Abstract
Nuclear and cytoplasmic protein glycosylation is a widespread and reversible posttranslational modification in eukaryotic cells. Intracellular glycosylation by the addition of N-acetylglucosamine (GlcNAc) to serine and threonine is catalyzed by the O-GlcNAc transferase (OGT). This “O-GlcNAcylation” of intracellular proteins can occur on phosphorylation sites, and has been implicated in controlling gene transcription, neurofilament assembly, and the emergence of diabetes and neurologic disease. To study OGT function in vivo, we have used gene-targeting approaches in male embryonic stem cells. We find that OGT mutagenesis requires a strategy that retains an intact OGT gene as accomplished by using Cre-loxP recombination, because a deletion in the OGT gene results in loss of embryonic stem cell viability. A single copy of the OGT gene is present in the male genome and resides on the X chromosome near the centromere in region D in the mouse spanning markers DxMit41 and DxMit95, and in humans at Xq13, a region associated with neurologic disease. OGT RNA expression in mice is comparably high among most cell types, with lower levels in the pancreas. Segregation of OGT alleles in the mouse germ line with ZP3-Cre recombination in oocytes reveals that intact OGT alleles are required for completion of embryogenesis. These studies illustrate the necessity of conditional gene-targeting approaches in the mutagenesis and study of essential sex-linked genes, and indicate that OGT participation in intracellular glycosylation is essential for embryonic stem cell viability and for mouse ontogeny.
Intracellular protein glycosylation is ubiquitous in eukaryotic cells yet is less studied than other types of posttranslational modifications such as phosphorylation. This is, in part, because of the relatively recent discovery that serine and threonine residues of many cytoplasmic and nuclear proteins are modified by the addition of an O-linked N-acetylglucosamine (O-GlcNAc) (1, 2). Moreover, this modification originally was difficult to detect before the development of new approaches. O-GlcNAc formation, also termed O-GlcNAcylation, has been found on nuclear pore proteins, RNA polymerase II, and many transcription factors, including Sp1, serum response factor, and the estrogen receptors. In addition, cytoskeletal proteins such as Tau, vinculin, talin, ankyrin, neurofilaments, cytokeratins, and clathrin assembly protein AP-3; and viral or oncogene proteins, such as the cytomegalovirus UL32 protein, myc, fos, jun, simian virus 40 T-antigen, and p53 are also O-GlcNAcylated (refs. 3–20 and reviewed in ref. 21).
UDP-N-acetyl-d-glucosamine:protein β-N-acetyl-d-glucosaminyltransferase (OGT) (EC 2.4.1.94) catalyzes the addition of O-GlcNAc to the polypeptide chain. All higher eukaryotic cell types studied contain OGT activity. Purified rat liver OGT is a heterotrimer with two catalytic subunits of 110 kDa and a 78-kDa subunit of unknown function (22). The 78-kDa form is structurally and immunologically related to the 110-kDa protein and may result from an internal translation start site or from an alternate OGT transcript. The gene encoding OGT has been isolated recently from Caenorhabditis elegans, rat, and human genomes (23, 24) and is highly conserved among these organisms, sharing over 65% amino acid identity. OGT contains multiple tandem tetratricopeptide repeat sequences and is itself modified by O-GlcNAc, as well as by tyrosine phosphorylation.
O-GlcNAcylation is a reversible modification, as suggested in studies of lymphocyte activation and confirmed by the rapid turnover O-GlcNAc on cytokeratins and αB-crystallin (7, 18, 25). The removal of O-GlcNAc is performed by an N-acetyl-β-d-glucosaminidase (O-GlcNAcase) that purifies from rat kidney as a heterodimer composed of a 54-kDa α-subunit and a 51-kDa β-subunit (26). The O-GlcNAcase is ubiquitous and also is localized to nuclear and cytoplasmic compartments. An apparent reciprocity of O-GlcNAcylation and phosphorylation has been noted with the same amino acids of some proteins such as RNA polymerase II and c-myc, existing as either GlcNAcylated or phosphorylated isoforms (27). The potential for O-GlcNAc involvement in intracellular signal transduction seems high. To investigate this potential, we mutated the OGT gene in embryonic stem (ES) cells and in mice to study the metabolic and physiologic events regulated by O-GlcNAcylation.
