Abstract
Neuroserpin isolated from inclusion bodies in the brain of a patient with a neurodegenerative disease was characterized biochemically. The protein consisted of residues 20 to 410 of the neuroserpin precursor deduced from its cDNA sequence indicating the entire molecule was deposited. A minor amount started with residue 19 of the precursor, and the carboxyl terminus was heterogeneous ending at residues 405, 407, 409, and 410. Arg was present at position 52. No normal Ser52 was found indicating that only mutant neuroserpin was present in the inclusion bodies. The three potential Asn glycosylation sites all contained carbohydrate. DNA sequence analysis of exons 2 to 9 of the neuroserpin gene in the proband showed the published normal neuroserpin sequence except for the presence of both adenine and cytosine at the first position of codon 52, that indicates heterozygosity for both the normal Ser(AGT) and variant Arg(CGT) at this position in the expressed protein. Restriction fragment length polymorphism analysis of a polymerase chain reaction product from exon 2 revealed the propositus and his affected sibling both were heterozygous for the mutation whereas 100 unaffected controls were negative. Chemical characterization of the variant neuroserpin will significantly enhance the understanding of this protein in both normal physiology and neurodegenerative diseases.
Neurodegenerative disease with intraneuronal inclusions containing neuroserpin has recently been described. 1,2 This disease, that has been reported thus far in two families, is characterized by the adult-onset of progressive dementia leading to death within 10 to 20 years. In one family the syndrome included epilepsy in addition to cognitive decline. 3 At postmortem examination inclusions of neuroserpin, a serine protease inhibitor expressed predominantly in the central nervous system, are found within neurons of the cerebral cortex and subcortical nuclei. Pathology has been linked to mutations in the gene for neuroserpin that are predicted to give mutant forms of the protein. 1
Several questions raised by this finding are apparent. Is protein structure or its effect on protein function the immediate cause of neurodegeneration? In this autosomal dominant disease is neuroserpin protein coded by the normal allele involved in the disease process and present in the intraneuronal inclusions in conjunction with the mutant neuroserpin? Are there identifiable changes in the composition of the mutant neuroserpin, eg, glycosylation, that render the protein resistant to normal degradation or to extracellular transport?
To begin to address these questions we have isolated and biochemically characterized a mutant form of neuroserpin from a patient with intraneuronal protein deposits associated with myoclonic epilepsy and dementia that started at age 24 and resulted in death at age 43.
Materials and Methods
Clinical Summary
The proband was a 43-year-old Caucasian male. His clinical and pathological details have been reported elsewhere. 4 In brief, he started to have generalized seizures and thereafter myoclonus at the age of 24 years. His performance at work as an architectural drafter deteriorated and he had difficulties with memory, occasionally being unable to write even his own name. At the age of 27 years, seizures composed of myoclonic, complex partial, and tonic-clonic reappeared. Despite aggressive treatment with several pharmacological agents, his seizures were difficult to control and there were several episodes of status epilepticus. He could not continue to work after age 28. On neurological examination at age 29, he showed slow speech, vertical and horizontal nystagmus, dysarthria, and myoclonus in the extremities. At age 30, he was slow in mental processing and calculation. Symptoms of cognitive abnormality gradually worsened. Although his score on the Mini Mental State Examination at age 34 was 24 of 30 indicating mild cognitive impairment, that score at age 38 dropped to 10 of 30 with advanced dementia. As the disease progressed generalized seizures, myoclonus of the face and extremities, and dementia worsened and were intractable. At the age of 43 years he died from aspiration pneumonia. His mother and his living brother also expressed similar clinical symptoms. This family has lived in Indiana for several generations, and there is no known relation to two previously reported families with mutations in the neuroserpin gene. 1-3
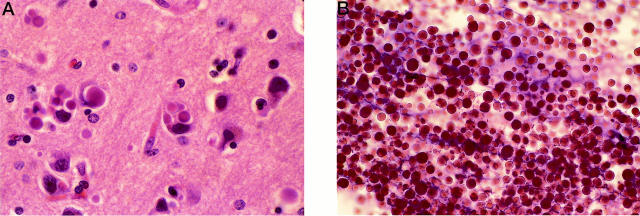
At postmortem examination, his brain weighed 980 g and showed diffuse atrophy that was most evident at the level of the frontal lobes. On microscopic examination, the most striking histological finding was the presence of intraneuronal eosinophilic homogenous bodies in the neuronal perikaryon and the neuropil in most gray matter areas of the brain and spinal cord (Figure 1A) ▶ . The diameter of these inclusions ranged from 1.5 to 25 μm. When a major portion of the perikaryon was occupied by one or more inclusions, the nucleus was eccentric and often deformed. In addition to being strongly eosinophilic, these bodies were positive to periodic acid-Schiff (PAS) staining. Neuronal loss was moderate in the frontal, temporal, parietal, and occipital cortices and mild in the hippocampus, amygdala, basal ganglia, and thalamus.
