Abstract
The interaction of 5-formyltetrahydrofolate analogs with murine methenyltetrahydrofolate synthetase (MTHFS) was investigated using steady-state kinetics, molecular modeling, and site-directed mutagenesis. MTHFS catalyzes the irreversible cyclization of 5-formyltetrahydrofolate to 5,10-methenyltetrahydrofolate. Folate analogs that cannot undergo the rate-limiting step in catalysis were inhibitors of murine MTHFS. 5-formyltetrahydrohomofolate was an effective inhibitor of murine MTHFS (Ki = 0.7 μM), whereas 5-formyl, 10-methyltetrahydrofolate was a weak inhibitor (Ki = 10 μM). The former, but not the latter, was slowly phosphorylated by MTHFS. 5-formyltetrahydrohomofolate was not a substrate for murine MTHFS, but was metabolized when the MTHFS active site Y151 was mutated to Ala. MTHFS active site residues do not directly facilitate N10 attack on the on the N5-iminium phosphate intermediate, but rather restrict N10 motion around N5. Inhibitors specifically designed to block N10 attack appear to be less effective than the natural 10-formyltetrahydrofolate polyglutamate inhibitors.
INTRODUCTION
The polyglutamate forms of tetrahydrofolate (THF) are metabolic cofactors that carry and chemically activate one-carbon units for the synthesis of purines and thymidylate, and for the remethylation of homocysteine to methionine (Figure 1)[1]. Methionine can be adenylated to form S-adenosylmethionine (AdoMet); AdoMet is a methyl donor for numerous cellular reactions including methylation of histones, DNA, RNA, and phospholipids [2, 3]. Disruptions in this metabolic network can result from single nucleotide polymorphisms or from low intracellular folate concentrations and/or other environmental factors. Aberrations in folate metabolism increase risk for pathologies including epithelial cancers, developmental anomalies, and cardiovascular disease[4]. This metabolic network has also been a successful target for the development of antiproliferative agents, and a focus for the development of new anticancer therapies [5, 6].
Figure 1.
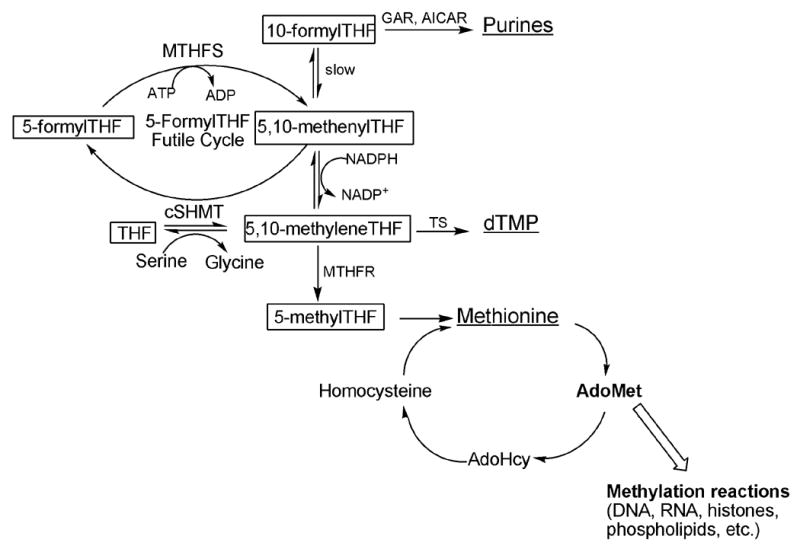
The folate-dependent one-carbon metabolic network in the cytoplasm. Folate cofactors and derivatives are indicated in boxes, and the products of one-carbon metabolism are underlined. Folate-mediated one-carbon metabolism is emphasized, highlighting the 5-formylTHF futile cycle. MTHFS, methenyltetrahydrofolate synthetase; cSHMT cytoplasmic serine hydroxymethyltransferase; AdoMet, S-adenosylmethionine. GARFT, glycinamide ribonucleotide formyltransferase; AICARFT, phosphoribosylaminoimidazole carboxamide formyltransferase; TS, thymidylate synthase; AdoMet, S-adenosylmethionine.
Folate cofactors contain three chemical moieties: reduced pteridine, p-aminobenzoate and a glutamate polypeptide that contains 1 to 9 glutamate residues linked through γ-peptide linkages (Figure 2). Polyglutamylation serves both to retain folates within the cell and to increase the affinity of folate derivatives for the enzymes that utilize them [1]. Folates serve as cofactors by carrying and chemically activating one-carbon units on N5, N10 or both; the single carbons are carried at the oxidation level of formate (i.e., 10-formylTHF), formaldehyde (i.e., 5,10-methyleneTHF) or methanol (5-methylTHF).
Figure 2.