Materials and Methods
OGT Gene Mutagenesis in ES Cells and the Mouse Germ Line.
Isolation and characterization of mouse OGT genomic DNA clones was accomplished from a 129/SvJ phage library (Stratagene) using the rat cDNA as probe (23). Targeting constructs were produced by using vectors as described (Fig. 1 and ref. 28). R1 ES cells (29) were electroporated with the linearized loxP-bearing targeting vector (NotI digest) or the conventional vector (BamHI digest), and G418 (150 μg/ml) resistant clones were analyzed by PCR to detect homologous recombinants. ES cells bearing the OGTF[tkneo] allele were electroporated with 10 μg of Cre expression plasmid, and subclones exhibiting Cre-recombined alleles were then identified. OGTΔ-bearing ES cells were detected by PCR at indicated time points (Fig. 1F) with the oligonucleotide primers rlox (5′-CAACTTTGGGCAAGCCTCCTCCC-3′) and RS-14 (5′-CTCGAATTGATCCCCCGGTACCG-3′). Chimeric mice were generated from selected OGTF ES cell clones and then bred with C57BL/6 mates to obtain heterozygous offspring. The latter were crossed with ZP3-Cre transgenic animals to produce mice carrying both OGTF and the ZP3-Cre transgene.
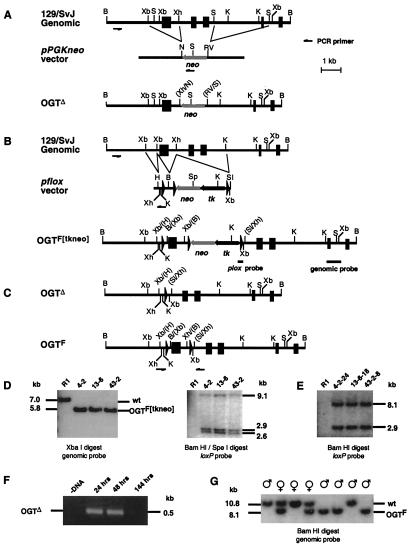
Figure 1.
OGT mutagenesis in ES cells and mice. A mouse OGT genomic clone bearing five exons (filled boxes) was used to construct two types of gene-targeting vectors. (A) The first vector would be expected to produce a null mutation by deletion of two OGT exons with insertion of a neomycin phosphotransferase cistron (neo). (B) The second vector incorporates loxP sites that initially would generate the OGTF[tkneo] allele. (C) ES cells bearing the OGTF[tkneo] allele that are subjected to transient Cre expression should yield both OGTΔ (type I) and OGTF (type II) alleles. (D) Homologous recombinants were obtained with the loxP-containing vector shown in B and were characterized by Southern blotting with probes of OGT genomic (Left) or loxP (Right) sequences. (E) Only OGTF alleles are present among subclones, as represented, in over 200 analyzed specimens. (F) The OGTΔ allele is detectable by PCR in polyclonal ES cell extracts only at early times after Cre expression. (G) OGT alleles in offspring of chimeric male (♂) mice bred to C57BL/6 females (♀) were analyzed by Southern blotting. Female offspring are either homozygous wild type or heterozygous for the OGTF allele, whereas male offspring are either homozygous wild type or appear homozygous for the OGTF allele. wt, wild type.
Production of ZP3-Cre transgenic mice required cloning the previously published ZP3 allele (30) by screening the mouse genomic DNA library with a PCR-derived ZP3 probe. The ZP3-Cre transgene was constructed by linking the 6-kb SalI-PvuII fragment, encompassing the promoter sequence 5′ to the Cre gene as well as the human growth hormone splicing and polyadenylation signals as previously described (31). Cre recombination was found to be restricted to oocytes in female mice (data not shown).
RNA Blotting.