Figure 1.
A: H&E-stained section of frontal cortex of proband showing neuronal inclusions and neuronal cell loss (original magnification, ×780). B: H&E-stained preparation of pellet after treatment with detergent showing many intact inclusion bodies (original magnification, ×270).
In immunohistochemical analysis, the intraneuronal inclusion bodies did not react with antibodies against glial fibrillary acidic protein, α-synuclein, neurofilament proteins, amyloid β-protein, tubulin, tau, and ubiquitin. By electron microscopy, the inclusions appeared to be composed of a fine granular material. These were surrounded by an irregularly shaped, electron lucent space that was limited by a unit membrane, that was most likely the rough endoplasmic reticulum.
Isolation of Inclusion Bodies
Cortical gray matter, 15 g, was homogenized in 120 ml of 250 mmol/L sucrose and 10 mmol/L ethylenediaminetetraacetic acid, buffered to pH 7.4 with 10 mmol/L HEPES containing one tablet of protease inhibitor cocktail (Boehringer Mannheim, Indianapolis, IN). The homogenate was filtered through gauze and centrifuged at 1,000 × g for 10 minutes. The supernatant was decanted and the pellet resuspended in 30 ml of 0.5% N-lauroylsarcosine (Sigma Chemical Co., St. Louis, MO) in homogenization medium. The suspension was incubated at 37°C for 30 minutes and then centrifuged again at 25,000 × g for 60 minutes. The pellet was resuspended in 30 ml of 1% sarcosyl solution, incubated at 37°C for 30 minutes, and then centrifuged at 25,000 × g for 60 minutes. Verification that the inclusion bodies were not lost was accomplished by visualization using light microscopy after hematoxylin and eosin (H&E) staining of an air-dried sample of the pellet on a glass slide followed by fixing for 1 minute in absolute ethanol before staining (Figure 1B) ▶ .
Chromatography
Isolated inclusions were solubilized in 5 ml of 6 mol/L guanidine HCl, 0.5 mol/L Tris, pH 8.2, containing 50 mg of dithiothreitol with stirring for 2 days, alkylated with 121 mg of iodoacetic acid, and then centrifuged at 25,000 × g for 60 minutes. The supernatant was applied to a Sepharose CL6B column (2.5 × 90 cm). Proteins were eluted with 4 mol/L guanidine HCl, 0.05 mol/L Tris, pH 8.2, and fractions were pooled according to OD280 nm, dialyzed against distilled water, and lyophilized.
Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE) Analysis
Protein samples were dissolved in 0.125 mol/L Tris-HCl, pH 6.8, buffer containing 4% SDS, 10% glycerol, and 5% 2-mercaptoethanol and boiled for 5 minutes. The samples were applied to a Bio-Rad (Hercules, CA) 4 to 20% gradient Readygel in Tris/glycine/SDS buffer, electrophoresed at 100 volts for 90 minutes, and the gel stained with 0.25% Coomassie brilliant blue solution.