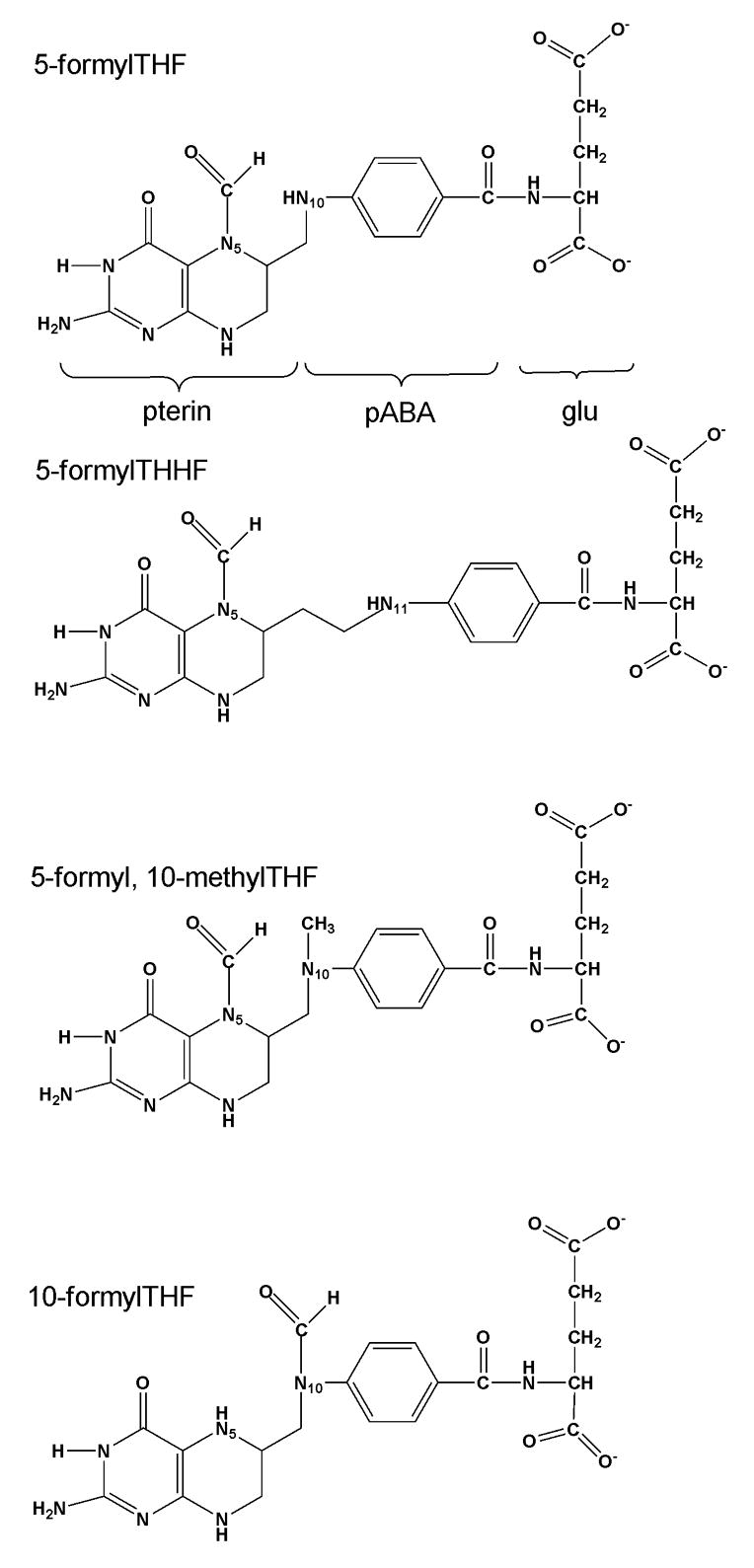
Structure of the substrate, 5-formyltetrahydrofolate, and MTHFS inhibitors described in the text. pABA, p-aminobenzoate; glu, glutamate.
There are three naturally-occurring folate derivatives that carry the one-carbon unit at the oxidation level of formate: 10-formylTHF, 5-formylTHF, and methenylTHF. 10-formylTHF is the cofactor that supplies the #2 and #8 carbons of the purine ring; 5-formylTHF and methenylTHF are also present in cells but are not metabolic cofactors (Figure 1). 5-formylTHF is the most stable form of reduced folate and its polyglutamate derivatives are tight-binding inhibitors of folate dependent enzymes including the serine hydroxymethyltransferase (SHMT) isozymes, and phosphoribosylaminoimidazole carboxamide formyltransferase (AICARFT). SHMT catalyzes the interconversion of serine and THF to glycine and methyleneTHF, a reaction that generates one-carbon units for purine, thymidine and methionine biosynthesis. AICARFT catalyzes the incorporation of formate into the C-2 position of the purine ring. Therefore, intracellular 5-formylTHF concentrations are believed to regulate the flow of one-carbon units through the folate-dependent one-carbon network [7].
MTHFS has been suggested as a target for the development of antiproliferative agents because it affects 5-formylTHF concentrations [8, 9]. Cellular concentrations of 5-formylTHF are regulated by a futile cycle (Figure 1). 5-FormylTHF is synthesized from methenylTHF in a second reaction catalyzed by SHMT. 5,10-Methenyltetrahydrofolate synthetase (MTHFS, EC 6.3.3.2), the only enzyme that metabolizes 5-formylTHF, catalyzes the ATP-dependent and irreversible cyclization of 5-formylTHF to 5,10-methenylTHF. The MTHFS-catalyzed reaction exhibits product inhibition; the product of the MTHFS reaction, methenylTHF, exists in chemical equilibrium with 10-formylTHF, which binds to MTHFS tightly [10].
