SUMMARY
Adduct-induced conformational heterogeneity complicates the understanding of how DNA adducts exert mutation. A case in point is the N-deacetylated AF lesion [N-(2′-deoxyguanosin-8-yl)-2-aminofluorene], the major adduct derived from the strong liver carcinogen N-acetyl-2-aminofluorene. Three conformational families have been previously characterized and are dependent on the positioning of the aminofluorene rings: B is in the ‘B-DNA’ major groove, S is ‘stacked’ into the helix with base-displacement, and W is ‘wedged’ into the minor groove. In the present study, we conducted 19F NMR, CD, Tm, and modeling experiments at various primer positions with respect to a template modified by a fluorine tagged AF-adduct (FAF). In the first set, the FAF-G was paired with C and in the second set it was paired with A. The FAF-G:C oligonucleotides were found to preferentially adopt the B- or S-conformers while the FAF-G:A mismatch ones preferred the B- and W-conformers. The conformational preferences of both series were dependent on temperature and complementary strand length; the largest differences in conformation were displayed at lower temperatures. The CD and Tm results are in general agreement with the NMR data. Molecular modeling indicated that the aminofluorene moiety in the minor groove of the W-conformer would impose a steric clash with the tight-packing amino acid residues on the DNA binding area of the Bacillus fragment (BF), a replicative DNA polymerase. In the case of the B-type conformer, the carcinogenic moiety resides in the solvent-exposed major groove throughout the replication/translocation process. The present dynamic NMR results, combined with previous primer extension kinetic data by Miller and Grollman, support a model in which adduct-induced conformational heterogeneities at positions remote from the replication fork affect polymerase function through a long-range DNA-protein interaction.
Keywords: Aminofluorene-DNA adducts, Conformational heterogeneity, Long-range effect, Tranlesion synthesis
INTRODUCTION
DNA adduct formation is a signature hallmark of mutation, ultimately leading to the initiation of chemical carcinogenesis (1,2). Arylamines and their nitro derivatives are a major group of mutagens and carcinogens (3,4). The environmental carcinogen 2-nitrofluorene and its amino derivatives are the prototype arylamine carcinogens (5,6). Upon activation in vivo, they react with cellular DNA to form two major C8-substituted dG adducts: N-(2′-deoxyguanosin-8-yl)-2-acetyaminofluorene (AAF)1 and N-(2′-deoxyguanosin-8-yl)-2-aminofluorene (AF)(Fig. 1a), along with the minor 3-(2′-deoxyguanosin-N2-yl)-2-acetylaminofluorene (dG-N2-AAF)(7). These bulky DNA lesions, if not repaired, can produce mutations during replication that initiate the process of carcinogenesis. In bacteria AF-adducts mostly induce G→T transversions, whereas AAF-adducts cause mostly deletion mutations (3). In mammalian cells, however, both adducts promote sequence dependent base substitution mutations (8).
Fig. 1.
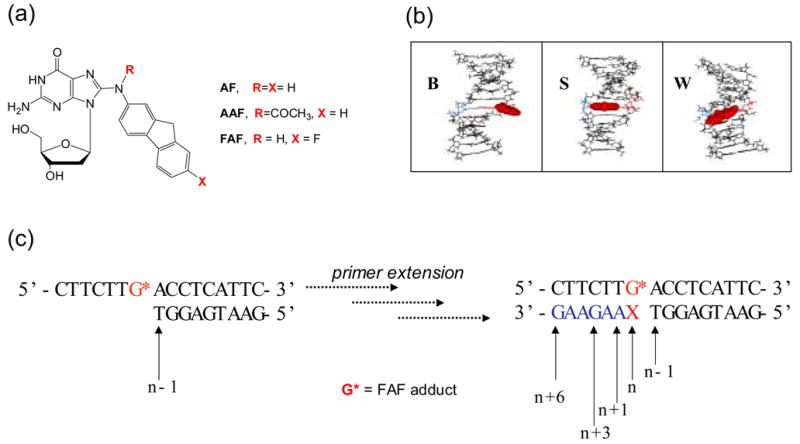
(a) Structures of C8-substituted dG-aminofluorene adducts. (b) Views from the minor-groove of a duplex for the three conformational motifs of the AF-modified DNA: B-, S-, and W-conformers. The modified dG and the complementary base (dC for B and S; dA for W) residues are shown in red and blue lines, respectively and the AF-moiety is highlighted with red CPK. In B conformer, anti-[AF]dG maintains Watson-Crick hydrogen bonds, thereby placing the aminofluorene ring in the major groove with displacement of the modified dG and partner dC. The aminofluorene moiety of the S-conformer stacks into the helix with the modified dG in the syn-glycosidyl conformation (base-displaced). The modified dG of the W-conformer adopts syn-configuration; however, the steric constraint at the lesion site allows the aminofluorene ring wedged into the surface of the narrow minor groove. (c) Sequence context studied (G* = FAF-adduct; X = C, FAF-G:C match series; X = A, FAF-G:A mismacth series).
The bulky AF and AAF adducts have long been used as model systems for investigation of the structure/functionion relationship in the arylamine mutagen family (1–4). However, the molecular understanding of how they exert mutation in vivo is generally poor and is complicated by adduct-induced conformational heterogeneities. The AAF-adduct exists predominantly in the base-displaced “stacked” (S) conformation (9–11). Because it lacks Watson-Crick base pairs at the lesion site, the AAF-adduct produces a major structural distortion in DNA. In contrast, the N-deacetylated AF-adduct possesses flexiblility around the glycosidyl bond (χ), enabling it to adopt multiple conformational motifs depending upon the location of the aminofulorene moiety, including a major-groove binding “B-type” (B) conformation and a minor-groove binding “wedged” (W) conformation in addition to the S conformation (Fig. 1b)(12-18). In general, fully base-paired DNA duplexes that have incorporated AF are present in an S/B equilibrium in which the tendency for the molecules to favor the S or the B conformation is dependent upon the flanking-sequence context (17–19). Consistent with this observation, the mutational specificity and frequency of the AF-adduct in mammalian cells vary depending upon the sequence context in which it is embedded (8). The W-conformer has only been observed in AF-modified duplexes with dA- and dG-mismatches at the lesion site (20–22); these mispairings apparently underlie G→T and G→C transversions, respectively.
Normally, DNA replication in a replicative polymerase proceeds with high fidelity and processivity on a natural DNA template (23–26). The process, however, is interupted significantly when a damaged base is present in the template. Continued replication of the adduct-containing template is termed as translesion synthesis (TLS), which is a major source of point mutations (27,28). The TLS events are modulated by various factors including the lesion-induced conformational changes of the template, its surrounding base sequence context, and by the nature of DNA polymerases (28,29). The slowing of replication in a replicative polymerase is now understood to predominantly produce switch to a lesion bypass polymerase in vivo which is frequently error prone. However, many specific details of this paradigm remains unknown, i.e., how does the polymerase switch at a site of DNA damage really occur? (27) Mutagenic outcomes in replicative polymerases may also contribute to the overall mutagenic burden in vivo (30).
Although the steady-state kinetics of nucleotide insertion opposite the lesion have been studied extensively for many DNA adducts (31–34), data dealing with the long-range effects of a lesion on TLS are limited (35–39). Lindsley and Fuchs (36) have shown that the rate of primer extension by T7 DNA polymerase both at (n) and adjacent to (n+1) the lesion site with a DNA template containing an AF-lesion is reduced significantly (~10−4) relative to unmodified control DNA. A greater rate reduction (~10−6) was observed in the presence of an AAF-adduct. Miller and Grollman (37) subsequently extended the scope of the investigation, probing the long-range effects of AF- and other DNA adducts on the exo− Klenow fragment of Escherichia coli DNA polymerase I. Polymerase activity was affected as far as four bases downstream (n ~ n+3)(10−4 ~ 10−6) of the dG[AF]:dA mismatch lesion. The effect was much less pronounced (10−1 ~ 10−2) for extension of dG[AF]:dC. Similar results were obtained with Pol II & III (39). The slowed replication rate even after incorporation of bases opposite the lesion provides sufficient time for template alignment, a general model to predict deletion mutations by various bulky DNA adducts including AF- and AAF-adducts (28,31).
In the present study we have used the well-defined AF-induced B/S/W-conformational heterogeneity model (17,40) as a basis for investigating the adduct structures at various positions relative to the primer terminus during simulated AF-induced TLS. Temperature dependent 19F NMR spectra were obtained for two distinct TLS models: extension of dG[FAF]:dC-match and dG[FAF]:dA mismatch. The dynamic NMR results coupled with the modeling work with the thermophilic DNA polymerase I Bacillus Fragment (BF)(41) were examined in the context of previous kinetic parameters (36,37) in order to gain insight into the long-range effects of the conformationally flexible AF-adduct on DNA polymerase function.
RESULTS
The synthesis and purification of an FAF-modified 12-mer oligodeoxynucleotide and time-of-flight-mass spectrometry characterization were carried out using the procedures described elsewhere (19). The modified template strand was annealed with appropriate primers in order to produce various TLS models of the primer extension reaction (n−1, n, n+1, n+3, and n+6, n = primer terminus)(Fig. 1c). Figures 2 and 3 show the dynamic 19F NMR results for the dC-match and dA-mismatch extension models, respectively. We have recently demonstrated the utility of 19F NMR/CD procedure for probing the AF-induced B-S-W-conformational heterogeneities (40). The 19F NMR method takes advantage of the sensitivity of the fluorine nucleus to the tertiary structure of DNA, thereby requiring fluorine-tagged FAF as a model probe (Fig. 1a)(12). Induced circular dichroism in the 290–360 nm FAF-absorbing range (ICD290–360nm) serves as a sensitive conformational marker for probing AF-induced B-S-W heterogeneities (19,40).