Trypsin Digestion and Peptide Fractionation
Pool III from CL6B chromatography was digested with trypsin (2% by weight, N-tosyl-L-phenylalanine chloro-methyl ketone (TPCK)-treated; Worthington Biochemical Corp., Lakewood, NJ) in 0.1 mol/L ammonium bicarbonate at room temperature overnight and recovered by freezing and lyophilization. The digest was dissolved in 0.5 ml of 25% acetic acid, insoluble material removed by centrifugation, and fractionated on a Beckman Ultra-sphere ODS high-pressure liquid chromatography column (0.46 × 25 cm) (Beckman-Coulter, Inc., Fullerton, CA), equilibrated with 0.1% trifluoro acetic acid (TFA) in water, and eluted with a 0 to 60% acetonitrile gradient. Eluant was monitored by absorbance at 215 nm. Separated peaks were recovered by drying in a Savant Speed Vac Concentrator (Savant Instruments, Farmingdale, NY).
Amino Acid Sequence Analysis
Samples were analyzed by Edman degradation on an Applied Biosystems Model 473A Protein Sequencer using the manufacturer’s standard cycles (Applied Biosystems, Foster City, CA).
DNA Sequencing
DNA was obtained from the brain of the proband and analyzed by direct sequencing of exons 2 to 9 of neuroserpin. 5 The same intronic primers based on the human neuroserpin genomic sequence, reported by Davis and colleagues 1 in 1999, were used for amplification and sequencing. Standard amplification reactions were done with 20 ng/μl of genomic DNA. The amplified products were gel-purified and then asymmetrically amplified to generate single-stranded template for sequencing. The reactions were subjected to QIAquick PCR purification spin columns (Qiagen, Valencia, CA) that remove remaining primers and deoxynucleotides.
Standard dideoxynucleotide sequencing was performed using the U.S. Biochemicals Sequenase kit, 35S-dATP (Amersham, Arlington Heights, IL) and modified T7 DNA polymerase (Sequenase Version 2.0, U.S. Biochemicals/Amersham). The reactions were loaded and run on a 6% polyacrylamide/8 mol/L urea sequencing gel. The gel was dried and exposed to X-ray film. Sequences were compared to that of normal controls and to the published neuroserpin coding sequence.
Genomic DNA was isolated from peripheral blood using phenol/chloroform. DNA samples from 100 unrelated patients were used as controls for RFLP analysis.
RFLP Analysis
In the absence of a spontaneous creation of an endonuclease restriction site by the A to C mutation, a PCR-induced mutation restriction analysis (IMRA) was performed. 6 A mutation primer (NSz) corresponding to nucleotides 236 to 260 of the neuroserpin cDNA sequence with a G instead of A at the second position from the 3′ end (5′ ATCATTCCCATTGCAAGAGCAATGC 3′) was used to create a restriction site for AviII (TGCGCA) only in the mutant gene PCR products. 7 PCR was performed with NSz and NSE2F (5′ GGACTCTTCTCTTTGCTG 3′) primers in a total volume of 50 μl 10 × PCR buffer (100 mmol/L Tris-HCl, pH 8.3, 500 mmol/L KCl, 12 mmol/L MgCl2), 8 μl of 1.25 mmol/L dNTP, 15 pmol of each primer, 2.5 U Taq polymerase, and 100 ng DNA. Amplification was performed using a Perkin-Elmer Thermal Cycler (Perkin-Elmer, Emeryville, CA) for 30 cycles consisting of denaturation at 95°C for 30 seconds, annealing at 60°C for 1 minute, and extension at 72°C for 30 seconds. After purification of the products by the QIAquick PCR purification kit, 12.5 μl of amplified DNA was added to 1 μl (5 U) of AviII and 1.5 μl of its specific buffer and incubated for 2 hours at 37°C. The samples were electrophoresed through 2% (w/v) Nusieve GTG agarose gel, stained with ethidium bromide, and photographed under UV light.