Total RNA from 8-week-old male mice of indicated strains was prepared and visualized by Sybergreen (Molecular Probes) staining as described (28). The RNA blots were hybridized overnight at 42°C with a 955-bp DraI fragment of the rat OGT cDNA clone. After hybridization, membranes were washed in 0.1× SSC (1× SSC = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7)/O.1% SDS and autoradiographed for 48 h at −80°C on Kodak Biomax-MS film.
Fluorescence in Situ Hybridization.
Peripheral blood lymphocytes (normal human male) synchronized in metaphase were prepared by standard methods. DNA was isolated by the Qiagen (Chatsworth, CA) Maxiprep method. By using the OGT cDNA clone as the probe, 1 μg of plasmid DNA was labeled by nick translation (BRL) with digoxigenin (Boehringer Mannheim). Probe (200 ng) was coprecipitated with 2 μg of sheared herring sperm DNA, then resuspended in 10 μl of hybridization solution [50% formamide in 2× SSC/10% (wt/vol) dextran sulfate]. The probe solution was denatured for 5 min at 75°C and held at 42°C before hybridization. Slides were denatured for 3 min at 75°C before hybridization at 37°C for 18 h. The slides were washed at low stringency for 5 min in 50% formamide, for 3 min in 2× SSC at 37°C, and then for 1 min in 4× SSC/0.05% Tween-20 at room temperature. The OGT genomic clone also was prepared as a probe with the exception that 5 μg of mouse cot-1 DNA was used in the coprecipitation step. This probe was denatured as above and preannealed for 1 h at 37°C before hybridization at 37°C for 18 h. Slides then were washed at high stringency (twice for 7 min in 50% formamide in 2× SSC at 42°C), followed by two 5-min washes in 2× SSC at 42°C. For each experiment, the signals were detected by using rhodamine-conjugated anti-digoxigenin according to manufacturer's instructions (Ventana Medical Systems, Tucson, AZ). Chromosomes were counterstained with the A-T binding fluorochrome 4′,6-diamidino-2-phenylindole (DAPI) and viewed with a Zeiss Axioskop equipped with a SenSys cooled charge-coupled device camera (Photometrics, Tuscon, AZ) and quips smartcapture vp imaging software (Vysis, Downersgrove, IL).
Chromosomal Localization in the Mouse by Radiation Hybrid (RH) Mapping.
The mouse T31 RH panel (RH04.02; Research Genetics, Huntsville, AL) consisting of 100 hybrid cell lines was used. To design a mouse-specific OGT genomic probe, the intron 3′ end of the first OGT coding exon was sequenced by using the OGT genomic clone as the template. For probe production, PCR was performed with the OGT A1 5′ intronic RH primer, 5′-GAC TAA AAT GTG GCA ATA GCT TCC-3′, and the OGT A1 3′ intronic RH primer 2, 5′-CTC ACA ACC ATC CGT AAC GAG ATC-3′, according to the RH assay protocol from Stanford Human Genome Center (http://www-shgc.stanford.edu/Mapping/rh/procedure/rhassaynew.html), and it produced a 378-bp product in the presence of the mouse OGT gene. The results were submitted to The Jackson Laboratory (http://www.jax.org/resources/documents/cmdata/rhmap/rhsubmit.html) for statistical analysis.
Results and Discussion
OGT Gene-Targeting in ES Cells Requires Conditional Mutagenesis.
A genomic DNA clone containing five OGT encoding exons was isolated and used to produce gene-targeting vectors. The first gene-targeting vector was constructed by using the classical approach that inactivates the gene as a result of homologous recombination with insertion of the Neo cistron cassette and, in this case, with simultaneous deletion of two OGT exons (Fig. 1A). After gene transfer into ES cells, PCR was used to detect homologous recombinants among G418 resistant clones. Although the PCR detection strategy was sufficiently sensitive as judged in parallel with a control vector, no homologous recombinants were obtained (data not shown).
