1F | 5′-TCC TAG AGT AAG CCA GGG CTT-3′ | Exon 9 283 base pair |
| CK9.3R | 5′-TGG TAG ACA GAG CCT AAA CAT CC-3′ | |
| CK10.4F | 5′-CCA GAG TGC TCT AAT GAC TG-3′ | Exon 11 284 base pair |
| CK11.3R | 5′-AGC CCC TGT TTC ATA CTG AC-3′ | |
| CK13.1F | 5′-GCT TGA CAT CAG TTT GCC AG-3′ | Exon 13 193 base pair |
| CK13.2R | 5′-AAA GGC AGC TTG GAC ACG GCT TTA-3′ |
Results
Clinicopathological Features
The study consisted of 188 primary and 12 recurrent or metastatic GISTs. Primary tumors represented 10 esophageal, 79 gastric, 42 small intestinal, 19 colonic, 25 rectal, three mesenteric, and three omental GISTs. In seven cases, with the large abdominal mass at presentation, the primary localization could not be established. Nine intraabdominal recurrences and three liver metastasis represented two gastric, four small intestinal, one colonic, and five GISTs of unknown primary location. Of the 200 patients, 116 were male and 84 were female. The age of the patients ranged from 17 to 90 years (median, 60 years). GISTs were diagnosed based on previously published criteria. 1 All analyzed tumors showed GIST-specific immunophenotype; KIT protein expression was documented in all cases. Seventy-four percent of cases showed co-expression of CD34. Forty-one percent and 20% of cases expressed α-smooth muscle actin and S100 protein, respectively. All but four GISTs were desmin-negative. Examples of the histological and immunohistochemical features are shown in Figure 1, A–E ▶ .
Figure 1.
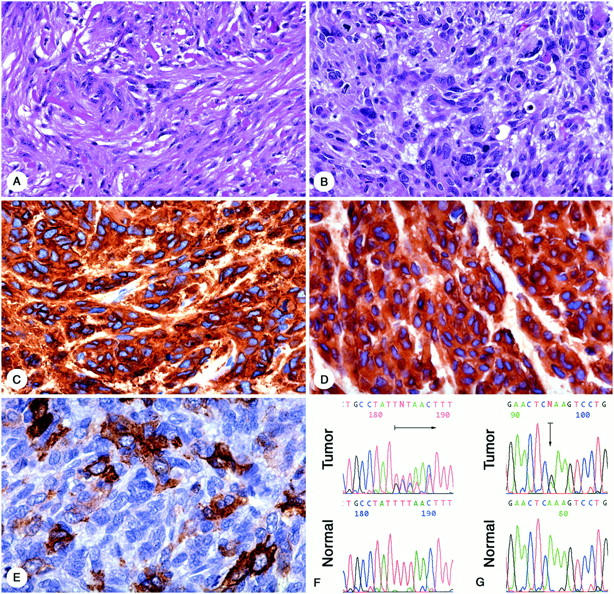
A: Case 1, spindle-cell GIST from small intestine with so-called skeinoid fibers. B: Case 8, pleomorphic GIST from stomach. C: Case 8, positive for CD117. D: Case 8 positive for CD34. E: Case 8, focally positive for α-smooth muscle actin. F: Case 1, sequence analysis of exon 9 from the tumor and corresponding normal tissue. Arrow indicates heterozygous 1530ins6 mutation (GCC TAT). G: Case 7, sequence analysis of exon 13 from tumor and corresponding normal tissue. Arrow indicates heterozygous 1945A>G mutation.
Evaluation of c-kit Mutation
Two hundred GISTs were evaluated for the presence of c-kit mutations. One hundred three (52%) GISTs had mutation in exon 11 of the c-kit gene. Mutation in exon 9, representing insertion of GCC TAT and resulting in duplication of amino acid residues Ala502 and Tyr503, was seen in six GISTs. The exon 13 1945A>G mutation resulting in substitution of a Glu to Lys642 was seen in two GISTs. All mutations found in exon 9 and 13 were heterozygous. Examples of mutated sequences are shown in Figure 1, F and G ▶ . None of the analyzed tumors revealed the presence of more than one type of c-kit mutation. No mutations were found in polymerase chain reaction products amplified from microdissected normal tissue. Exon 9 1530ins6 mutation was found in 6% and exon 13 1945A>G mutation was found in 2% of GISTs that lacked mutation in exon 11. Together mutations in exons 9, 11, or 13 were found in 56% of all analyzed GISTs.
Correlation between Pathological and Genetic Features
Four of six tumors with mutation in exon 9 were localized in the small intestine. In two GISTs (cases 5 and 6), the primary localization could not be clearly established because of the large size and infiltration of different organs. However, in neither case was the small intestine ruled out as a primary location. Six of eight tumors with duplication in exon 9 or point mutation in exon 13 had an aggressive clinical behavior. Clinical follow-up was not available in case 1 who had a 5-cm-large small-intestinal GIST with low mitotic index. The clinicopathological data of GISTs carrying mutation in exons 9 and 13 are summarized in Tables 2 and 3 ▶ ▶ .