Fig. 2.
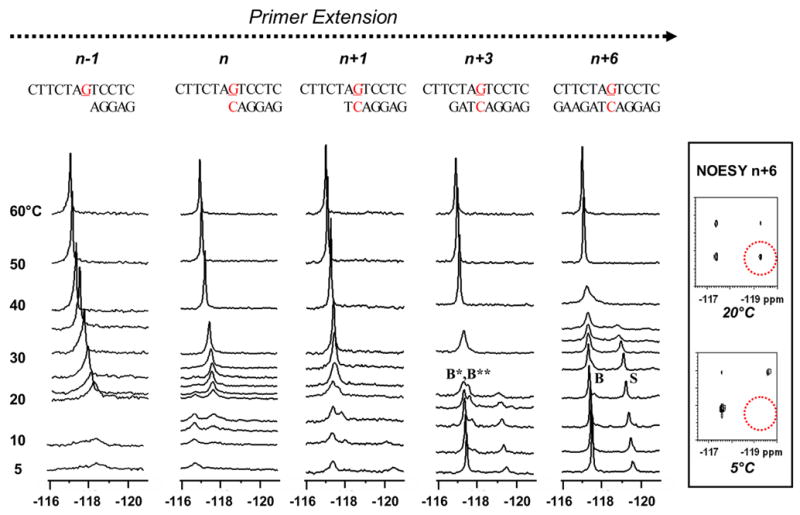
Dynamic 19F NMR spectra (−116 ~ −121 ppm) of various FAF-G:C match template/primer models at n−1, n, n+3, and n+6 positions (n=primer terminus; G=FAF-adduct). Spectra of all sequences obtained at seven standard temperatures (labeled as 5, 10, 20, 30, 40, 50, and 60 °C). Additional temperatures were recorded for n−1 (23, 25 °C), n (12, 15, 21, 22, 23, 25 °C), n+1 (15, 22, 25 °C), n+3 (15, 18 °C), and n+6 (15, 25, 33, 35 °C). Inset shows NOESY/exchange spectra of the n+6 dC-match duplex at 5 and 20 °C (dotted red circles indicate the areas for possible off-diagonal cross peaks). Tm (10−4 M) of the n+3 template-primer is 24 ~ 29 ºC. Tm (10−4) of the n+6 dC template-primer is 45.6±0.4 ºC
Fig. 3.
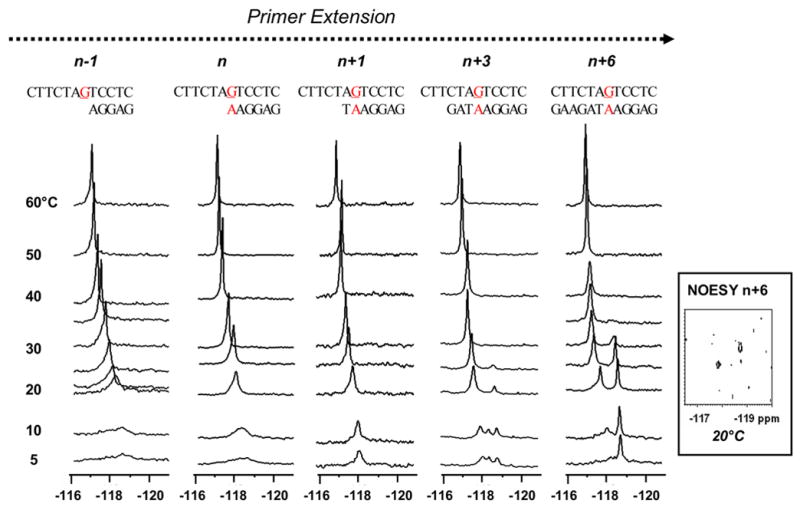
Dynamic 19F NMR spectra (−116 ~ −121 ppm) of various FAF-G:A mismatch template/primer models at n−1, n, n+3, and n+6 positions (n=primer terminus; G=FAF-adduct). Spectra of all sequences obtained at seven standard temperatures (labeled as 5, 10, 20, 30, 40, 50, and 60 °C). Additional temperatures were recorded for n−1 (23, 25 °C), n (25 °C), n+1 (25 °C), n+3 (25 °C), and n+6 (25, 35 °C). Inset shows NOESY/exchange spectrum of the n+6 dA-mismatch duplex at 20 °C. Note the lack of off-diagonal cross peaks. Tm (10−4 M) of the n+3 template-primer is 24 ~ 29 ºC. Tm (10-4) of the n+6 dA template-primer is 41.4±0.4 ºC
General Assignment Strategy
19F NMR signal assignments were made on the basis of the H/D isotopic shielding effect, nuclear Overhauser effect spectroscopy (NOESY) and adduct-induced CD experiments, as well as consideration of the sequence at the lesion site. The rationale for the H/D isotope effect is that the 19F resonance of the exposed FAF residue in a B- or W-conformer should be more susceptible to solvent-induced shielding (usually > 0.2 ppm) than the buried FAF in an S-conformer (usually < 0.1 ppm) when the deuterium content is increased from 10 to 100% (12,42). While the ICD290–360nm of a B-conformer is mostly negative, W- and S-conformers are characterized by positive ICD290–360nm values with much greater intensity for the former (40). While the sequence-dependent S/B equilibrium is a common conformational theme for fully paired DNA duplexes (19), the W-conformer has been observed exclusively in sequences that contain a purine mismatch opposite the lesion (20–22). Strong temperature dependence of off-diagonal NOESY contours indicates a transfer of magnetization from one conformer to another (12). In addition, dynamic coalescence behavior was taken as evidence for exchanging conformers. Lineshape analyses of the dynamic 19F NMR spectra were performed to determine various thermodynamic and kinetic parameters (ΔG≠, kC and τ)(43).
The FAF-lesions at (n) or adjacent to (n−1, n+1) the primer terminus generally exhibited broad NMR signals, indicating their conformational flexibility in the solvent-exposed environment. These duplexes mostly exist in single strand form at an ambient temperature due to their lower Tm values (<20 ºC). Therefore, the adduct conformation at these sites could not be defined clearly, thus considered as being in an “intermediate state” (marked with an asterisk: B*, S*, and W*).
n+6 dC-match
This fully paired n+6 dC-match duplex represents a simulated TLS model, in which the template would have undergone six replication/translocation processes from the FAF-lesion site (Fig. 2). We studied previously a similar 12-mer duplex with a reversed flanking sequence context (-TG[FAF]A-)(12). The imino proton spectrum at 5°C exhibited more than 12 proton signals with varying intensities, consistent with conformational heterogeneity (Supplementary Fig. S1). The 19F NMR spectrum at the same temperature exhibited two well-resolved signals at −117.6 and −119.6 ppm in a ratio of approximately 6:4 (Fig. 2). These signals were identified as B- and S-conformers, respectively, on the basis of the H/D isotope shielding effects (+0.17 and +0.10 ppm, respectively). Figure 2 inset shows the contour plots of NOESY/exchange spectrum recorded at 20°C, in which the presence of off-diagonal cross peaks indicates the interconverting nature of the two conformers. In contrast, no such cross peaks were present when a spectrum was obtained at 5°C. Furthermore, the two signals followed line-shape changes characteristic of a two-site exchange with coalescence occurring around ~40°C. Upon raising the temperature further to 60°C, the coalescent signal became narrowed, which is a result of a rapidly rotating FAF moiety in a single strand 12-mer.
Complete line-shape analysis yielded an activation energy value ΔG≠ of 14.6 kcal/mol, and a rate of interconversion (k = 200 s−1) at 30 ºC which is equivalent to a chemical exchange lifetime of 5 ms. The dynamics of the S/B-equilibrium can also be gleaned from the rate constant at the coalescence temperature (kC = 2.22 × Δν), at which rapid chemical shift averaging between the two conformers occurs (Fig. 2). Δν reflects the separation in frequency in Hz between the two conformer signals at 5°C, when dynamic exchange is minimal. The lower limit for kC was determined to be 1715 s−1, which is much faster than the rates of spontaneous base pair opening in a regular B-DNA molecule (44,45). The situation resembles base flipping by the DNA repair enzyme uracil DNA glycosylase (46).
n+6 dA-mismatch
Unlike the n+6 dC-duplex described above, 19F NMR spectrum of the dA-mismatch duplex revealed a single prominent resonance at −118.9 ppm at 5°C (Fig. 3). This signal revealed a +0.25 ppm H/D effect, indicative of an exposed fluoro atom. The imino proton spectrum at 5°C showed well-resolved signals for a single conformation (i.e., W-conformation see below). The duplex exhibited a strong positive ICD290–360nm (Fig. 4), which is a characteristic marker for W-conformation (40). These results are in agreement with previous findings that duplexes containing purine bases (A or G) opposite the AF-lesion adopt a W-conformation (20–22).
Fig. 4.