An alternate approach to abrogating OGT gene function was undertaken that incorporated conditional mutagenesis to excise an OGT exon. The exon deleted encompasses amino acids 206–232 of the complete 1,037 aa OGT sequence as derived from the rat cDNA. Any OGT RNA expressed by this mutant allele would encode a highly truncated and aberrant form bearing the first 206 amino acids of OGT followed by 27 unrelated amino acids before translational termination. A gene-targeting vector was constructed that incorporated Cre-loxP recombination, thereby enabling the production of ES cell subclones bearing either a deleted (Δ) allele or a functional loxP-flanked (F) allele, the latter being likely nondeleterious but further susceptible to Cre recombination and exon deletion (Fig. 1 B and C). Use of this vector was successful in establishing multiple ES cell clones that had undergone homologous recombination at a frequency of approximately 1/200 G418-resistant cells. Curiously, all targeted ES cells appeared homozygous for the OGTF[tkneo] allele (Fig. 1D). Three distinct targeted ES cell clones bearing the OGTF[tkneo] allele were subjected to transient Cre expression and gancyclovir selection to obtain subclones bearing the OGTΔ and OGTF alleles for chimeric mouse production (reviewed in ref. 32). DNA Southern analysis of over 200 subclones failed to identify cells bearing the OGTΔ allele; only the OGTF allele was present (Fig. 1E).
To determine whether Cre recombinase was capable of acting on the OGTF allele, Cre expression was transferred into ES cells bearing the OGTF allele, and PCR was used to detect the Δ allele at various times. The OGTΔ allele was detected in the cell population by 24 and 48 h after Cre gene transfer; however, viable ES cells present at 144 h were all of the OGTF genotype (Fig. 1 E and F). These results indicate that Cre recombinase can operate to excise the loxP-flanked OGT exon, but that this deletion in the OGT gene is deleterious to ES cells.
Chimeric male mice were obtained at a normal frequency from ES clones bearing the OGTF allele, and these mice were bred to C57BL/6 females to produce offspring heterozygous for the OGTF allele. The OGTF allele was transmitted through the germ line to offspring, but segregated in a manner consistent with an X chromosomal location. Females heterozygous for the F and wild-type alleles were present as expected; however, male offspring were either wild type or appeared homozygous for the F allele (Fig. 1G). Heterozygous females were bred to wild-type males, and OGT alleles were analyzed in the offspring. Male offspring contained only the OGTF allele, whereas female offspring were either wild type or heterozygous for the OGTF allele (Table 1). These results indicate that a single OGT allele is present in ES cells and that male mice contain a single copy of the OGT gene that segregates during meiosis, which is consistent with a position on the X chromosome.
Table 1.
Segregation of the OGTF allele
| Parental genotype | Offspring
|
|||
|---|---|---|---|---|
| Sex | OGT genotypes
|
|||
| wt only | F and wt | F only | ||
| wt male × F/wt female | Male | 12 | 0 | 18 |
| Female | 12 | 19 | 0 | |
Numbers represent total offspring from four different breeding pairs producing a total of eight litters. Each litter produced at least five progeny. wt, wild type.
Chromosomal Localization of the OGT Gene.
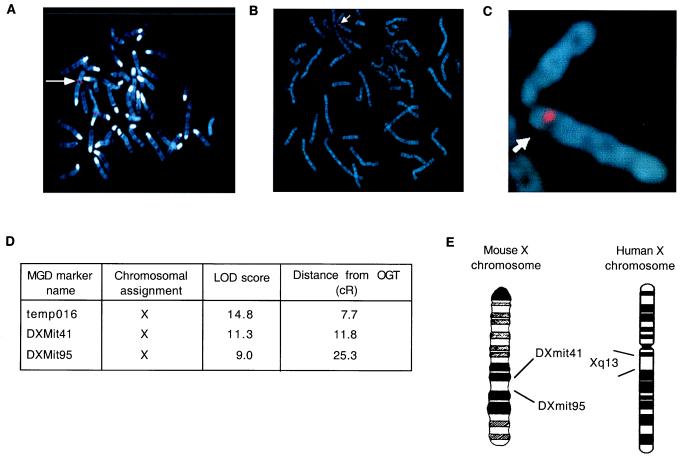
The chromosomal location of the human and mouse OGT genes was determined directly by fluorescence in situ hybridization and analysis of a mouse/hamster radiation hybrid cell line DNA panel. Fluorescence in situ hybridization mapping of the human OGT gene was performed by using both a rat OGT cDNA probe, which is >95% identical to human OGT sequence, and by using the mouse OGT genomic DNA clone as a probe. With either probe, one prominent signal was observed centromeric on the long arm of human chromosome X in region q13 (Fig. 2A–C).