Table 2.
Clinicopathological Data of Five GISTs Containing 1530ins6 Mutation in Exon 9
| Case | Age/Sex | Ethnicity | Localization | Tumor size | Cell type | Mitoses/10 HPF | Immunophenotype** | Status and follow-up in months | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CD117 | CD34 | SMA | DES | S100 | ||||||||
| 1 | 35 /F | Hispanic | Small intestine | 5 cm | Spindle | 0 | 100% | 5% | 0% | 0% | 0% | NA |
| 2 | 68 /M | Caucasian | Small intestine | 7× 6 cm | Spindle | 17 | 100% | 0% | 0% | 0% | 0% | DOD (36) |
| 3 | 54 /M | Caucasian | IAR, primary tumor in small intestine | 5 cm | Spindle | 2 | 100% | 0% | 0% | 0% | 0% | Abdominal wall metastasis (48) |
| 4 | 48 /M | Caucasian | Small intestine | 6–7 cm | Epithelioid | 3 | 100% | 0% | 0% | 0% | 60% | DOD (15) |
| 5* | 67 /F | Caucasian | Retroperitoneal mass | 15 cm | Epithelioid | 5 | 100% | 50% | 0% | 0% | 0% | Liver metastasis at operation |
| 6* | 68 /F | Caucasian | Large abdominal mass | 16× 14× 12 | Spindle | 4 | 100% | 40% | 0% | 0% | 0% | DOD (45) |
*Localization of primary tumor could not be clearly established; **, percentage of cells showing positive staining. NA, not available; DOD, died of disease; IAR, intra-abdominal recurrence.
Table 3.
Clinicopathological Data of Two GISTs Containing 1945A>G Mutation in Exon 13
| Case | Age/Sex | Ethnicity | Localization | Tumor size | Cell type | Mitoses/10 HPF | Immunophenotype** | Status and follow-up in months | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CD117 | CD34 | SMA | DES | S100 | ||||||||
| 7 | 70 /F | Caucasian | Esophagus | 25 cm | Spindle | 5 | 100% | 100% | 80% | 0% | 0% | DOD (18) |
| 8 | 53 /M | unknown | Stomach | 3.5 cm | Spindle/epithelioid/ pleomorphic | 29 | 100% | 100% | 15% | 0% | 0% | IAR (6) |
*Localization of primary tumor could not be clearly established; **, percentage of cells showing positive staining. NA, not available; DOD, died of disease; IAR, intra-abdominal recurrence.
Discussion
Activating mutations in the KIT gene have been found in the spectrum of different tumors. The kinase domain of the KIT gene is mutated in mast cell neoplasms and seminomas, 10,11 whereas extracellular and transmembrane domains are mutated in acute myeloid leukemia and in myeloproliferative disorders. 9 In the first study on GISTs mutations were mapped exclusively to exon 11, the KIT juxtamembrane domain. 13
Approximately 40 to 50% of GISTs, mostly the malignant variants, have mutations in the juxtamembrane domain of the c-kit gene, 16,17,19 although some studies found mutations in only 15% 20 or others in as many as 80% 21 of analyzed cases. Lack of mutations in exon 11 in a significant portion of GISTs may suggest that mutations may occur in other domains of the KIT gene. However, no mutations were found in exon 17 (kinase domain) in a large group of GISTs studied; 19 this is the area where KIT mutations occur in mastocytoma 10 and seminoma. 11
Recently, new mutational hotspots were identified as a result of comprehensive sequencing of KIT cDNA obtained from 13 GISTs, that were negative for exon 11 (juxtamembrane domain) mutations. Mutations in exon 9 and exon 13 were detected in six (46%) and two (15%) of 13 cases suggesting that such mutations may be relatively common in GISTs. 21
In this study, we analyzed 200 GISTs, including 97 cases that did not have mutations in exon 11 of c-kit. Mutations in exon 9 were found in six cases. These mutations were all two codon duplications (Ala502 and Tyr503), similar to those described by Lux et al. 21 Exon 13 mutation (missense mutation leading to substitution of Glu to Lys642) was seen in only two cases. Our results confirm that some GISTs may have mutations in the previously described hotspot areas in exon 9 and exon 13 of c-kit, but indicate that such mutations are rare. Therefore, these mutations do not offer a KIT mutation-based pathogenetic explanation for GIST oncogenesis for any substantial portions of those GISTs that are negative for exon 11 mutations.