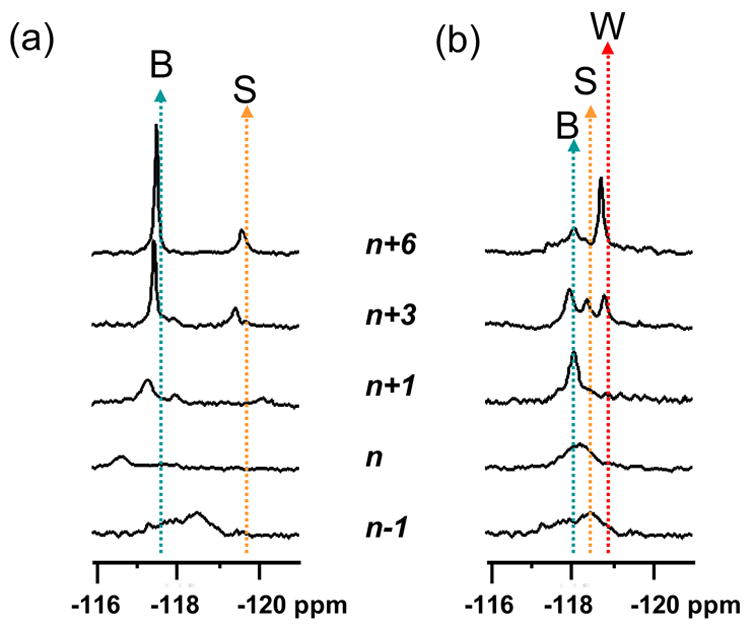
Stack plots of 19F NMR spectra of various (a) dC-match and (b) dA-mismatch FAF-template/primer series at 10°C. Dotted arrows specify conformer assignments.
A new signal (−118.2 ppm) appeared at 10°C and it gained intensity with increasing temperatures (i.e., 20 ~ 30°C). The emerging heterogeneity is evidenced by the appearance of varying intensities of proton signals at 20°C in the 1H NMR spectrum, a situation which was distinct from that observed at 5°C (Supplementary Fig. S2). At 20°C, the downfield and upfield signals exhibited deuterium-induced shielding of 0.10 and 0.41 ppm, respectively. The unusually large isotope effect observed for the upfield signal is consistent with the fully exposed fluoro at C7 of the FAF-moiety, which is sandwiched within the walls of the narrow minor groove. The NOESY spectrum of the n+6 dA duplex at 20°C (Fig. 3 inset) lacked discernable off-diagonal cross peaks, indicating that it does not exhibit conformeric exchange. This is in contrast with the afore-mentioned dC-duplex data (Fig. 2a, inset) obtained at the same temperature, which adopted a well-defined S/B-equilibrium (see above). The shape and chemical shifts of the upfield W-conformer signal did not change substantially until the temperature reached 30°C, and then it collapsed in the narrow 30 – 35°C range. This behavior appeared more like that of a duplex melting rather than a two-site dynamic exchange. Taken together, these results suggest a local denaturation at the lesion site, which represents a transition from a W-conformer to the thermodynamically less stable S- and B-conformers and eventually to single strands (Tm = 41.4°C).
Duplex Stability
We were unable to accurately determine Tm values for the n−1, n, and n+1 template-primers. The Tm values for the n+3 dC or n+3 dA template-primers were determined to be in the 24 ~ 29 ºC range. The fully paired 12-mer duplexes are a mixture of multiple conformers and exist as a mixture of single and double stranded forms at ambient temperatures. As for the n+6 dC-match duplex, the FAF-adduction decreased the thermal (ΔTm = −7.2°C) and thermodynamic (ΔΔG° = 1.8 kcal/mol) stabilities (Table 1). This is a typical trend for S/B-conformeric duplexes (19). In contrast, the values observed for the n+6 dA duplex were comparable (ΔTm = 0.9°C, ΔΔG = ~ 0 kcal/mol) to those of the control duplex (Table 1). Similar duplex stability has been observed for other W-like conformeric duplexes (17,18). This is also reminiscent of “another” exclusively W-conformeric dG-N2-AAF-modified 11-mer duplex, whose thermal (ΔTm = 6.3°C) and thermodynamic stability (ΔΔG° = −1.8 kcal/mol) is greater than the control (47). In this case, the acetylaminofluorene-moiety is sandwiched very tightly within the walls of the minor groove and maintains Waston-Crick hydrogen bonds at the lesion site. The entropic component of the Gibb’s free enegy has been shown to be responsible for the stability of this unique W-conformation. It should be noted, however, that this is not true for all minor-minor groove binding structures (18). Thus, it appears that the duplex stability of minor groove conformers depends on the curvature of the helix and the depth of the minor groove, which in turn are influenced by the size, planarity, shape and stereochemistry (in the case of PAH-adducts) of the carcinogenic adduct.
TABLE 1.
Effects of FAF-modification on the thermal and thermodynamic stability of the n+6 dC-match and n+6 dA-mismatch 12-mer duplexes
| Sequencea | −ΔGb (kcal/mol) | −ΔHb (kcal/mol) | Tmc (ºC) | ΔΔGd (kcal/mol) | ΔΔHe (kcal/mol) | ΔTmf (ºC) |
|---|---|---|---|---|---|---|
| -AG*T-
-TC A- |
8.4 (10.2) | 76.8 (68.6) | 45.6 (52.8) | 1.8 | 8.2 | −7.2 |
| -AG*T-
-TA A- |
7.4 (7.4) | 66.5 (74.8) | 41.4 (40.5) | 0 | −8.3 | 0.9 |
The central trimer portion of the 12-mer duplex (G* = FAF-adduct, see Fig. 1).
The results of curve fit and Tm-lnCt dependence were within ±15% of each other and therefore these numbers are average of the two methods (19). The average standard deviations for − ΔG, − ΔH°, and Tm are ± 0.2, ± 6.3, and ± 0.4 respectively. Values in parenthesis are for the controls.
Tm values at 10−4 M taken from the 1/Tm − lnCt/4 Meltwin plots.
ΔΔG = ΔG° (FAF-modified) − ΔG° (control).
ΔΔH = ΔH° (FAF-modified) − ΔH° (control).
ΔTm= Tm (FAF-modified) − Tm (control).
n-1
The n-1 sequence represents a 12/5-mer TLS model, in which replication of the modified strand would have proceeded prior to the lesion. The 19F signals in the 5 ~ 10°C range were very broad, reflecting the extensive conformational mobility at the lesion site (Figs. 2 and 3). Upon increasing temperature, the duplex portion (5 nt) of the model melted quickly (Tm < 17 °C) to give rise to a sharp signal, which shifted gradually downfield. The narow signals represent the mobile FAF residue of the denatured single strand template. Previous 1H NMR studies have shown that a similar n−1 model AF-modified 13/9-mer adopts AF-stacked S-like conformation, reminiscent to the intercalated (+)-anti-trans-N2-[BP]dG adducts in the same sequence context (48). In addition, the 1H NMR results revealed the existence of the AF-rotamers with respect to the β’ torsion angle. In concordance, further evidence of mobility of the carcinogen moiety is supported the observation of a large (+0.21 ppm at 20°C) H/D isotope effect. Therefore, the signals below 20°C appear to indicate the presence of S*- or S*-like conformers with possible AF-β’-rotamers. The existence of a mixed S*/B*-equilibrium, however, cannot be ruled out.
n
In the n model replication would have proceeded up to the adduct site with either the normal partner dC (Fig. 2) or the mismatch dA (Fig. 3) directly opposite of the lesion. There were not many differences between the n and n−1 dA-mismatch models in terms of spectral patterns and H/D solvent effect (+0.19 ppm at 20°C)(Fig. 3). Thus, definite conformational assignment for the dA mismatch model was difficult (Tm < 20 ºC), although it could be considered a mixture of S*/B*-equilibrium. On the other hand, the spectrum of the n dC-match model (Fig. 2) revealed two signals at −116.8 and −117.8 ppm in a 3:7 ratio at 20°C, which were coalesced at ~30°C (Fig. 2). The downfield −116.8 ppm signal, which was dominant in the lower temperatures, exhibited an H/D isotope shift (+0.19 ppm at 20°C). This is contrasted with the small effect (+0.02 ppm at 20°C) observed for the upfield signal. The distinctive resonance patterns observed for the two n models are presumably due to the fact that dC, but not dA, can form Watson-Crick base pairs at the lesion site. These results support the presence of a B* (−116.8 ppm)/S* (−117.8 ppm) equilibrium. The S-conformer is generally more susceptible to a ring current effect, which enables it to be shielded relative to the B-conformer (12).