The MgATP-dependent MTHFS reaction is essentially irreversible (Keq = 66 to 500M). The proposed mechanism proceeds with a nucleophilic attack by the 5-formyl oxygen on the γ-phosphate of ATP, forming an N5-iminium phosphate intermediate (Figure 3, Intermediate 1). This intermediate undergoes cyclization via nucleophilic attack by N10 to form a phosphoimidazolidine tetrahedral intermediate (Figure 3, Intermediate 2); this has been suggested to be the rate-limiting step in catalysis. Breakdown of this intermediate through phosphate elimination, the only irreversible step in the reaction, generates the methenylTHF product [8].
Figure 3.
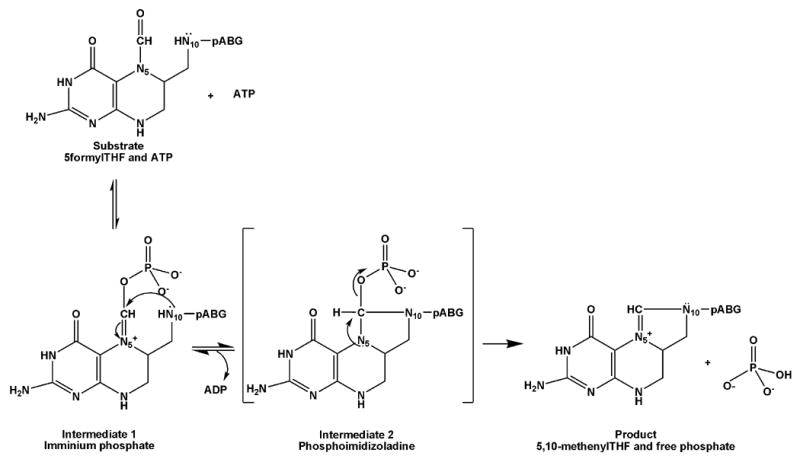
Reaction intermediates of MTHFS catalysis. The iminium phosphate (Intermediate 1) results from the transfer of the ATP γ-phosphate to 5-formylTHF; the phosphoimidazolidine (Intermediate 2) results from N10 nucleophilic attack to form a species with a tetrahedral carbon, which then breaks down rapidly to eliminate phosphate and form product, methenylTHF. pABG indicates p-aminobenzoylglutamate.
Previous studies have shown that MTHFS purified from rabbit liver is inhibited by the folate analog, 5-formyltetrahydrohomofolate (5-formylTHHF) [8]. Furthermore, MCF-7 cells exposed to 5-formylTHHF accumulate 5-formylTHF and are auxotrophic for purines [11]. 5-formylTHHF contains an additional methylene group between the pterin and p-aminobenzoate moiety of 5-formylTHF (Figure 2); the extra methylene group impairs N11 (corresponding to THF N10) attack on the N5-iminium phosphate intermediate [8]. However, its clinical utility is expected to be limited, as it has been shown to be slowly metabolized to 5,11-methenylTHHF by MTHFS purified from rabbit liver [8]. In this study, the interaction of murine MTHFS with 5-formylTHF analogs that impair N10/N11 attack was investigated using steady-state kinetics, molecular modeling, and site-directed mutagenesis.
METHODS
Materials
MES, HEPES, and Tris were purchased from Sigma. ATP was purchased from Roche. [6S]-5-formylTHF (the natural isomer) and [6R,S]-5-formylTHF were a generous gift from Eprova AG. 5-formyltetrahydrofolate triglutamate was from Schircks Laboratories (Switzerland). Homofolate (NSC 79249) was the generous gift of Dr. Roy Kisliuk, Tufts University. All other materials were of high quality and obtained from various commercial vendors.
Synthesis of folate derivatives
5-formyl, 10-methylTHF was synthesized by incubating 5-formylTHF, formaldehyde and sodium cyanoborohydride for 12 h as described elsewhere [12]. 5-Formyltetrahydrohomofolic acid was synthesized from homofolic acid as follows: Homofolic acid was reduced to tetrahydrohomofolic acid by reduction over palladium(II) oxide hydrate in water; formylation at N5 was achieved by the immediate addition of formic acid and N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide at pH 4.0 as described previously [13]. All synthesized compounds were purified by anion exchange chromatography and desalted by gravity filtration on a 0.5 in. × 64 in. G-10 Sephadex column using water as the mobile phase, as described elsewhere [14]. Samples were lyophilized and their identity verified by UV spectroscopy and 1H NMR.
Generation of mutant MTHFS enzymes
Three separate amino acid substitution mutants were generated using the Stratagene Quick Change Site Directed Mutagenesis Kit, following the manufacturer’s protocol. The Y151A mutant was prepared using the following primers: forward, 5′-GGG CGG GGT AAG GGG GCA TAT GAC ACC TAC CTG-3′ and reverse, 5′-CAG GTA GGT GTC ATA TGC CCC CTT ACC CCG CCC-3′. The Y152A mutant was prepared using the following primers: forward, 5′-CGG GGT AAG GGG TAC GCG GAC ACC TAC CTG AAG-3′ and reverse, 5′-CTT CAG GTA GGT GTC CGC GTA CCC CTT ACC CCG-3′. Finally the Y152F mutant was created using the following primers: forward, 5′-GGG CGG GGT AAG GGG TAC TTT GAC ACC TAC CTG-3′ and reverse, 5′-CAG GTA CCT GTC AAA GTA CCC CTT ACC CCG CCC-3′. The underlined codon was targeted in the site directed mutation. All mutated plasmids generated were sequence verified.