Malignant clinical behavior of the tumor was documented in seven of eight cases that had mutation in exons 9 and 13, suggesting that similar to mutations in exon 11 of c-kit, those in exons 9 and 13 predominantly occur in malignant versus benign GISTs. 16,17,19
1530ins6 mutation in exon 9 seems to occur preferentially in malignant small intestinal tumors and may be the marker of malignant behavior for the GISTs of this location. However, a larger group of small intestinal GISTs with clinical follow-up should be evaluated to establish the statistical significance of this observation.
The apparent difference in the frequency of exon 9 and exon 13 mutation (combined 8% in this study versus 61% a previous study 21 ) has several potential explanations. They most importantly include case selection bias, as larger malignant tumors may have been included in the frozen-tumor bank collection available for cDNA analysis in the earlier study. Another study showed that larger tumors more frequently revealed mutations in exon 11 of the c-kit gene than the smaller ones. 19
Geographic genetic differences cannot be completely ruled out, as our material was predominantly composed of individuals of Caucasian ethnicity (80%) from northern and central Europe. An extensive study on GISTs from ethnically different populations has to be performed to exclude such a possibility.
Technical factors, including normal tissue contamination, could impair the detection of mutation. In this study, tumor tissue was carefully microdissected to maximize the tumor content in the samples to be analyzed for mutation. Also, frequent detection of mutation in exon 11, using the same strategy, provided an internal positive control in our study.
In summary, the present study showed that mutations in exons 9 and 13 of c-kit may occur in GISTs, predominantly in malignant tumors. However, such mutations seem to be rare. Other structural or functional genetic alterations may activate KIT in GISTs, or other genetic mechanism may be operational.
Footnotes
Address reprint requests to Jerzy Lasota M.D. or Markku Miettinen, M.D., Department of Soft Tissue Pathology, Armed Forces Institute of Pathology, 14th Street and Alaska Avenue, N.W., Washington, DC 20306-6000.
This study was partially supported by the Grant from the Polish Committee for the Scientific Research (4PO5A07117) and by the American Registry of Pathology.
The opinions and assertions contained herein are the expressed views of the authors and are not to be construed as official or reflecting the views of the Departments of the Army or Defense.
Agnieszka Wozniak is a research fellow at the Department of Soft Tissue Pathology, Armed Forces Institute of Pathology, Washington, DC.
References
- 1.Miettinen M, Sarlomo-Rikala M, Lasota J: Gastrointestinal stromal tumors—recent advances in understanding of their biology. Hum Pathol 1999, 30:1213-1220 [DOI] [PubMed] [Google Scholar]
- 2.Yarden Y, Kuang WS, Yang-Fend T, Coussens L, Munemitsas S, Dull TJ: Human proto-oncogene kit, a new cell surface-receptor tyrosine kinase for an unidentified ligand. EMBO J 1987, 6:3341-3351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Williams DE, Eisenman J, Baird A, Rauch C, van Ness K, March CJ, Park LS, Martin U, Mochizuki DY, Boswell HS, Burgess GS, Cosman D, Lyman SD: Identification of a ligand for the c-kit proto-oncogene. Cell 1990, 63:167-174 [DOI] [PubMed] [Google Scholar]
- 4.Zsebo KM, Williams DA, Geissler EN, Broudy WC, Martin FH, Atkins HL, Hsu R-Y, Birkett NC, Okino KH, Murdock DC, Jacobsen FW, Langley KE, Smith KA, Takeishi T, Cattanach BM, Galli SJ, Suggs SV: Stem cell factor is encoded at the SI locus of the mouse and is ligand for the c-kit tyrosine kinase receptor. Cell 1990, 63:214-224 [DOI] [PubMed] [Google Scholar]
- 5.Vliagoftis H, Worobec AS, Metcalfe DD: The proto-oncogene c-kit and c-kit ligand in human disease. J Allergy Clin Immunol 1997, 100:435-440 [DOI] [PubMed] [Google Scholar]
- 6.Maeda H, Yamagata A, Nishikawa S, Yoshinaga K, Kobayshy S, Nishi K, Nishikawa S: Requirement of c-kit for development of intestinal pacemaker system. Development 1992, 116:369-375 [DOI] [PubMed] [Google Scholar]
- 7.Tsuura Y, Hiraki H, Watanabe K, Igarashi S, Shimamura K, Fukuda T, Suzuki T, Seito T: Preferential localization of c-kit product in tissue mast cells, basal cells of skin, epithelial cells of breast, small cell lung carcinoma and seminoma/dysgerminoma in human: immunohistochemical study of formalin-fixed, paraffin-embedded tissues. Virchows Arch 1994, 424:135-141 [DOI] [PubMed] [Google Scholar]