The observation that the n dC model adopts an S/B-equilibrium is not in line with previous 1H NMR results (49), which showed the presence of an S-type conformer for the AF-modified n-model 13/10-mer regardless of whether dC or dA was present opposite of the lesion. The (+)-anti-trans-N2-[BP]dG adducts in the same sequence context exhibited the S-type structural feature as well (50). The reason for the discrepancy is not clear, but could be due to the fact that the 13/10-mer sequence possesses a longer duplex (10 vs. 6 bp) segment and a shorter single strand segment (3 vs. 6 nt) as well as different flanking sequence context (e.g., -CG[AF]C). It is also worth mentioning that the authors pointed out the presence of an envelope of broad imino signals centered around 11.0 ppm (49). They assigned only one of those to the imino proton for the modified dG of the dG[AF]:dC match sequence. This is contrasted with the sharp resonance detected for the same proton of the same 13/10-mer containing the dG[AF]:dA mismatch, thus hinting multiple conformations for the dG[AF]:dC match sequence.
n + 1 ~ n + 3 dC-match
The dynamic profile of the n+3 dC-match model below 10°C was analogous to that observed for the n+6 dC duplex (Fig. 2). The signals at −117.5 and −119.4 gave H/D isotope effects of +0.38 and +0.01 ppm at 10°C, as expected for B- and S-conformers, respectively. Unlike the fully paired n+6 duplex, however, the downfield B-conformer signal split up into two signals (B* and B**) at 15°C and coalesced at 30°C. The H/D isotope shielding of the split signals was found to be in the 0.34 ~ 0.38 ppm range, indicating the presence of a fully solvent-exposed FAF residue (i.e., B-conformer). In contrast, a small shift (+0.01 ppm) was measured for the upfield S-conformer −119.4 ppm signal. Figure 4a shows the stacked plots of all dC-extension models at 10°C, demonstraing a clear progression of conformational distinction with increasing length of the primer. The pattern observed for the n+1 model is similar to that of the n+3 model, except for the absence of a well-defined S*-conformer signal. The nature of the B-conformer split is not clear, although the conformational flexibility at the lesion site could in principle produce a number of low-energy structures including rotamers with respect toα’ and β’ tortional angles.
n + 1 ~ n + 3 dA-mismatch
The dynamic NMR profile of the n+1 dA-mismatch model did not differ substantially from those of the n−1 and n models except that the signals were narrowed significantly at lower temperatures. The adduct conformation was well-defined at the n+3 position, in which three distinctive resonances were observed at -118.1, −118.5, and −118.9 ppm in the 5 – 10°C range. These signals merged into the most shielded one (−118.9 ppm) at n+6, which was identified as a W-conformer (see above). The two downfield signals (−118.1 and −118.5 ppm) were identified as B* and S*-conformers, respectively, on the basis of the chemical shift and consideration of the ring current effect (12). Figure 4b shows the stack plot of all six dA-mismatch models at 10°C, which illustrates the dramatic S*-B*/S*/W*-W conformational transition. A rapid disappearance of the S*-conformer at 20°C indicated its intermediary nature in the B*/S*/W*-equilibrium. Various S*/B*, S*/W* and B*/W* equilibria presumably exist in the 5 ~ 20°C range. The persistence of the W*-conformer at n+3 confirm the greater thermal and thermodynamic stability of W- over S- and B-conformers.
ICD290–360 nm
Figure 5 shows an overlay of the CD spectra of the n+3 and n+6 models for both the dC- and dA-series. The n+6 dC duplex exhibited mostly negative ICD290–360 nm because of its high B-conformer content (~64%)(19). In contrast, the highly positive ICD290–360 nm observed for the corresponding n+6 dA duplex must be due to the exclusive presence of the W-conformer (40). The ellipticity intensities of ICD290–360 nm in both dC and dA series were decreased significantly in the n+3 TLS models, reflecting conformational mobility at the lesion site (Figs. 2 and 3). The CD results are in good agreement with the dynamic 19F NMR data.
Fig. 5.
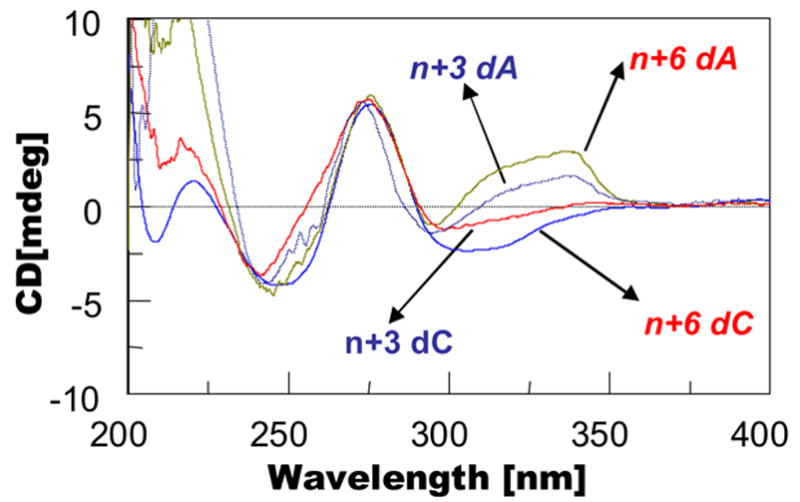
Overlay of CD plots of dC-match and dA-mismatch series at n+3 and n+6 positions.
DISCUSSION
19F NMR Probing of the AF-Conformational Heterogeneity
The FAF-adduct conformation was found to be influenced greatly by the extent of the primer extension (n−1 ~ n+6) with respect to the template lesion. We also found that the conformational diversity at a given primer position is modulated by temperature as well as by the nature of the base pair at the lesion site. For example, while the dC-match series adopted an S/B-equilibrium (Fig. 2), a more complex B/S/W-heterogeneity (Fig. 3) was observed for the dA-mismatch series. Our dynamic 19F NMR results indicated that conformational flexibilities (“intermediate states”) exist for those sequences containing the AF-adduct at (n) or flanking (n−1, n+1) the primer-template junction. However, conformational distinction improved gradually as the length of primer sequences increased. This phenomenon was observed for both dC- and dA-series in a specific manner (Fig. 2 and 3) and appeared to be driven by the greater thermal and thermodynamic stabilities of the W-conformer relative to the S- and B-conformers. Thus, the extension of dG[FAF]:dA-mismatch underwent a progressive conformational transition from a B*/S*- (n−1 ~ n+1), B*/S*/W*- (n+3)-equilibrium to an exclusive W-conformation (n+6) (see Figs. 3 and 4b). The transition for the dC-match series consisted of a B*- or S*- (n−1), S*/B*- (n), B*/B**- (n+1), B*/B**/S*-equilibrium (n+3) to an S/B-equilibrium (n+6) (Figs. 2 and 4a). Table 2 summarizes the structure of DNA duplex, the conformational snapshots of FAF-adducts at various primer positions, and kinetics of extension available form previous studies (36,37).
TABLE 2.
Conformational heterogeneities observed at various FAF-modified primer-templates and their relative nucleotide insertion frequencies by KFexo−
| Position of primer terminus with respect to lesiona | Conformation for dG*:dC matcha | Conformation for dG*:dA mismatcha | Relative frequencyb for nucleotide (base insertion catalyzed by KFexo− opposite) |
|---|---|---|---|
| n−1 | S*/B*a | S*/B* | 0.37 |
| n | B(30%), S(70%) | S* | 0.36 (dC);d 1.1 × 10−5 (dA) |
| n+1 | B*, B** | S* | 0.032 (dC); 7.46 × 10−5 (dA) |
| n+3 | B*, B**(80%), S(20%) | B* (80%)/W* (20%)
B* (38%)/S* (24%)/W* (37%)c |
0.16 (dC); 5.8 × 10−4 (dA) |
| n+6 | B(60%), S(40%) | S (56%) /W (44%) | NAe |
dG* = FAF adduct (Figure 1). Conformation population at 20 °C otherwise indicated.
Data taken from reference 37. Fext = (kcat/Km)lesion template/(kcat/Km)control template.
@ 10 °C.
Preferred nucleotide inserted at lesion site is dC.
NA: not available
We observed the predominant presence of the W-conformer for the n+6 dA-mismatch duplex at 5°C, which was quickly unraveled into single strands in the 5 – 30°C range (Fig. 3). These results imply the existence of a Wsyn/Banti-equilibrium, the conversion of which could take place through a two-step process; i.e., an initial Wsyn/Ssyn transition followed by an Ssyn/Banti one. The dynamic energy barriers of the B/S/W-equilibrium for duplexes with a purine mismatch at the lesion site are not yet known. However, the interconversion energy for the Ssyn/Banti-equilibrim of fully paired duplexes was found to be in the range of ΔG≠ = 14 – 15 kcal/mole (Meneni et al., unpublished results). The Wsyn/Ssyn-equilibrium is expected to occur readily since both conformers maintain very closeχ,α’, andβ’ torsion angles (21,22).
Long-range Conformational Distortion Effects
Replication via high fidelity replicative DNA polymerases proceeds by threading DNA through the active site in a well-orchestrated manner (23–26). Using BF, Johnson and Beese (51) characterized the crystal structures of all 12 possible mismatches captured at the growing primer terminus in the active site of the polymerase. They found that mismatch-induced distortion is not just localized to the mismatch site, but extends up to six base pairs downstream through the polymerase ‘DNA binding region.’ This so-called ‘short-term memory of replication errors’ is conceptually analogous to the ‘long-range distortion effects” observed previously by AF-adducts on T7 and KF polymerases (36,37). Thus, inhibition of polymerase function is not only at the site of the lesion but also positions up to at least 3 nucleotides past the adduct site. The rates of primer extension for dG[AF]:dC mispair is known to be ~104-fold slower compared to the dG[AF]:dC match (Table 2).