Protein expression and purification
The MTHFS protein and mutants thereof (Y151A, Y152A, Y152F) were expressed and purified as described elsewhere[15]. For the mutant proteins, all purification and dialysis buffers contained 10mM MgATP to prevent denaturation of the protein.
Steady-State kinetics
MTHFS activity was monitored by measuring the increase in methenylTHF production with time at 37°C in 1mL cuvettes using a spectrophotometer; methenylTHF has a unique absorbance maximum at λ=355 nm (ε=25,100). When 5-formylTHHF was used as a substrate, the absorbance of methenylTHHF at λ=320 nm was monitored. Reaction mixtures contained 100mM MES, pH 6.0, 20mM MgATP. Reactions were initiated by the addition of 70nM MTHFS. Km and kcat values were determined from Lineweaver-Burke plots and reported values are the average of three experiments. For all inhibition experiments, 70nM MTHFS was preincubated with the inhibitors for 2–4 minutes in the reaction mixture; the reaction was then initiated by adding [6S]-5-formylTHF. Ki values were determined from Dixon plots, and reported values are the average of at least two independent determinations.
ATP utilization assay
MTHFS activity was also monitored by measuring the generation of ADP. The reaction buffer contained 100mM MES pH7.2, 10mM MgATP, 1mM phosphoenolpyruvate, 200 μM NADPH, 3U/mL pyruvate kinase, 3U/mL lactate dehydrogenase, and 5U/ml C1 tetrahydrofolate synthase purified from rabbit liver. This system generates 2 equivalents of NADP+ when 5-formylTHF is converted to methyleneTHF, or 1 equivalent of NADP+ if the reaction is unable to proceed beyond the formyl phosphorylation step. The folate or folate analog substrate was added to a cuvette containing 1mL reaction buffer and allowed to incubate for 15 minutes at 37°C. The reaction rate was quantified by following A340 for 2 minutes following the addition of 70nM MTHFS.
Molecular modeling
A homology model of the mouse MTHFS protein was constructed using the crystal structure of Mycoplasma pneumoniae MTHFS with ADP, phosphate, and 5-formylTHF bound and is described elsewhere [10]. To visualize the MTHFS-mediated conversion of 5-formylTHF to methenylTHF, a possible series of conformational intermediates in the reaction of ATP with 5-formylTHF, to produce first a phosphorylated THF intermediate and then the products, was generated by molecular modeling. Several intermediate conformations of the ATP-THF species were built manually, using the program O [16], keeping the protein atoms fixed except for small side-chain motions. Energy minimization was performed on each of these intermediates using CNS [17], allowing both protein and ligands to relax. Finally, transitions between the intermediates were smoothed using the Indie morphing server (http://molbio.info.nih.gov/structbio/indie_morph.html). The resulting series of structures was used to generate images (using RasMol [18]), which were combined into a movie (Online Supporting Material) using gifsicle (http://www.lcdf.org/~eddietwo/gifsicle/index.html).
Results and Discussion
Mechanism-based inhibition of MTHFS
The mechanism of MTHFS inhibition by 5-formylTHF analogs (Figure 2) was investigated using recombinant murine MTHFS protein. The ability of folate analogs that cannot undergo the rate-limiting nucleophilic attack by N10 on the N5-iminium phosphate intermediate to inhibit MTHFS was investigated. Nucleophilic attack by N10 can be impaired by N10 substitution (i.e. methylation of N10) and/or by increasing the distance between the N5-iminium phosphate and N10 (i.e. 5-formylTHHF)(Figure 2). As expected, [6R,S]-5-formyl, 10-methylTHF was demonstrated to be a competitive inhibitor of MTHFS (Ki=10 μM (Table 1)), but was not as effective as [6R,S]-5-formylTHHF (Ki = 0.7 μM). Surprisingly, N11 methylation of 5-formylTHHF diminished its effectiveness nearly 10-fold; [6R,S]-5-formyl,11-methylTHHF was a competitive inhibitor with a Ki=6.0 μM (Table 1).
Table 1.
Inhibition of MTHFS by 5-formylTHF analogs. All reactions were carried out at 37°C in 100mM MES, pH6.0.
| Folate analog | Ki(μM) |
|---|---|
| [6R,S]-5-formyl,10-methylTHF | 10 ± 1 |
| [6R,S]-5-formyl,10-methylTHFGlu3 | 20 ± 2 |
| [6R,S]-5-formylTHHF | 0.7 ± 0.3 |
| [6R,S]-5-formyl,11-methylTHHF | 6.0 ± 3 |
Previously, we have shown that MTHFS exhibits increased affinity for the polyglutamate forms of the substrate [10]. The Km of [6R,S]-5-formylTHFGlu3 (Km=0.4 μM) is decreased by over 96% compared to [6R,S]-5-formylTHF (Km= 10 μM) [10]. However, the Ki of [6R,S]-5-formyl, 10-methylTHFGlu3 (Ki=20 μM) was 2-fold higher than for the monoglutamate (Table 1). This is unusual as the addition of polyglutamates to natural and unnatural folate derivatives generally increases their affinity for folate-dependent enzymes [1]. Tight binding of the polyglutamate moiety may make it difficult for 5-formyl,10-methylTHFGlu3 to occupy the active site because of a steric clash between its 10-methyl group and ATP’s γ-phosphate. The model with 5-formylTHF in position to react with ATP places N10 only 2.8 A from one of the γ-phosphate oxygens, suggesting that insertion of the N10-methyl group would require movement of THF somewhat out of the binding pocket.