Figure 2.
Regional localization of the OGT gene on the X chromosome in mice and humans. (A–C) Fluorescence in situ hybridization using both OGT cDNA (A) and genomic probes (B and C) localizes the OGT gene to the long arm of human X chromosome near the centromere (arrows). (D) Radiation-hybrid analysis of OGT gene localization in the mouse placed OGT on the X chromosome at the indicated centirads (cR) from the markers temp016, DXMit4, and DXMit95. (E) Statistical analysis placed OGT on the mouse X chromosome between markers DXMit41 and DXMit95 and in a region syntenic with human Xq13. LOD, logarithm of odds; MGD, mouse genome database.
Chromosomal localization of the mouse OGT gene was obtained by PCR mapping using the T31 mouse radiation hybrid panel. Mouse-specific OGT primers used for PCR amplification corresponded to the intron region that immediately flanked the 3′ end of targeted exon of the OGT gene. Specificity of the amplified 378-bp product was confirmed by the presence of the product in the parental 129-aa mouse donor cell line and not in the A23 hamster recipient cell line (data not shown). The PCR product was amplified in DNA from 16 of 100 hybrid cell lines, and these results were submitted and processed at The Jackson Laboratory Mouse Radiation Hybrid Database (http://www.jax.org/resources/documents/cmdata/). The consequent statistical analysis is summarized in Fig. 2D and indicated that the mouse OGT gene mapped 7.7 centirad distal to the temp016 locus [logarithm of odds (lod) score of 14.8) (Fig. 2D). The next closest locus was DXMit41 (lod score of 11.8), which is 4.1 centirad on the other side of temp016. The OGT gene also mapped 25.3 centirad away from DXMit95 (lod score of 9.0) in the other direction. Another loci, DXMit230 (lod score of 12.0) also mapped near the OGT, but differences between the scorings for DXMit41 and DXMit230 did not distinguish their order.
Comparison of genes in region D on the mouse X chromosome residing between markers DXMit41 and DXMit95 with genes in region q13 on the human X chromosome showed a good agreement in syntenic genes. The schematic diagram in Fig. 2E illustrates the chromosomal placement of the OGT gene at a relative distance to framework markers in The Jackson Laboratory radiation hybrid map of the mouse genome and its corresponding syntenic position on the human X chromosome.
OGT Gene Expression.
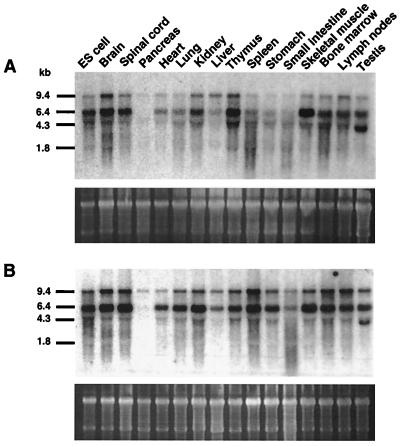
OGT enzyme activity is widespread among tissues, and especially high levels of OGT RNA have been found in the human pancreas (24). This finding may be noteworthy because the GlcNAc analog streptozotocin is an inhibitor of the O-GlcNAcase and induces diabetes in rodents, including specific strains of mice (33). We surveyed tissues and cells to detect levels of OGT RNA in both streptozotocin-sensitive (C57BL/6J) and streptozotocin-resistant (C3H/HeJ) mouse strains. All tissues and cells exhibited high levels of OGT RNA with multiple transcripts, as observed in rat and human tissues. However, much lower OGT RNA levels were found in the pancreas. This pattern of expression was the same regardless of the strain surveyed (Fig. 3).
Figure 3.