Recent crystal studies have provided insight into how incorporation rates might be reduced drastically at the n and n+1 positions. Hsu et al (52) presented snapshot structures of the AF-adduct docked in the active site of a BF that undergoes one round of replication both in solution and as a crystal. The syn-AF-adduct occupied the preinsertion site with minimal perturbation of the active site. With an incoming dCTP, however, the modified dG switched its conformation to anti-, enabling Watson-Crick hydrogen bonds to form. A subsequent chemical reaction allowed the aminofluorene-moiety to project into the solvent-exposed major groove, reminiscent of the B*-conformeric n dG[FAF]:dC match TLS model described in the present study. The newly formed dG[AF]:dC base pair, however, blocks the next template base from occupying the template preinsertion site, thereby stalling DNA synthesis. Dutta et al (53) have studied a crystal from the T7 DNA polymerase treated with ddCTP or ddATP and found sketchy electron density around the template AF-adduct. The observed disorder reflects the dynamic mobility of the AF-adduct within the active site of a crystalline polymerase and is consistent with our 19F NMR finding, indicating that the n dC-match model adopts an S*/B*-equilibrium (Fig. 2). Thus, the dG[AF]-adduct at the replication fork adopted either an anti-conformation to form Watson-Crick hydrogen bonds (i.e., B*-conformer) or a syn-conformation to produce an S*-like conformer and the process evolved into a well-defined S/B-mixture. Similarly, population of the W-conformer increases with increasing the length of primers (Fig. 3). Conformational status is well-establshed at n+6, yet the nucleotide insertion rates in both cases were found to be the slowest between n and n+3 and normal activities resumed at n+5 and beyond (37).
To understand the mechanisms of this long-range effect, we modeled several AF-template-primers (n+1, n+3, n+6) with known coordinates of BF, a high-fidelity DNA polymerase with extensive sequence and structural homology to the KF polymerase (41). No suitable crystal structure of a ternary complex for KF is currently available. Figure 6 shows the BF polymerase complexed with the AF-modified n+3 TLS model with either a B- (dC-match) or a W- (dA-mismatch) conformer as illustrative purposes. In the B-conformation the bulky aminofluorene-moiety (shown in red CPK) remains in the solvent-exposed major groove; thus much less disruption of replication is expected as the DNA binding region translocates through the polymerase. On the other hand, the AF-moiety in the minor groove of the W-conformer limits not only the mobility of the DNA, but also imposes a serious steric clash with the polymerase throughout the replication/translocation process. As a result, the replication rate is dramatically reduced. This finding is reminiscent of the minor groove binding (+)-trans-anti-BP-DNA adduct, in which the bulky benzo[a]pyrene imposes major disruption in the duplex binding site (54,55). Similar situations were observed for the n and n+1 models. At the n+6 position (Fig. 6), however, the AF-lesion is located near the tip of the duplex binding region of the polymerase binding region, and therefore exerts less interference upon protein-DNA interactions. The adverse steric impact of the AF-moiety in the S-conformation is expected to be in between those observed with the W- and B-conformations.
Fig. 6.
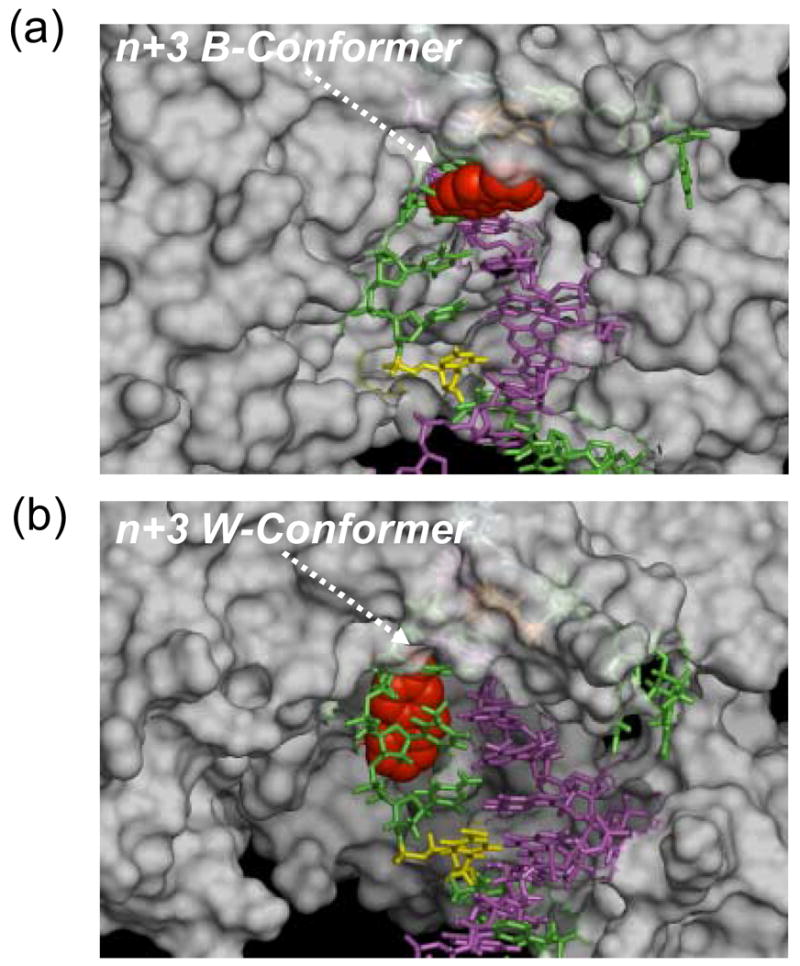
Example n+3 TLS models of BF complexed with AF-modified template/primer DNA: Arrows indicate (a) the n+3 dC-match (B-type conformer) and (b) the n+3 dA-mismatch (W-conformer) template-primers. The carcinogenic aminofluorene moiety is shown in red CPK. Template and primer sequences are colored green and magenta, respectively. For illustrative purpose, the n+1 and n+6 positions relative to the primer terminus n in the template strand are colored orange (under the surface) and yellow, respectively.
These results are consistent with the steady state kinetic data described previously by Miller and Grollman (36,37)(Table 2). The dramatic differences in the rates of nucleotide insertion (~104-fold) observed for dG[FAF]:dC match and dG[FAF]:dA mismatch emphasize the importance of adduct-induced steric constraints in the DNA binding area, especially the minor groove interaction between the template-primer DNA and the polymerase, for determining replication fidelity (52–55). Altering the adduct-induced conformational equilibria is important, but the complex features of enzyme structure and interaction with the lesion must be taken into consideration.
Mutational Implication of the Long-range AF-Conformational Heterogeneity
The fact that retardation of DNA synthesis persists even after incorporation of the base immediately opposite of the lesion has an important implication for the mechanisms of mutagenesis (56–62). Such a delay during TLS increases the propensity to produce a family of misaligned slipped mutagenic intermediates (SMI), a model that is known to lead to frameshift mutations by a variety of bulky DNA adducts including AF and AAF. The sequence context is a critical factor, which can be used to predict the nature of various targeted or semi-targeted deletions. For example, it has been shown in Escherichia coli that insertion of a correct dCMP (most frequent) opposite of the AAF-lesion can produce one-, two- or longer-base deletions with high frequency when the appropriate complementary base match is available 5’ to the lesion (59–62). Analogous insertion-misalignment mechanisms could also be applied to mammalian cells (57). It has recently been reported that the structure of the AAF-adduct in the Escherichia coli NarI hot-spot sequence in the polymerase active site is very different from that of a non-NarI sequence, highlighting the importance of proper active site binding (63).
dAMP is the second most common nucleotide to be incorporated opposite of the AF-lesion (57). The resulting dA-mismatch adducts, whose structures have been studied in both fully paired duplexes (20,21) and at primer-template junctions (58), provide the mechanistic rational for G → T transversions, which represent the major mutation observed for an AF-adduct. As described above, elongation is significantly hampered by several nucleotides 5′ of the lesion, and the effect is much greater when a dA is incorrectly inserted than when a dC is correctly inserted opposite of the lesion. This effect can be attributed to a growing W-conformer whose steric clash with the protein is transmitted to the active site through long-range distortions. No such steric interference is expected for the B-conformer once the polymerase has progressed past the lesion. Thus, primer extension over the dG[AF]:dA-mismatch is expected to be more mutagenic synergistically by not just base mutations, but also various deletions in the appropriate sequence contexts.
The mutagenic consequences of lesion bypass and replicative polymerases are very different, both structurally and functionally. Primer-extension studies indicate that the AF adduct causes partial blocking both at -1 and n positions with Escherichia coli DinB and human pol κ Δc (62). Zhang et al (64) have modeled nucleotide incorporation opposite the major adduct derived from the food carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) in Dpo4, the DinB family polymerase. Like the AF-adduct, the PhIP adduct exhibits an S/B conformational heterogeneity in solution (65). The results indicate that the PhIP ring system is pushed into the minor groove (as in W-type conformer) to accommodate an incoming dNTP at the active site of the enzyme, which seriously disturbs its geometry, regardless of the identity of dNTP. The B-conformeric PhIP ring on the spaceous major groove in Dpo4 allows the active site to accommodate dCTP, dTTP or dATP reasonaly well; however. This is contrasted with the same adduct in the replicative polymerase RB69, which experiences considerable active site distortion when partnered by C or A. Polymerase stalling in vivo could lead to switch to an error-prone bypass polymerase.
In conclusion, adduct- or sequence-induced conformational heterogeneity has become a signature property for many bulky carcinogen-DNA adducts. The results from the dynamic 19F NMR, CD, and modeling experiments provide conformational insight into how AF-induced heterogeneity at positions distant from the primer terminus can influence the downstream polymerase activity, a novel aspect of the present study. The S/B conformational equilibria depend on the adduct position in relation to the single strand/double strand junction. The dramatic differences in the rates of nucleotide insertion observed for dG[FAF]:dC match and dG[FAF]:dA mismatch can be attributed to the differences in the adduct conformation at the lesion site and their long range distortion effects on inducing replication blockage in the active site of the polymerase. The energy differences among the conformers are small, such that their relative conformeric populations could be readily altered in the active sites of an enzyme. Therefore, the ‘enzyme-induced conformational heterogeneity’ must be taken into consideration in order to establish a meaningful ‘conformation-function’ relationship. In addition, the findings in the present studies further reinforce the utility of the dynamic 19F NMR/CD procedure in investigating adduct-induced conformational heterogeneities.