- 8.Sanders KM: A case for interstitial cells of Cajal as pacemakers and mediators of neurotransmission in the gastrointestinal tract. Gastroenterology 1996, 111:492-515 [DOI] [PubMed] [Google Scholar]
- 9.Nakata Y, Kimura A, Katoh O, Kawaishi K, Hyodo H, Abe K, Kuramoto A, Satow Y: C-kit point mutation of extracellular domain in patients with myeloproliferative disorders. Br J Haematol 1995, 91:661-663 [DOI] [PubMed] [Google Scholar]
- 10.Nagata H, Worobec AS, Oh CK, Chowdhury BA, Tannenbaum S, Suzuki Y, Metcalfe DD: Identification of a point mutation in the catalytic domain of the protooncogene c-kit in peripheral blood mononuclear cells of patients who have mastocytosis with an associated hematologic disorder. Proc Natl Acad Sci USA 1995, 92:10560-10564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tian Q, Frierson HF, Krystal GW, Moskaluk CA: Activating c-kit mutations in human germ cell tumors. Am J Pathol 1999, 154:1643-1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gari M, Goodeve A, Wilson G, Winship P, Langabeer S, Linch D, Vandenberghe E, Peak I, Reilly J: C-kit proto-oncogene exon 8 in-frame deletion plus insertion mutations in acute myeloid leukaemia. Br J Haematol 1999, 105:894-900 [DOI] [PubMed] [Google Scholar]
- 13.Hirota S, Isozaki K, Moriyama Y, Hashimoto K, Nishida T, Ishiguro S, Kawano K, Hanada M, Kurata A, Takeda M, Tunio GM, Matsuzawa Y, Kanakura Y, Shinomura Y, Kitamura Y: Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science 1998, 279:577-580 [DOI] [PubMed] [Google Scholar]
- 14.Nishida T, Hirota S, Taniguchi M, Hashimoto K, Isozaki K, Nakamura H, Kanakura Y, Tanaka T, Takabayshi A, Matsuda H, Kitamura Y: Familial gastrointestinal stromal tumours with germline mutation of the KIT gene. Nat Genet 1998, 19:323-324 [DOI] [PubMed] [Google Scholar]
- 15.Furitsu T, Tsujimura T, Tono T, Ikeda H, Kitayama H, Koshimizu U, Sugahara H, Butterfielf JH, Ashman LK, Kanayama Y, Matsuzawa Y, Kitamura Y, Kanakura Y: Identification of mutations in the coding sequence of the proto-oncogene c-kit in a human mast cell leukemia cell line causing ligand-independent activation of c-kit product. J Clin Invest 1993, 92:1736-1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ernst SI, Hubbs AE, Przygodzki RM, Emory TS, Sobin LH, O’Leary TJ: KIT mutation portends poor prognosis in gastrointestinal stromal/smooth muscle tumors. Lab Invest 1998, 78:1633-1636 [PubMed] [Google Scholar]
- 17.Lasota J, Jasinski M, Sarlomo-Rikala M, Miettinen M: C-kit mutations occur preferentially in malignant vs. benign gastrointestinal stromal tumors and do not occur in leiomyomas and leiomyosarcomas. Am J Pathol 1999, 154:53-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakurai S, Fukasawa T, Chong JM, Tanaka A, Fukayama M: C-kit gene abnormalities in gastrointestinal stromal tumors (tumors of interstitial cells of Cajal). Jpn J Cancer Res 1999, 90:1321-1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taniguchi M, Nishida T, Hirota S, Isozaki K, Ito T, Nomura T, Matsuda H, Kitamura Y: Effect of c-kit mutation on prognosis of gastrointestinal stromal tumors. Cancer Res 1999, 59:4297-4300 [PubMed] [Google Scholar]
- 20.Moskaluk CA, Tian Q, Marshall CR, Rumpel CA, Franquemont DW, Frierson HF, Jr: Mutations of c-kit JM domain are found in a minority of human gastrointestinal stromal tumors. Oncogene 1999, 18:1897-1902 [DOI] [PubMed] [Google Scholar]
- 21.Lux ML, Rubin BP, Biase TL, Chen C-J, Maclure T, Demetri G, Xiao S, Singer S, Fletcher CDM, Fletcher JA: KIT extracellular and kinase domain mutations in gastrointestinal stromal tumors. Am J Pathol 2000, 156:791-795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sarlomo-Rikala M, Kovatich AJ, Barusevicius A, Miettinen M: CD117: a sensitive marker for gastrointestinal stromal tumors that is more specific than CD34. Mod Pathol 1998, 11:728-734 [PubMed] [Google Scholar]
- 23.Lasota J, Jasinski M, Debiec-Rychter M, Szadowska A, Limon J, Miettinen M: Detection of the SYT-SSX fusion transcripts in formaldehyde-fixed, paraffin-embedded tissue: a reverse transcription polymerase chain reaction amplification assay useful in the diagnosis of synovial sarcoma. Mod Pathol 1998, 11:626-633 [PubMed] [Google Scholar]