OGT RNA expression among normal mouse tissues. (A and B) Total RNA (6 μg) was probed for OGT expression among ES cells and normal tissues of male mice either streptozotocin-sensitive (C57BL/6J) (A) or streptozotocin-resistant (C3H/HeJ) (B). Lower gels in A and B show that RNA is intact and comparable levels are present as detected by Sybergreen II staining.
OGT Gene Deletion Is Lethal in Ontogeny.
Male offspring containing the single X-linked OGTF allele were found to be normal and fertile. When they were bred with heterozygous female mice bearing the F and wild-type OGT alleles, female offspring homozygous for the OGTF allele were obtained (Table 2, first row). These females appeared normal and were fertile (Table 2, second row). To study the effects of OGT gene deletion in the mouse germ line, we developed a line of mice bearing the ZP3-Cre transgene in which Cre expression and recombination is targeted to the developing oocyte, as described (see Materials and Methods and ref. 34).
Table 2.
Mice bearing the OGTF allele are viable and fertile, whereas the OGTΔ allele is lethal
| Parental OGT genotype | Offspring
|
||||||
|---|---|---|---|---|---|---|---|
| Sex | OGT genotype
|
||||||
| wt/Y | F/Y | wt/wt | F/wt | F/F | Δ/Y, F/Δ, wt/Δ, Δ/Δ | ||
| F/Y male × F/wt female | Male | 10 | 11 | — | — | — | — |
| Female | — | — | — | 8 | 10 | — | |
| F/Y male × F/F female | Male | — | 15 | — | — | — | — |
| Female | — | — | — | — | 12 | — | |
| wt/Y male × F/wt female ZP3-Cre | Male | 11 | 0 | — | — | — | 0 |
| Female | — | — | 10 | 0 | — | 0 | |
| F/Y male × F/wt female ZP3-Cre | Male | 12 | 0 | — | — | — | 0 |
| Female | — | — | — | 14 | 0 | 0 | |
Offspring were derived from two or more different breeding pairs producing a total of four or more litters. Each litter contained at least five progeny. Dashes indicate genotypes not possible, and not observed, from the specific parental genotypic crosses indicated. wt, wild type.
Female mice were bred to heterozygosity for the OGTF allele and hemizygosity for the ZP3-Cre transgene. These mice were then bred with males that were either wild type or bore the OGTF allele. Genotypes of offspring obtained are shown in Table 2 (rows 3 and 4). From these matings, wild-type male or female offspring were obtained as expected. However, neither males of the F/Y OGT genotype nor females bearing the F allele were present, as they were in the absence of ZP3-Cre, suggesting that Cre recombination was taking place in the oocyte as expected. The presence of F/wild-type female offspring derived from the F/Y paternal genome occurs by acquisition of the OGTF allele from the male pronucleus, which is not accessible to Cre recombination in the developing oocyte. Although efficient Cre recombination in the oocyte appeared to have occurred, no male or female offspring were obtained that bore the deleted OGTΔ allele. These results show that an intact OGT gene is required for mouse ontogeny and that females bearing a single OGTΔ allele do not survive to term. These findings do not discount the possibility that a dominant-negative form of OGT has been produced; however, preliminary studies indicate that embryos bearing OGTΔ alleles may develop past embryonic day 4 and implantation. The mechanisms of how mutation of the OGT gene disables embryogenesis, and at which particular stages in males and females, require further study.
Conclusions
OGT is on the X chromosome in region D in mice and in the syntenic region at Xq13 in humans. An intact OGT gene is necessary for the viability of ES cells, which explains why the classical gene disruption approach proved unsuccessful because male XY ES cells were used. Mice bearing the deleted OGTΔ allele have not been observed from crosses of appropriate parental genomes, which indicates that mouse ontogeny requires fully intact OGT alleles. The expression pattern of OGT RNA is ubiquitous in the mouse tissues studied; however, expression appears differentially regulated in the pancreas, with much lower levels of RNA evident. This finding does not indicate lack of a role in the etiology of diabetes, but the position of the OGT locus in humans at Xq13 does not map to known diabetes susceptibility loci. Rather, this chromosomal position is associated with other human maladies, including neurologic disease. Intriguingly, recent studies have found that neurofilaments, several microtubule-associated proteins (including Tau), and synapsin 1 are extensively O-GlcNAcylated, and that reduced levels of O-GlcNAc are found on clathrin assembly proteins in Alzheimer's disease (8, 12, 16, 17, 35, 36). Furthermore, other studies suggest that O-GlcNAc modulates the degradation of the amyloid precursor protein (37).¶ Collectively, these findings should promote further investigations aimed at elucidating the roles of O-GlcNAc in embryogenesis as well as in normal and abnormal neural physiology.