The role of N10 substitution on formation of the N5 iminium phosphate
Structural modeling studies revealed that the MTHFS active site can accommodate a number of natural and synthetic folates, including folic acid, 5-formylTHHF,11-formylTHHF, and 5-formyl, 10-methylTHF (Figure 4). The site is not tightly constricted, so a variety of folate analogs can be positioned to form favorable interactions between the pterin moiety and residues lining the binding site, and between the glutamate moiety and surface hydrophilic residues. In agreement with aforementioned kinetic data, 5-formyl, 10-methylTHF was not predicted to bind to the active site as tightly as 5-formylTHF in the presence of ATP, due to interference between the methyl group and the ATP γ-phosphate. Therefore, 5-formyl, 10-methylTHF may not be able to form the iminium phosphate intermediate. To test this prediction, the ability of recombinant mouse MTHFS to phosphorylate its inhibitors (form Intermediate 2 in Figure 3) was examined by monitoring the generation of ADP from ATP using a coupled enzymatic assay (Materials and Methods). This assay yielded a similar Km for 5-formylTHF (10.5±2.5 μM) as previously obtained using the standard spectrophotometric assay (Km=10±2 μM)(Table 2). The inhibitors 10-formylTHF and 5-formyl,10-methylTHF did not form the N5-iminium phosphate, as evidenced by the lack of ADP generated above background (Table 2). This result affirms the modeling predictions described above, and previous structural modeling studies that indicated that the formyl group of 10-formylTHF does not assume proper orientation for nucleophilic attack on ATP γ-phosphate[10]. When 5-formylTHHF was incubated with MTHFS and MgATP, ADP formation occurred at a rate 5% of that observed for 5-formylTHF, suggesting that 5-formylTHHF is slowly phosphorylated to form the N5-iminium phosphate. Since 5-formylTHHF is not a substrate for mouse MTHFS (as described below), this indicates that the conversion of 5-formylTHHF to 5,11-methenylTHHF by MTHFS is impaired by an inability of N11 to attack the N5-iminium phosphate. Although both mouse MTHFS and rabbit liver MTHFS are capable of phosphorylating 5-formylTHHF, only rabbit liver MTHFS can also slowly metabolize 5-formylTHHF to 5,11-methenylTHHF [8].
Figure 4.
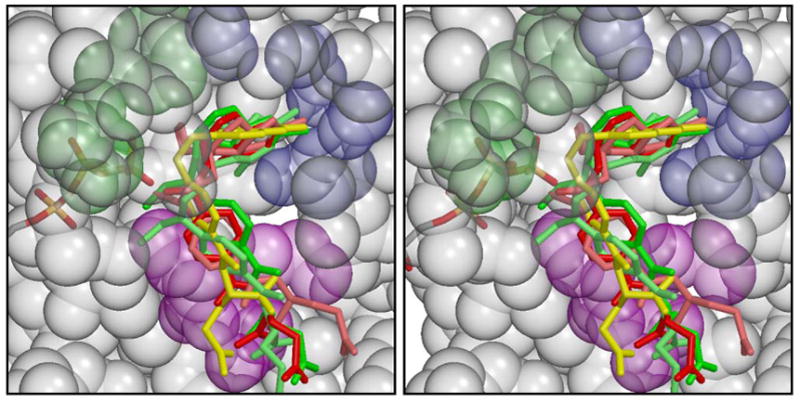
Stereo figure modeling of inhibitors bound in the active site of MTHFS in the presence of ATP. Inhibitors include: 5-formyl,10-methylTHF (red), 5-formyl,11-methylTHHF (light red), 10-formylTHF (green), 10-formyl THHF (light green), folic acid (yellow). MTHFS is shown in semitransparent spacefilling form, colored gray, except for residues coordinating the pterin ring, which are partially blue, those which potentially make H-bonds to the glutamate moiety, whose side-chains are magenta, and tyrosines 151 and 152, whose side-chains are green. Of the blue residues, it appears that Glu 64 and the main chain O of Asp 59 (on the right side of the figure) can interact with all the inhibitors, while Ser 97 and Gln 58 only coordinate some of them. The figure was rendered using Molscript [20] and Raster3D [21].
Table 2.
MTHFS catalyzed formation of the N5 iminium phosphate intermediate. The ability of MTHFS to catalyze formation of the N5 iminium phosphate was determined as described in Materials and Methods. Results shown as average and standard deviation of three experiments. The reaction buffer consisted of 100mM MES pH7.2, 10mM MgATP, 1mM phosphenolpyruvate, 200 μM NADPH, 3U/mL pyruvate kinase, 3U/mL lactate dehydrogenase, and 5U/ml C1 tetrahydrofolate synthase purified from rabbit liver. Activity is expressed as μM NADPH consumed per minute.