MATERIALS AND METHODS
Crude oligodeoxynucleotides (10–15 μmol scale) in desalted form were purchased from Sigma-Genosys (The Woodlands, TX) and purified by reverse phase high performance liquid chromatography (RP-HPLC). All HPLC solvents were purchased from Fisher Inc (Pittsburgh, PA). The HPLC system consisted of a Hitachi EZChrom Elite HPLC system with a L2450 diode array as a detector and a Luna column (10 × 150 mm, 5μ) (Phenomenex, Torrance, CA). The mobile phase system involved a 30-min linear gradient profile of 5–15% acetonitrile/0.1 M ammonium acetate buffer (pH 7.0) with a flow rate of 2.0 mL/min. Sorvall Evolution RC Ultracentifuge (Thermo Electron, Waltham, MA) was used to filter NMR samples.
Preparation of the FAF-modified template
An FAF-modified 12-mer DNA template (5′-CTTCTAG[FAF]TCCTC-3′) was prepared according to a previously published procedure (12,19). Briefly, an unmodified oligo was treated with N-Acetoxy-N-trifluoroacetyl-7-fluoro-2-aminofluorene, an activated derivative of 7-fluoro-2-aminofluorene. The modified strand was purified by reverse HPLC and characterized by mass spectrometric procedure described previously (Calculated, 3724.70; Found, 3724.82).
NMR Experiments
Approximately 60 OD of pure modified oligos were annealed with primer sequences to produce appropriate template-primers (see Fig. 1c for sequences). The samples were filtered by ultracentrifugation using a Pall Microsep MF centrifugal device (Yellow, MW cutoff = 1,000). The centrifuged samples were dissolved in 300 μL of a neutral buffer (10% D2O/90% H2O containing 100 mM NaCl, 10 mM sodium phosphate, and 100 μM tetrasodium EDTA, pH 7.0), filtered through a 0.2 μm membrane filter, and placed in a Shigemi tube for NMR experiments.
All 1H and 1H-decoupled 19F NMR results were recorded using a dedicated 5-mm 19F/1H dual probe on a Bruker DPX400 Avance spectrometer operating at 400.0 and 376.5 MHz, respectively. Imino proton spectra were obtained using phase sensitive jump-return sequences at 5°C and referenced relative to DSS. 19F NMR spectra were referenced to CFCl3 by assigning external hexafluorobenzene in C6D6 at −164.90 ppm. One-dimensional 19F NMR spectra at 5 – 60°C were obtained by collecting 65536 points using a 37664-Hz sweep width and a recycle delay of 1.0 s between acquisitions. A total of 1600 scans were acquired for each spectrum. The spectra were processed by zero-filling, exponential multiplication using 20 Hz line broadening and Fourier transformation. The peak areas were base-line corrected and integrated using XWIN NMR software (Bruker, Billerica, MA). Two-dimensional NOESY/exchange 19F NMR spectra were carried out in the phase-sensitive mode using a NOESY pulse sequence: sweep width 4529 Hz, number of complex data points in t2 1024, number of complex free induction decays in t1 256, number of scans 96, dummy scanes 16, recycle delays 1.0 s, and mixing time 400 ms. The data were subjected to sine bell apodization using 2 Hz line broadening in both dimensions and then zero-filled before Fourier transformation of the 1024 × 256 data matrix. The data were not symmetrized. Complete line-shape analysis (43) was carried out using WINDNMR-Pro (version 7.1.6, J. Chem. Educ. Software Series; Reich, H. J., University of Wisconsin, Madison, WI).
Circular Dichroism (CD) Spectra
CD measurements were conducted on a Jasco J-810 spectropolarimeter equipped with a variable Peltier temperature controller. Typically, 2 OD of a FAF-modified template strand were annealed with 1 equivalent of primer strands. The samples were dissolved in 400 μL of a neutral buffer [0.2 M NaCl, 10 mM sodium phosphate, 0.2 mM EDTA, pH 7.0] and placed in a 1-mm path-length cell. The sample was heated at 85°C for 5 min and then cooled to 15°C over a 10 min period to ensure duplex formation. Spectra were measured from 200 to 400 nm at a rate of 50 nm/min; the final data were averaged from 10 accumulations. Data points were acquired every 0.2 nm with a 2-s response time.
UV-Thermomelting Experiments
Melting experiments were conducted on a Beckman DU 800 UV/VIS spectrophotometer equipped with a 6-chamber, 1-cm path length Tm cell. Sample cell temperatures were controlled by a built-in Peltier temperature controller. Duplex solutions with a total concentration in the range of 0.2 – 14 μM were prepared in solutions containing 0.2 M NaCl, 10 mM sodium phosphate, and 0.2 mM EDTA at pH 7.0. The concentration of each oligo sample was estimated based on UV absorbance at a wavelength of 260 nm. Thermomelting curves were constructed by varying the temperature of the sample cell (1°C/min) and monitoring the absorbance of the sample at 260 nm. A typical melting experiment consisted of forward/reverse scans and was repeated three times. Thermodynamic parameters for bimolecular melting reactions of the duplexes were obtained from melting curve data using the program MELTWIN® version 3.5. The margins of the parameters derived from the curve fit data and from Tm−1 - lnCt plots were found to be within ± 15%, therefore average values from the triplicated experiments were used as described previously (19).
Modeling of the BF-DNA Complex
The Biopolymer module of Insight II (Accelrys Inc.) was used to construct models. The molecular models of the ternary FAF-DNA/dNTP/BF structures were generated using the coordinates obtained from the Protein Data Bank (PDB ID: 1UA0 (n−1)(52) and the unit A of the crystal structure 1LV5 within the DNA polymerase BF (41). The DNA sequence in the crystal structures was adjusted as needed to match those in the present work. For the n−1 model, an anti-dG[AF] model was also constructed in addition to the syn-dG[AF] observed in the crystal structure. This was achieved by rotating the dG[AF] glycosidic bond by ~180°. Representative anti- and syn-dG[AF] structures were selected by varying the the linkage site torsion angles α‘ and β‘ over their 360° range and evaluating steric collisions. For the n+3 and n+6 W-conformer models, the linkage site geometry observed by Norman et al. (20) in the NMR solution structure of the dG[AF]:dA mismatch was used. The models were subjected to minimization, equilibration, and 700 ps dynamics simulation with AMBER8 (67). The structures in Figure 6 were prepared using PyMOL (68).
Supplementary Material
Acknowledgments
We thank Dr. Paul Chiarelli for providing TOF-MS spectra of the FAF-modified 12-mer template. We are grateful to the NIH (R01CA098296) for their financial support of this work. This research was also made possible in part by the use of the Research Core Facility supported by the NCRR/NIH (P20 RR016457).