Results
Protein Purification
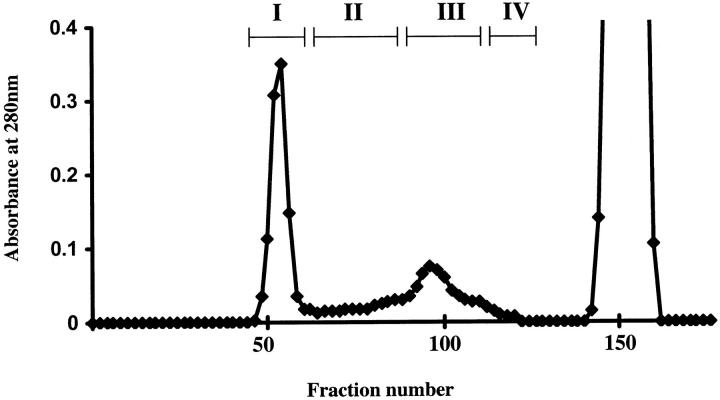
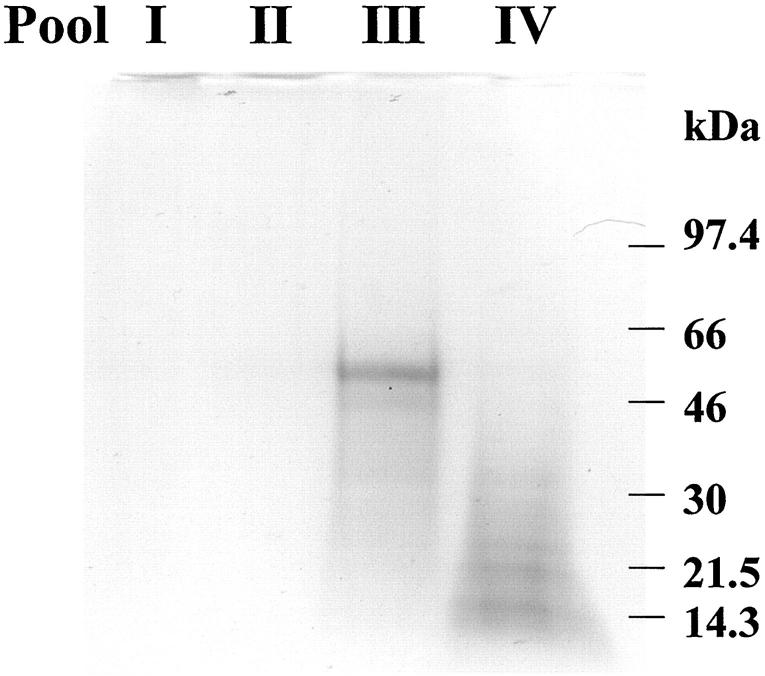
Neuroserpin inclusions were isolated from frozen cerebral cortex by homogenization in a buffered sucrose solution and centrifugation. The pellet was then treated with detergent to release the inclusions that were isolated by high-speed centrifugation (Figure 1) ▶ . The inclusions were solubilized in 6 mol/L guanidine HCl in the presence of dithiothreitol and proteins fractionated on a Sepharose CL6B column (2.5 × 90 cm) (Figure 2) ▶ . Only one major retarded peak (pool III) was found. SDS-PAGE of the solubilized inclusion preparation before fractionation revealed a major protein band migrating at a molecular mass of ∼50 kd. This band was present in pool III of the fractionated proteins (Figure 3) ▶ .
Figure 2.
Chromatogram of inclusion bodies solubilized in 6 mol/L of guanidine HCl on a Sepharose CL6B column (2.6 × 90 cm) equilibrated and eluted with 4 mol/L of guanidine HCl, 0.05 mol/L Tris, pH 8.2. Fractions were pooled as indicated by the bars.
Figure 3.
SDS-PAGE analysis of Sepharose CL6B pools. A major band migrating at a molecular mass of ∼50 kd is present in pool III. Positions of molecular weight standards are indicated on the right.
Protein Characterization
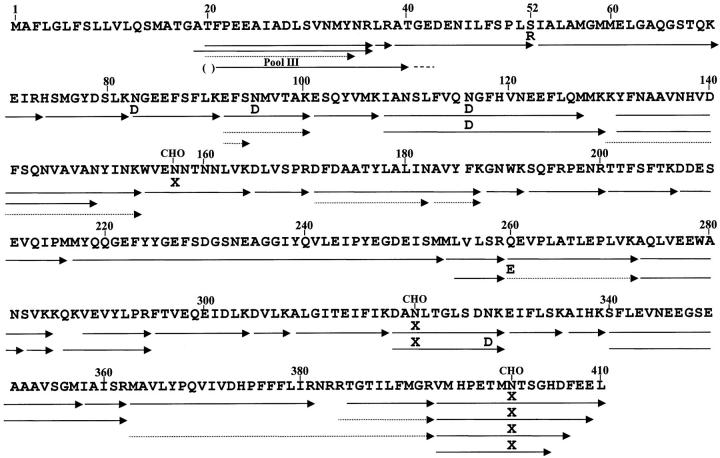
Amino terminal sequence analysis of pool III gave a major sequence starting with position 20 of human neuroserpin (Figure 4) ▶ . The first two residues could not be unambiguously identified because of high background in the first few cycles.