| Substrate | [S] (μM) | Activity (μM/min) |
|---|---|---|
| 5-formylTHF | 10 | 0.99 ± 0.03 |
| 5-formylTHHF | 10 | 0.05 ± 0.02 |
| 10-formylTHF | 10 | < 0.01 |
| 80 | < 0.01 | |
| 5-formyl, 10-methylTHF | 10 | < 0.01 |
| 80 | < 0.01 |
Role of the MTHFS active site in the conversion of the N5-iminium phosphate (intermediate 1) to the phosphoimidazolidine (intermediate 2)
The role of the MTHFS active site in facilitating N10 movement was investigated. The 5-formylTHF binding site consists of residues 58-64, 97-105, 133-136 and 144-152 [10]. Glycines at positions 148 and 150 allow proper folding of the 147-150 loop which defines one side of the 5-formylTHF-binding cavity. The tyrosines at positions 151-152 (shown in green in Figures 4 and 5) appear to be important in defining the shape of the active site and are conserved as Y or F in all MTHFS primary sequences. The position of these residues indicates that they are likely to play a role in the conversion of intermediate 1 to intermediate 2 (Figure 3). A tyrosine could direct the nucleophilic attack of N10 on the N5-iminium phosphate through ring stacking interactions with the pABA moiety of the substrate, as is the case with 5-formylTHF bound to cSHMT, a reaction that also involves an N10 nucleophilic attack [19]. Previous studies have shown that the pABA moiety of the substrate is essential for binding to MTHFS (5-formyltetrahydropterin does not inhibit mouse MTHFS). However, the homology model clearly shows that there is no ring stacking interaction with either of these tyrosine residues and the pABA moiety (Figure 5).
Figure 5.
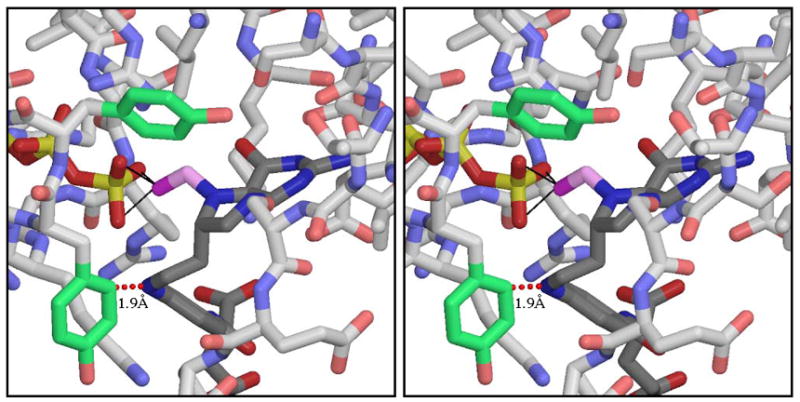
5-formylTHHF, in position to attack the γ-phosphate of ATP. THHF and ATP are shown with dark CPK coloring and MTHFS with lighter CPK coloring, except for Y151 (on the left) and Y152 (at the top), whose side-chains are colored green. The THHF formyl group is shown in magenta. The distance between THHF N11 and Y151 Cδ1 is shown to indicate that this placement of THHF is only possible if the side-chain of Y151 is removed. The figure was rendered using Molscript and Raster3D.
Alternatively, tyrosines 151 and 152 may facilitate N10 attack on the N5-iminium phosphate by forming a pocket that constrains the motion of N10. The position of the Y152 side-chain is well-determined by hydrophobic interactions with a number of neighboring residues; the Y151 side-chain is more exposed, but a possible H-bond from its OH to the main chain oxygen of residue 102 may help secure it in place. The tyrosines were changed by site-directed mutagenesis to investigate the effects of altering the integrity of the active site on substrate binding and catalysis. The Y151A mutant protein exhibited similar Michaelis-Menten kinetics parameters as the wild-type MTHFS (Table 3), indicating that this side-chain does not make major contributions to substrate binding or catalysis. The Y152A mutant protein, however, was impaired in both binding and catalysis; Km for 5-formylTHF was elevated 300-fold and kcat reduced by greater than 99% (Table 3). To ascertain whether the pABA ring or the hydroxyl of the tyrosine side-chain was required for effective substrate binding and catalysis, Y152 was changed to a phenylalanine. The Y152F mutation increased the Km by only 4-fold and did not affect kcat (Table 3), indicating that the hydrophobic/aromatic properties of Y152 are essential in defining the architecture of the active site and for effective substrate binding and catalysis.
Table 3.
Steady-State Kinetic Parameters for MTHFS Mutants. All reactions were carried out in 100mM MES, pH 6.0 at 37°C.