Acknowledgments
We thank our colleagues Greg Odorizzi, Robert Campbell, Michael Durnin, Eileen Westerman, Zhengyi Ye, Lisa Kreppel, and Holly Pieck for technical assistance and helpful discussions. Funding for this research was provided in part by National Institutes of Health Grants DK 48247 to J.D.M. and HD 13563 to G.W. H., and by the Juvenile Diabetes Foundation and Fifty 50 Foods. J.D.M. is an Investigator of the Howard Hughes Medical Institute. G.W.H. acknowledges that he was a consultant for Monsanto Corporation during some of this work.
Abbreviations
- OGT
O-GlcNAc transferase
- ES cells
embryonic stem cells
- RH
radiation hybrid
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.100471497.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.100471497
Mäß, C., Gollner, U. F., Thelen, K., Griffith, L. S., Koch, J. & Schmitz, B. (1999) Glycoconjugate J. 16, S45 (abstr.).
References
- 1.Torres C-R, Hart G W. J Biol Chem. 1984;259:3308–3317. [PubMed] [Google Scholar]
- 2.Holt G D, Hart G W. J Biol Chem. 1986;261:8049–8057. [PubMed] [Google Scholar]
- 3.Hanover J A, Cohen C K, Willingham M C, Park M K. J Biol Chem. 1987;262:9887–9894. [PubMed] [Google Scholar]
- 4.Jackson S P, Tjian R. Cell. 1988;55:125–133. doi: 10.1016/0092-8674(88)90015-3. [DOI] [PubMed] [Google Scholar]
- 5.Starr C M, Hanover J A. J Biol Chem. 1990;265:6868–6873. [PubMed] [Google Scholar]
- 6.Haltiwanger R S, Holt G D, Hart G W. J Biol Chem. 1990;265:2563–2568. [PubMed] [Google Scholar]
- 7.Chou C-F, Smith A J, Omary M B. J Biol Chem. 1992;267:3901–3906. [PubMed] [Google Scholar]
- 8.Dong D L-Y, Xu Z-S, Chevrier M R, Cotter R J, Cleveland D W, Hart G W. J Biol Chem. 1993;268:16679–16687. [PubMed] [Google Scholar]
- 9.Kelly W G, Dahmus M E, Hart G W. J Biol Chem. 1993;268:10416–10424. [PubMed] [Google Scholar]
- 10.Murphy J E, Hanover J A, Froehlich M, DuBois G, Keen J H. J Biol Chem. 1994;269:21346–21352. [PubMed] [Google Scholar]
- 11.Greis K D, Gibson W, Hart G W. J Virology. 1994;68:8339–8349. doi: 10.1128/jvi.68.12.8339-8349.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cole R N, Hart G W. J Neurochem. 1999;73:418–428. doi: 10.1046/j.1471-4159.1999.0730418.x. [DOI] [PubMed] [Google Scholar]
- 13.Chou T-Y, Dang C V, Hart G W. Proc Natl Acad Sci USA. 1995;92:4417–4421. doi: 10.1073/pnas.92.10.4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chou T-Y, Hart G W, Dang C V. J Biol Chem. 1995;270:18961–18965. doi: 10.1074/jbc.270.32.18961. [DOI] [PubMed] [Google Scholar]
- 15.Heese-Peck A, Cole R N, Borkhsenious O N, Hart G W, Raikhel N V. Plant Cell. 1995;7:1459–1471. doi: 10.1105/tpc.7.9.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arnold C S, Johnson G V W, Cole R N, Dong D L-Y, Lee M, Hart G W. J Biol Chem. 1996;271:28741–28744. doi: 10.1074/jbc.271.46.28741. [DOI] [PubMed] [Google Scholar]