Abbreviations
- AF-adduct
N-(2′-deoxyguanosin-8-yl)-2-aminofluorene
- AAF-adduct
N-(2′-deoxyguanosin-8-yl)-2-acetylaminofluorene
- BF
Bacillus fragment
- CD
circular dichroism
- FAF
7-fluoro-2-aminofluorene
- ICD290–360 nm
induced circular dichroism at 290–360 nm
- SMI
Slipped Mutagenic Intermediates
- TLS
translesion synthesis
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Luch A. Nature and nature-lessons from chemical carcinogenesis. Nature Rev Cancer. 2005;5:113–125. doi: 10.1038/nrc1546. [DOI] [PubMed] [Google Scholar]
- 2.Garner RC. The role of DNA adducts in chemical carcinogenesis. Mutat Res. 1998;402:67–75. doi: 10.1016/s0027-5107(97)00283-2. [DOI] [PubMed] [Google Scholar]
- 3.Heflich RH, Neft RE. Genetic toxicity of 2-acetylaminofluorene, 2-aminofluorene and some of their metabolites and model metabolites. Mutat Res. 1994;318:73–114. doi: 10.1016/0165-1110(94)90025-6. [DOI] [PubMed] [Google Scholar]
- 4.Kriek E. Fifty years of research on N-acetyl-2-aminofluorene, one of the most versatile compounds in experimental cancer research. J Cancer Res Clin Oncol. 1992;118:481–489. doi: 10.1007/BF01225261. [DOI] [PubMed] [Google Scholar]
- 5.Beije B, Moller L. 2-Nitrofluorene and related compounds: prevalence and biological effects. Mutat Res. 1988;196:177–209. doi: 10.1016/0165-1110(88)90019-x. [DOI] [PubMed] [Google Scholar]
- 6.Ueda O, Kitamura S, Ohta S. Metabolism of 2-nitrofluorene, an environmental pollutant, by liver preparations of sea bream. Pagrus major Xenobiotica. 2002;32 :667–682. doi: 10.1080/00498250210144839. [DOI] [PubMed] [Google Scholar]
- 7.Beland FA, Kadlubar FF. In: Handbook of Experimental Pharmacology. Cooper CS, Grover PL, editors. Springer-Verlag and Heidelberg; 1990. pp. 267–325. [Google Scholar]
- 8.Shibutani S, Suzuki N, Tan X, Johnson F, Grollman AP. Influence of flanking sequence context on the mutagenicity of acetylaminofluorene-derived DNA adducts in mammalian cells. Biochemistry. 2001;40:3717–3722. doi: 10.1021/bi0027581. [DOI] [PubMed] [Google Scholar]
- 9.Cho BP, Zhou L. Probing the conformational heterogeneity of the acetylaminofluorene-modified 2′-deoxyguanosine and DNA by 19F NMR spectroscopy. Biochemistry. 1999;38:7572–7583. doi: 10.1021/bi990182d. [DOI] [PubMed] [Google Scholar]
- 10.O’Handley SF, Sanford DG, Xu R, Lester CC, Hingerty BE, Broyde S, Krugh TR. Structural characterization of an N-acetyl-2-aminofluorene (AAF) modified DNA oligomer by NMR, energy minimization, and molecular dynamics. Biochemistry. 1993;32:2481–2497. doi: 10.1021/bi00061a005. [DOI] [PubMed] [Google Scholar]
- 11.Milhe C, Fuchs RP, Lefevre JF. NMR data show that the carcinogen N-2-acetylaminofluorene stabilises an intermediate of -2 frameshift mutagenesis in a region of high mutation frequency. Eur J Biochem. 1996;235:120–127. doi: 10.1111/j.1432-1033.1996.00120.x. [DOI] [PubMed] [Google Scholar]
- 12.Zhou L, Rajabjadeh M, Traficante DD, Cho BP. Conformational heterogeneity of aryl amine-modified DNA: 19F NMR evidence. J Am Chem Soc. 1997;119:5384–5389. [Google Scholar]
- 13.Cho BP, Beland FA, Marques MM. NMR structural studies of a 15-mer DNA duplex from a ras protooncogene modified with the carcinogen 2-aminofluorene: conformational heterogeneity. Biochemistry. 1994;33:1373–1384. doi: 10.1021/bi00172a013. [DOI] [PubMed] [Google Scholar]
- 14.Eckel LM, Krugh TR. Structural characterization of two interchangeable conformations of a 2- aminofluorene-modified DNA oligomer by NMR and energy minimization. Biochemistry. 1994;33:13611–13624. doi: 10.1021/bi00250a012. [DOI] [PubMed] [Google Scholar]
- 15.Mao B, Hingerty BE, Broyde S, Patel DJ. Solution structure of the aminofluorene [AF]-intercalated conformer of the syn-[AF]-C8-dG adduct opposite dC in a DNA duplex. Biochemistry. 1998;37:81–94. doi: 10.1021/bi972257o. [DOI] [PubMed] [Google Scholar]
- 16.Mao B, Hingerty BE, Broyde S, Patel DJ. Solution structure of the aminofluorene [AF]-external conformer of the anti-[AF]-C8-dG adduct opposite dC in a DNA duplex. Biochemistry. 1998;37:95–106. doi: 10.1021/bi972258g. [DOI] [PubMed] [Google Scholar]
- 17.Cho BP. Dynamic conformational heterogeneities of carcinogen-DNA adducts and their mutagenic relevance. J Environ Sci Health C: Environ Carcinog Ecotoxicol Rev. 2004;22:57–90. doi: 10.1081/LESC-200038217. [DOI] [PubMed] [Google Scholar]
- 18.Geacintov NE, Cosman M, Hingerty BE, Amin S, Broyde S, Patel DJ. NMR solution structures of stereoisometric covalent polycyclic aromatic carcinogen-DNA adduct: principles, patterns, and diversity. Chem Res Toxicol. 1997;10:111–146. doi: 10.1021/tx9601418. [DOI] [PubMed] [Google Scholar]
- 19.Meneni SR, D’Mello R, Norigian G, Baker G, Gao L, Chiarelli MP, Cho BP. Sequence effects of aminofluorene-modified DNA duplexes: thermodynamic and circular dichroism properties. Nucleic Acids Res. 2006;34:755–763. doi: 10.1093/nar/gkj480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Norman D, Abuaf P, Hingerty BE, Live D, Grunberger D, Broyde S, Patel DJ. NMR and computational characterization of the N-(deoxyguanosin-8-yl) aminofluorene adduct [(AF)G] opposite adenosine in DNA: (AF)G[syn] A[anti] pair formation and its pH dependence. Biochemistry. 1989;28:7462–7476. doi: 10.1021/bi00444a046. [DOI] [PubMed] [Google Scholar]
- 21.Shapiro R, Hingerty BE, Broyde S. Minor-groove binding models for acetylaminofluorene modified DNA. J Biomol Struct Dyn. 1989;7:493–513. doi: 10.1080/07391102.1989.10508506. [DOI] [PubMed] [Google Scholar]
- 22.Abuaf P, Hingerty BE, Broyde S, Grunberger D. Solution conformation of the N-(deoxyguanosin-8-yl) aminofluorene adduct opposite deoxyinosine and deoxyguanosine in DNA by NMR and computational characterization. Chem Res Toxicol. 1995;8:369–378. doi: 10.1021/tx00045a008. [DOI] [PubMed] [Google Scholar]
- 23.Steitz TA. DNA Polymerases: Structural diversity and common mechanisms. J Biol Chem. 1999;274:17395–17398. doi: 10.1074/jbc.274.25.17395. [DOI] [PubMed] [Google Scholar]
- 24.Joyce CM, Benkovic SJ. DNA Polymerase fidelity: kinetics, structure, and checkpoints. Biochemistry. 2004;43:14317–14324. doi: 10.1021/bi048422z. [DOI] [PubMed] [Google Scholar]
- 25.Bebenek K, Kunkel TA. Functions of DNA polymerases. Adv Protein Chem. 2004;69:137–165. doi: 10.1016/S0065-3233(04)69005-X. [DOI] [PubMed] [Google Scholar]
- 26.Doublie S, Ellenberger T. The mechanism of action of T7 DNA polymerase. Current Opinion Struc Biol. 1998;8:704–712. doi: 10.1016/s0959-440x(98)80089-4. [DOI] [PubMed] [Google Scholar]
- 27.Guengerich FP. Interactions of carcinogen-bound DNA with individual DNA polymerases. Chem Rev. 2006;106:420–452. doi: 10.1021/cr0404693. [DOI] [PubMed] [Google Scholar]
- 28.Pages V, Fuchs RP. How DNA lesions are turned into mutations within cells? Oncogene. 2002;21:8957–8966. doi: 10.1038/sj.onc.1206006. [DOI] [PubMed] [Google Scholar]
- 29.Seo KY, Jelinsky SA, Loechler EL. Factors that influence the mutagenic patterns of DNA adducts from chemical carcinogens. Mutat Res. 2000;463:215–246. doi: 10.1016/s1383-5742(00)00047-8. [DOI] [PubMed] [Google Scholar]
- 30.LeLnne-Samuel N, Janel-Bintz R, Kolbanovskiy A, Geacintov NE, Fuchs RP. The processing of a benzo(a)pyrene adduct into a frameshift or a base substitution mutation requires a different set of genes in Escherichia coli. Mol Microbiol. 2000;38:299–307. doi: 10.1046/j.1365-2958.2000.02116.x. [DOI] [PubMed] [Google Scholar]
- 31.Shibutani S, Grollman AP. Molecular mechanisms of mutagenesis by aromatic amines and amides. Mutat Res. 1997;376:71–78. doi: 10.1016/s0027-5107(97)00027-4. [DOI] [PubMed] [Google Scholar]
- 32.Bassett E, Vaisman A, Havener JM, Masutani C, Hanaoka F, Chaney SG. Efficiency of extension of mismatched primer termini across from cisplatin and oxaliplatin adducts by human DNA polymerases β and η in vitro. Biochemistry. 2003;42:14197–14206. doi: 10.1021/bi035359p. [DOI] [PubMed] [Google Scholar]
- 33.Suzuki N, Yasui M, Santosh Laxmi YR, Ohmori H, Hanaoka F, Shibutani S. Translesion synthesis past equine estrogen-derived 2′-deoxycytidine DNA adducts by human DNA polymerases η and κ. Biochemistry. 2004;43:11312–11320. doi: 10.1021/bi049273n. [DOI] [PubMed] [Google Scholar]
- 34.Zang H, Harris TM, Guengerich FP. Kinetics of nucleotide incorporation opposite polycyclic aromatic hydrocarbon-DNA adducts by processive bacteriophage T7 DNA polymerase. Chem Res Toxicol. 2005;18:389–400. doi: 10.1021/tx049683c. [DOI] [PubMed] [Google Scholar]
- 35.Carver TE, Jr, Hochstrasser RA, Millar DP. Proofreading DNA: recognition of aberrant DNA termini by the Klenow fragment of DNA polymerase I. Proc Natl Acad Sci USA. 1994;91:10670–10674. doi: 10.1073/pnas.91.22.10670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lindsley JE, Fuchs RP. Use of single-turnover kinetics to study bulky adduct bypass by T7 DNA polymerase. Biochemistry. 1994;33:764–772. doi: 10.1021/bi00169a018. [DOI] [PubMed] [Google Scholar]
- 37.Miller H, Grollman AP. Kinetics of DNA polymerase I (Klenow fragment exo−) activity on damaged DNA templates: effect of proximal and distal template damage on DNA synthesis. Biochemistry. 1997;36:15336–15342. doi: 10.1021/bi971927n. [DOI] [PubMed] [Google Scholar]
- 38.Dzantiev L, Romano LJ. Differential effects of N-acetyl-2-aminofluorene and N-2-aminofluorene adducts on the conformational change in the structure of DNA polymerase I (Klenow Fragment) Biochemistry. 2000;39:5139–5145. doi: 10.1021/bi992571d. [DOI] [PubMed] [Google Scholar]
- 39.Fujii S, Fuchs RP. Defining the position of the switches between replicative and bypass DNA polymerases. EMBO J. 2004;23:4342–4352. doi: 10.1038/sj.emboj.7600438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang F, Meneni S, Cho BP. Induced circular dichroism characteristics as conformational probes for carcinogenic aminofluorene-DNA adducts. Chem Res Toxicol. 2006;19:1040–1043. doi: 10.1021/tx0601253. [DOI] [PubMed] [Google Scholar]
- 41.Johnson SJ, Taylor JS, Beese LS. Processive DNA synthesis observed in a polymerase crystal suggests a mechanism for the prevention of frameshift mutations. Proc Natl Acad Sci USA. 2003;100:3895–3900. doi: 10.1073/pnas.0630532100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hansen PE, Dettman HD, Sykes BD. Solvent-induced deuterium isotope effects on fluorine-19 chemical shifts of some substituted fluorobenzenes. Formation of inclusion complexes. J Mag Res. 1985;62:487–496. [Google Scholar]
- 43.Basic one- and two-dimensional NMR spectroscopy. Horst Friebolin, 3rd Ed.; Wiley-VCH, 1998. Chapter 11. Dynamic NMR Spectroscopy. pp 301–329.