Figure 4.
Amino acid sequence of human neuroserpin precursor deduced from cDNA sequence analysis is presented. 7 Putative glycosylation sites at Asn residues 157, 321, and 401 are indicated by CHO above the residues. The arrow labeled pool III gives the major sequence obtained from Edman degradation analysis of pool III from Sepharose CL6B. The first two residues indicated by parentheses were tentatively identified because of initial high background. Sequences obtained from tryptic peptides are indicated by arrows. Differences from the deduced cDNA sequence are indicated by single-letter amino acid code over the arrows. X at residues 157, 321, and 401 indicates no amino acid was found at these positions in the tryptic peptides suggesting that they contained Asn with attached carbohydrate. Dashed arrows indicate minor peptides.
The sequence of the isolated protein was obtained from analysis of tryptic peptides that identified all residues except numbers 1 to 18 (putative signal peptide), and number 285 (Figure 4) ▶ . Residue 52 was Arg instead of the normal Ser. No peptide with Ser at position 52 was found in the trypsin digest, indicating that normal neuroserpin is not present in the inclusion bodies. Although the major N-terminal tryptic peptide was residues 20 to 36, some residue 19 to 36 peptide was also found. From the recoveries of the peptides, ∼70% of the mutant neuroserpin in the inclusion bodies starts with residue 20 and 30% with residue 19. No tryptic peptides were found that started with residue 17 or 18. Several carboxyl terminal tryptic peptides starting with residue 394 and ending with residues 405, 407, 409, and 410 were found indicating that some C-terminal proteolysis of neuroserpin had occurred. Three potential Asn glycosylation sites at residues 157, 321, and 401 were predicted from the neuroserpin cDNA sequence. 7,8 No amino acid was found at these positions in the tryptic peptides containing these three residues suggesting that all three Asn are glycosylated. Also, peptides containing Asp instead of Asn at residues 83, 95, 116, and 328, and Glu instead of Gln at residue 260 were found indicating that some deamidation of Asn and Gln had occurred. Only Asp was found at residues 83 and 116, whereas residue 95 contained 87% Asp and 13% Asn, and residue 328 contained 70% Asn and 30% Asp. In addition to residues 92 to 100 peptides with Asp or Asn at residue 95, a peptide yielding only residues 92 to 94 was found (yield ∼20% that of residue 92 to 100 peptides). Also, an N-terminal residue 20 to 34 peptide was found in a yield ∼10% that of the residue 20 to 36 peptide. The fact that only four of 24 Asn and one of 16 Gln in neuroserpin have undergone deamidation may indicate these positions are in a tertiary structure especially susceptible to deamidation. A common nonenzymatic mechanism of Asn deamidation is via a cyclic imide formation that then hydrolyzes to Asp or iso-Asp. 9 The finding of tryptic peptides, that on Edman sequence analysis stopped at the residue before Asn35 and Asn95 (Figure 4) ▶ , may indicate the presence of iso-Asp at these positions. An iso-Asp residue would be resistant to Edman cyclization and cleavage.