| [6S] 5-formylTHF | [6R,S]-5-formylTHHF | ||||
|---|---|---|---|---|---|
| Km (μM) | kcat (min−1) | Km (μM) | kcat (min−1) | Ki (μM) | |
| WT-MTHFS | 10 ± 3 | 200 ± 50 | nd | nd | 0.7 ± 0.3 |
| Y151A | 9 ± 2 | 55 | 31 ± 2 | 0.2 | 0.3 ± 0.1 |
| Y152A | 3000 ± 500 | 1.2 ± 0.4 | nd | nd | -- |
| Y152F | 47 ± 15 | 258 ± 50 | -- | -- | -- |
nd – no activity detected
The effect of reduced active site integrity on N10 attack was investigated using the MTHFS inhibitor 5-formylTHHF. The MTHFS-catalyzed conversion of 5-formylTHHF to 5,11-methenylTHHF requires the formation of a six-membered ring rather than the five-membered ring present in methenylTHF. 5-formylTHHF is slowly metabolized by rabbit MTHFS; the kcat is one-thousandth of that observed for 5-formylTHF[8]. As previously stated, this reduced rate of catalysis has been attributed to steric constraints that impair nucleophilic attack by N11 of 5-formylTHHF (corresponding to N10 of 5-formylTHF) on the iminium phosphate intermediate. In contrast to the rabbit enzyme, 5-formylTHHF is not a substrate for wild-type murine recombinant MTHFS, but does inhibit MTHFS and MTHFS Y151A (Ki=0.7 μM and 0.3 μM respectively) (Table 3). Surprisingly, 5-formylTHHF was a substrate for the Y151A mutant (Km=31 μM and kcat=0.2 min−1), but not the Y152A mutant (Table 3, Figure 6). The differences in the Km and Ki values for 5-formylTHHF with Y151A MTHFS are explained by the strong product inhibition by 11-formylTHHF (Ki = 0.2 μM), which is a product of 5,11-methenylTHHF nonenzymatic hydrolysis [10]. Modeling 5-formylTHHF into the binding site, with the formyl group positioned for attack on ATP γ-phosphate (to create Intermediate 1 in Figure 3), reveals a steric clash between N11 of 5-formylTHHF and the side-chain of Y151, as shown in Figure 5. Mutation of tyrosine to alanine removes this obstacle, thereby permitting 5-formylTHHF phosphorylation and subsequent cyclization to create product. We conclude that steric restriction of N11 motion in the wild-type enzyme prevents 5-formylTHHF metabolism, suggesting that the MTHFS active site functions to constrain the movement of N10 (or N11 of THHF) near the N5-iminium phosphate during catalysis. Modeling of reaction intermediates (supplementary material) showed that the conformational changes to ATP and 5-formylTHF during the reaction can be accommodated with small adjustments to MTHFS. Much of the 5-formylTHF molecule is solvent-exposed, and so can flex freely without interacting with the protein. Energy-minimized structures showed backbone motions of 1 Å or less, concentrated in the region of residues 146-151. The glycines in this region (positions 146, 148, 150), which form one face of the 5-formylTHF binding site, facilitate adjustments to main-chain torsion angles without producing long-range disturbances. This model indicates that MTHFS does not have to undergo large-scale conformation changes during catalysis, and further supports the observation that the MTHFS active site mediates cyclization of 5-formylTHF by limiting, but not directing, the motion of N10.
Figure 6.
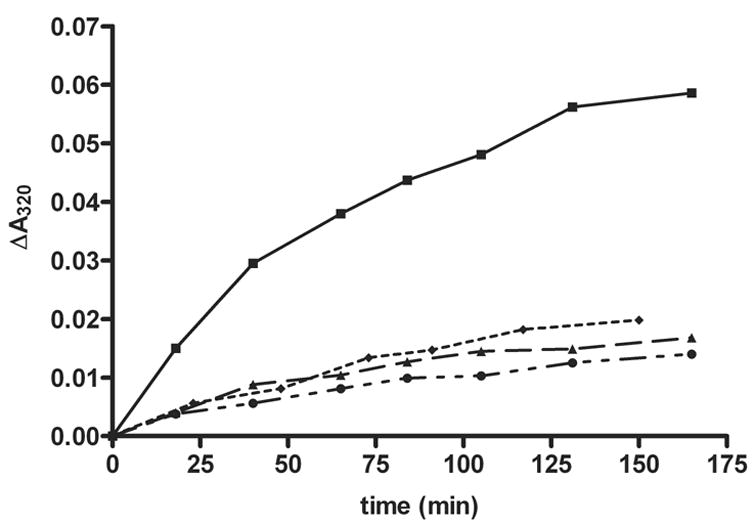
MTHFS-mediated catalysis of 5-formylTHHF. The MTHFS-catalyzed conversion of 5-formylTHHF to methenylTHHF was investigated by monitoring the increase in absorbance at λmax=320nm as described in Materials and Methods. (■) Y151A-MTHFS, (♦) Y152A-MTHFS, (▲) wild-type MTHFS, (●) control (no enzyme).
This study advances our understanding of the mechanism of inhibition by 5-formylTHHF and further informs the design of MTHFS inhibitors. Huang and Schirch showed that rabbit MTHFS phosphorylated and very slowly converted 5-formylTHHF to 5,11-methenylTHHF. They inferred that the observed inhibition by 5-formylTHHF resulted from the phosphorylated intermediate remaining tightly bound to the enzyme [8]. Here we show that 5-formylTHHF is also phosphorylated by mouse MTHFS and that removal of the Tyr 151 side-chain is necessary for metabolism of 5-formylTHHF. These data suggest that 5-formylTHHF, once phosphorylated, remains bound due to hindered N11 attack. Mechanistically, MTHFS-mediated catalysis appears to be the result of restricted, rather than directed, N10 motion. This study also indicates that synthetic 10-methyl substituted folates are not viable in vivo MTHFS inhibitors, since occupation of the polyglutamate binding site forces the N10-methyl into a sterically unfavorable position relative to ATP. As indicated previously [10], synthetic analogs of the natural feedback inhibitor 10-formylTHF (Ki=30 nM as a tri-glutamate), rather than inhibitors designed to block N10 attack, may prove to be the most effective MTHFS inhibitors.
Supplementary Material
Acknowledgments
This work was supported by PHS HD35687 to PJS. The authors would like to thank Bhumit Patel for protein purification.