- 44.Russu IM. Probing site-specific energetics in proteins and nucleic acids by hydrogen exchange and NMR spectroscopy. Methods Enzymol. 2004;379:152–175. doi: 10.1016/S0076-6879(04)79009-6. [DOI] [PubMed] [Google Scholar]
- 45.Gueron M, Leroy JL. Studies of base pair kinetics by NMR measurement of proton exchange. Methods Enzymol. 1995;261:383–413. doi: 10.1016/s0076-6879(95)61018-9. [DOI] [PubMed] [Google Scholar]
- 46.Jiang YL, McDowell LM, Poliks B, Studelska DR, Cao C, Potter GS, Schaefer J, Song F, Stivers JT. Recognition of an unnatural difluorophenyl nucleotide by uracil DNA glycosylase. Biochemistry. 2004;43:15429–15438. doi: 10.1021/bi0483864. [DOI] [PubMed] [Google Scholar]
- 47.Zaliznyak T, Bonala R, Johnson F, de Los Santos C. Structure and stability of duplex DNA containing the 3-(deoxyguanosin-N2-yl)-2-acetylaminofluorene (dG(N2)-AAF) lesion: a bulky adduct that persists in cellular DNA. Chem Res Toxicol. 2006;19:745–752. doi: 10.1021/tx060002i. [DOI] [PubMed] [Google Scholar]
- 48.Mao B, Gu Z, Gorin A, Hingerty BE, Broyde S, Patel DJ. Solution structure of the aminofluorene-stacked conformer of the syn [AF]-C8-dG adduct positioned at a template-primer junction. Biochemistry. 1997;36:14491–14501. doi: 10.1021/bi972206r. [DOI] [PubMed] [Google Scholar]
- 49.Gu Z, Gorin A, Hingerty BE, Broyde S, Patel DJ. Solution structures of aminofluorene [AF]-stacked conformers of the syn [AF]-C8-dG adduct positioned opposite dC or dA at a template-primer junction. Biochemistry. 1999;38:10855–10870. doi: 10.1021/bi991266p. [DOI] [PubMed] [Google Scholar]
- 50.Feng B, Gorin A, Hingerty BE, Geacintov NE, Broyde S, Patel DJ. Structural alignment of the (+)-trans-anti-benzo[a]pyrene-dG adduct positioned opposite dC at a DNA template-primer junction. Biochemistry. 1997;36:13769–13779. doi: 10.1021/bi970069s. [DOI] [PubMed] [Google Scholar]
- 51.Johnson SJ, Beese LS. Structure of mismatch replication errors observed in a DNA polymerase. Cell. 2004;116:803–816. doi: 10.1016/s0092-8674(04)00252-1. [DOI] [PubMed] [Google Scholar]
- 52.Hsu GW, Kiefer JR, Burnouf D, Becherel OJ, Fuchs RP, Beese LS. Observing translesion synthesis of an aromatic amine DNA adduct by a high-fidelity DNA polymerase. J Biol Chem. 2004;279:50280–50285. doi: 10.1074/jbc.M409224200. [DOI] [PubMed] [Google Scholar]
- 53.Dutta S, Li Y, Johnson D, Dzantiev J, Richardson CC, Romano LJ, Ellenberger T. Crystal structures of 2-acetylaminofluorene and 2-aminofluorene in complex with T7 DNA polymerase reveal mechanisms of mutagenesis. Proc Natl Acad Sci USA. 2004;101:16186–16191. doi: 10.1073/pnas.0406516101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alekseyev YO, Romano LJ. Effects of benzo[a]pyrene adducts on downstream DNA replication in vitro: evidence for different adduct conformations within the active site of DNA polymerase I (Klenow fragment) Biochemistry. 2002;41:4467–4479. doi: 10.1021/bi015850l. [DOI] [PubMed] [Google Scholar]
- 55.Hsu GW, Huang X, Luneva NP, Geacintov NE, Beese LS. Structure of a high fidelity DNA polymerase bound to a benzo[a]pyrene adduct that blocks replication. J Biol Chem. 2005;280:3764–3770. doi: 10.1074/jbc.M411276200. [DOI] [PubMed] [Google Scholar]
- 56.Kunkel TA. Misalignment-mediated DNA synthesis errors. Biochemistry. 1990;29:8003–8011. doi: 10.1021/bi00487a001. [DOI] [PubMed] [Google Scholar]
- 57.Shibutani S. Requirements for frame-shift deletion during translesion synthesis. Environ Mutagen Res. 2004;26:135–141. [Google Scholar]
- 58.Hoffmann GR, Fuchs RPP. Mechanisms of frameshift mutations: insight from aromatic amines. Chem Res Toxicol. 1997;10:347–359. doi: 10.1021/tx960128n. [DOI] [PubMed] [Google Scholar]
- 59.Shibutani S, Suzuki N, Grollman AP. Mechanism of frameshift (deletion) generated by acetylaminofluorene-derived DNA adducts in vitro. Biochemistry. 2004;43:15929–35. doi: 10.1021/bi048087e. [DOI] [PubMed] [Google Scholar]
- 60.Napolitano RL, Lambert IB, Fuchs RP. DNA Sequence determinants of carcinogen-induced frameshift mutagenesis. Biochmeistry. 1994;33:1311–1315. doi: 10.1021/bi00172a004. [DOI] [PubMed] [Google Scholar]
- 61.Suzuki N, Ohashi E, Hayashi K, Ohmori H, Grollman AP, Shibutani S. Translesional synthesis past acetylaminofluorene-derived DNA adducts catalyzed by human DNA polymerase κ and Escherichia coli DNA polymerase IV. Biochemistry. 2001;40:15176 –15183. doi: 10.1021/bi010702g. [DOI] [PubMed] [Google Scholar]
- 62.Tan X, Suzuki N, Grollman AP, Shibutani S. Mutagenic events in Escherichia coli and mammalian cells generated in response to acetylaminofluorene-derived DNA adducts positioned in the Nar I restriction enzyme site. Biochemistry. 2002;41:14255–14262. doi: 10.1021/bi0202878. [DOI] [PubMed] [Google Scholar]
- 63.Gill JP, Romano LJ. Mechanism for N-acetyl-2-aminofluorene-induced frameshift mutagenesis by Escherichia coli DNA polymerase I (Klenow fragment) Biochemistry. 2005;44:15387–15395. doi: 10.1021/bi051437s. [DOI] [PubMed] [Google Scholar]
- 64.Zhang L, Rechkoblit O, Wang L, Patel DJ, Shapiro R, Broyde S. Mutagenic nucleotide incorporation and hindered translocation by a food carcinogen C8-dG adduct in Sulfolobus solfataricus P2 DNA polymerase IV (Dpo4): modeling and dynamics studies. Nucleic Acids Res. 2006;34:3326–2237. doi: 10.1093/nar/gkl425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brown K, Hingerty BE, Guenther EA, Krishnan VV, Broyde S, Turteltaub KW, Cosman M. Solution structure of the 2-amino-1- methyl-6-phenylimidazo[4,5-b]pyridine C8-deoxyguanosine adduct in duplex DNA. Proc Natl Acad Sci USA. 2001;98:8507–8512. doi: 10.1073/pnas.151251898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang L, Shapiro R, Broyde S. Molecular dynamics of a food carcinogen-DNA adduct in a replicative DNA polymerase suggest hindered nucleotide incorporation and extension. Chem Res Toxicol. 2005;18:1347–1363. doi: 10.1021/tx050132b. [DOI] [PubMed] [Google Scholar]
- 67.Case DA, Darden DA, Cheatham TE, III, Cimmerling CL, Wang J, Duke RE, Luo R, Merz KM, Wang B, Perlman DA, Crowley M, Brozell S, Tsue V, Gohlke H, Mongan J, Hornak V, Cui G, Beroza P, Schafmeister C, Caldwell JW, Ross WS, Kollman PA. AMBER 8. University of California; San Francisco: 2004. [Google Scholar]
- 68.Delano WL. The PyMOL Molecular Graphic System. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