- 17.Dong D L-Y, Xu Z-S, Hart G W, Cleveland D W. J Biol Chem. 1996;271:20845–20852. doi: 10.1074/jbc.271.34.20845. [DOI] [PubMed] [Google Scholar]
- 18.Roquemore E P, Chevrier M R, Cotter R J, Hart G W. Biochemistry. 1996;35:3578–3586. doi: 10.1021/bi951918j. [DOI] [PubMed] [Google Scholar]
- 19.Shaw P, Freeman J, Bovey R, Iggo R. Oncogene. 1996;12:921–930. [PubMed] [Google Scholar]
- 20.Jiang M-S, Hart G W. J Biol Chem. 1997;272:2421–2428. doi: 10.1074/jbc.272.4.2421. [DOI] [PubMed] [Google Scholar]
- 21.Hart G W. Annu Rev Biochem. 1997;66:315–335. doi: 10.1146/annurev.biochem.66.1.315. [DOI] [PubMed] [Google Scholar]
- 22.Haltiwanger R S, Blomberg M A, Hart G W. J Biol Chem. 1992;267:9005–9013. [PubMed] [Google Scholar]
- 23.Kreppel L K, Blomberg M A, Hart G W. J Biol Chem. 1997;272:9308–9315. doi: 10.1074/jbc.272.14.9308. [DOI] [PubMed] [Google Scholar]
- 24.Lubas W A, Frank D W, Krause M, Hanover J A. J Biol Chem. 1997;272:9316–9324. doi: 10.1074/jbc.272.14.9316. [DOI] [PubMed] [Google Scholar]
- 25.Kearse K P, Hart G W. Proc Natl Acad Sci USA. 1991;88:1701–1705. doi: 10.1073/pnas.88.5.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong D L-Y, Hart G W. J Biol Chem. 1994;269:19321–19330. [PubMed] [Google Scholar]
- 27.Hart G W, Kreppel L K, Gomer F I, Arnold C S, Snow D M, Ye Z, Cheng X, DellaManna D, Caine D S, Earles B J, et al. Glycobiology. 1996;6:711–716. doi: 10.1093/glycob/6.7.711. [DOI] [PubMed] [Google Scholar]
- 28.Priatel J J, Sarkar M, Schachter H, Marth J D. Glycobiology. 1997;7:45–56. doi: 10.1093/glycob/7.1.45. [DOI] [PubMed] [Google Scholar]
- 29.Nagy A, Rossant J, Nagy W, Abramow-Newerly W, Roder J C. Proc Natl Acad Sci USA. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kinloch R A, Wassarman P M. Nucleic Acids Res. 1989;17:2861–2863. doi: 10.1093/nar/17.7.2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orban P C, Chui D, Marth J D. Proc Natl Acad Sci USA. 1992;89:6861–6865. doi: 10.1073/pnas.89.15.6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marth J D. J Clin Invest. 1996;97:1999–2002. doi: 10.1172/JCI118634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roos M D, Xie W, Su K, Clark J A, Yang X, Chin E, Paterson A J, Kudlow J E. Proc Assoc Am Physicians. 1998;110:422–432. [PubMed] [Google Scholar]
- 34.Lewandoski M, Wassarman K M, Martin G R. Curr Biol. 1997;7:148–151. doi: 10.1016/s0960-9822(06)00059-5. [DOI] [PubMed] [Google Scholar]
- 35.Yao P J, Coleman P D. J Neurosci. 1998;18:2399–2411. doi: 10.1523/JNEUROSCI.18-07-02399.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yao P J, Coleman P D. Neurosci Lett. 1998;252:33–36. doi: 10.1016/s0304-3940(98)00547-3. [DOI] [PubMed] [Google Scholar]
- 37.Griffith L S, Mathes M, Schmitz B. J Neurosci Res. 1995;41:270–278. doi: 10.1002/jnr.490410214. [DOI] [PubMed] [Google Scholar]