Abbreviations
- MTHFS
methenyltetrahydrofolate synthetase
- THF
tetrahydrofolate
- THHF
tetrahydrohomofolate
- SHMT
serine hydroxymethyltransferase
- AICARFT
phosphoribosylaminoimidazole carboxamide formyltransferase
- GARFT
glycinamide ribonucleotide formyltransferase
- FPGS
folylpolyglutamate synthetase
- PBS
phosphate-buffered saline
- AdoMet
S-adenosylmethionine
- pABA
p-aminobenzoate
- pABG
p-aminobenzoylglutamate
- NMR
nuclear magnetic resonance
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shane B. Folate Chemistry and Metabolism. In: Bailey LB, editor. Folate in Health and Disease. Marcel Dekker, Inc; New York: 1995. pp. 1–22. [Google Scholar]
- 2.Finkelstein JD. Homocysteine: a history in progress. Nutr Rev. 2000;58:193–204. doi: 10.1111/j.1753-4887.2000.tb01862.x. [DOI] [PubMed] [Google Scholar]
- 3.Clark S, Banfield K. S-adenosylmethionine -dependent methyltransferases. In: Carmel R, DW Jacobsen, editors. Homocysteine in Health and Disease. Cambridge University Press; Cambridge: 2001. pp. 63–78. [Google Scholar]
- 4.Suh JR, Herbig AK, Stover PJ. New perspectives on folate catabolism. Annu Rev Nutr. 2001;21:255–282. doi: 10.1146/annurev.nutr.21.1.255. [DOI] [PubMed] [Google Scholar]
- 5.Kisliuk RL. Deaza analogs of folic acid as antitumor agents. Curr Pharm Des. 2003;9:2615–2625. doi: 10.2174/1381612033453695. [DOI] [PubMed] [Google Scholar]
- 6.Zhao R, Goldman ID. Resistance to antifolates. Oncogene. 2003;22:7431–7457. doi: 10.1038/sj.onc.1206946. [DOI] [PubMed] [Google Scholar]
- 7.Stover P, Schirch V. The metabolic role of leucovorin. Trends Biochem Sci. 1993;18:102–106. doi: 10.1016/0968-0004(93)90162-g. [DOI] [PubMed] [Google Scholar]
- 8.Huang T, Schirch V. Mechanism for the coupling of ATP hydrolysis to the conversion of 5-formyltetrahydrofolate to 5,10-methenyltetrahydrofolate. J Biol Chem. 1995;270:22296–22300. doi: 10.1074/jbc.270.38.22296. [DOI] [PubMed] [Google Scholar]
- 9.Girgis S, Suh JR, Jolivet J, Stover PJ. 5-Formyltetrahydrofolate regulates homocysteine remethylation in human neuroblastoma. J Biol Chem. 1997;272:4729–4734. doi: 10.1074/jbc.272.8.4729. [DOI] [PubMed] [Google Scholar]
- 10.Field MS, Szebenyi DM, Stover PJ. Regulation of de novo purine biosynthesis by methenyltetrahydrofolate synthetase in neuroblastoma. J Biol Chem. 2006;281:4215–4221. doi: 10.1074/jbc.M510624200. [DOI] [PubMed] [Google Scholar]
- 11.Bertrand R, Jolivet J. Methenyltetrahydrofolate synthetase prevents the inhibition of phosphoribosyl 5-aminoimidazole 4-carboxamide ribonucleotide formyltransferase by 5-formyltetrahydrofolate polyglutamates. J Biol Chem. 1989;264:8843–8846. [PubMed] [Google Scholar]
- 12.Temple C, Jr, Bennett LL, Jr, Rose JD, Elliott RD, Montgomery JA, Mangum JH. Synthesis of pseudo cofactor analogues as potential inhibitors of the folate enzymes. J Med Chem. 1982;25:161–166. doi: 10.1021/jm00344a014. [DOI] [PubMed] [Google Scholar]
- 13.Moran RG, Keyomarsi K, Colman PD. Preparation of (6S)-5-formyltetrahydrofolate labeled at high specific activity with 14C and 3H. Methods Enzymol. 1986;122:309–312. doi: 10.1016/0076-6879(86)22185-0. [DOI] [PubMed] [Google Scholar]
- 14.Stover P, Schirch V. Synthesis of (6S)-5-formyltetrahydropteroyl-polyglutamates and interconversion to other reduced pteroylpolyglutamate derivatives. Anal Biochem. 1992;202:82–88. doi: 10.1016/0003-2697(92)90210-x. [DOI] [PubMed] [Google Scholar]
- 15.Anguera MC, Liu X, Stover PJ. Cloning, expression, and purification of 5,10-methenyltetrahydrofolate synthetase from Mus musculus. Protein Expr Purif. 2004;35:276–283. doi: 10.1016/j.pep.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 16.Jones TA, Zou JY, Cowan SW, Kjeldgaard Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. Pt 2. [DOI] [PubMed] [Google Scholar]
- 17.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallographica Section D-Biological Crystallography. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 18.Sayle RA, Milnerwhite EJ. Rasmol - Biomolecular Graphics for All. Trends in Biochemical Sciences. 1995;20:374–376. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
- 19.Szebenyi DM, Liu X, Kriksunov IA, Stover PJ, Thiel DJ. Structure of a murine cytoplasmic serine hydroxymethyltransferase quinonoid ternary complex: evidence for asymmetric obligate dimers. Biochemistry. 2000;39:13313–13323. doi: 10.1021/bi000635a. [DOI] [PubMed] [Google Scholar]
- 20.Kraulis P. A program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 21.Merritt E, Bacon D. Raster3D: photorealistic molecular graphics. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