Several major nontrypsin-like cleavages in neuroserpin were found in the tryptic digest of pool III (Figure 4) ▶ . The most prominent involved cleavages between adjacent Met residues and on the carboxyl side of Met. Complete cleavage of the Met216-Met217 and Met253-Met254 peptide bonds and ∼50% cleavage of the Met127-Met128 bond were found. Significant cleavage of the Met254-Leu255 and Met 357-Ile 358 peptide bonds were also observed, as well as cleavage after Asn at residues 149, 182, and 281. Proteolysis after Met is rare by trypsin, but can occur with chymotrypsin. TPCK-treated trypsin was used to minimize chymotryptic-like proteolysis. It is unlikely that these cleavages were because of contaminating chymotrypsin in the trypsin preparation because no usual chymotryptic-specific proteolysis after aromatic residues (>40 in neuroserpin) was observed in the high-pressure liquid chromatography separated digest. Possibly, the minor residue 20 to 34 sequence peptide could have resulted from a chymotrypsin-like cleavage of Tyr34-Asn35, or it could be because of cyclization of Asn and termination of Edman degradation as mentioned above. The data would suggest that the rare Met-Met sequence and Met peptide bonds in neuroserpin are unusually susceptible to trypsin cleavage.
Tryptic peptides from human α- and β-tubulin, β- and γ-actin, 2′, 3′-cyclic nucleotide-3′-phosphodiesterase, and creatine kinase B chain were also present in the tryptic digest of pool III in ≤10% of the molar amounts of neuroserpin. All are similar in molecular mass (∼40 kd to 50 kd) to neuroserpin and would elute from a molecular sieve column close to neuroserpin. It is highly unlikely that these proteins are part of the inclusion bodies, but rather contaminants from the cytosol incompletely removed during isolation of the inclusion bodies.
DNA Sequence
DNA sequence of neuroserpin exons 3 to 9 of the proband were as published for the normal human cDNA. 7 Sequence of exon 2 revealed both adenine and cytosine at the first position of codon 52 of neuroserpin. This sequence indicates heterozygosity for both the normal serine (AGT) and variant arginine (CGT) at this position in the expressed protein.
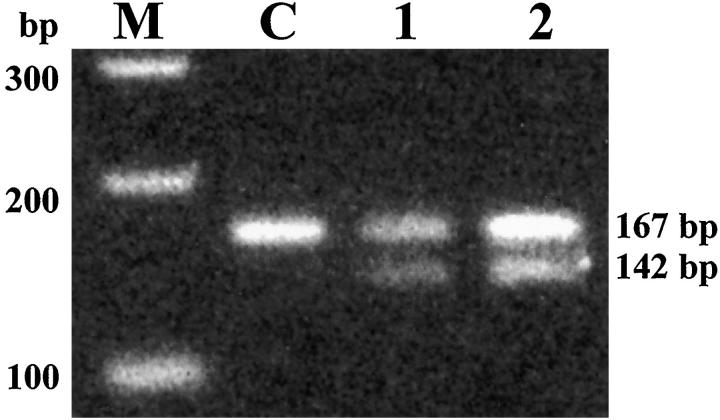
RFLP analysis of the 167-bp PCR product from NSE2F and NSz primers revealed that the propositus and his affected sibling both had the AviII recognition site associated with the adenine to cytosine mutation (Figure 5) ▶ . Electrophoresis of the digested PCR product gave a digestion band of 142 bp in addition to the normal 167-bp band indicating heterozygosity for the mutation. Nonaffected individuals (100 unrelated patients) did not show any digestion of PCR product.
Figure 5.
AviII restriction site analysis of neuroserpin exon 2 PCR products. The A to C transition at the first position corresponding to codon 52 of neuroserpin creates an AviII restriction site within the 167-bp PCR-amplified fragment when coupled with mutagenesis primer (NSz). Enzyme digestion gives fragments of 142 bp and 25 bp (the latter is not visible) for the mutant allele. The PCR-amplified fragments (167 bp) of the proband (lane 1) and his affected brother (lane 2) were incompletely digested, suggesting that they are heterozygous for this mutation. The fragment from a normal control is not digested with AviII (lane C). M: 100-bp molecular ruler DNA size standard (Bio-Rad).
Discussion
Neuroserpin was originally identified as a protein secreted by cultured chicken dorsal root ganglion cells. 10 Subsequently, isolation and purification from ocular vitreous fluid of 14-day-old chick embryos allowed determination of sufficient amino acid sequence to clone a neuroserpin cDNA from an embryonic chicken brain cDNA library. 11 The cDNA encoded a 410-residue protein that by sequence homology was assigned to the serpin superfamily of serine protease inhibitors. Expression of neuroserpin in embryonic neurons and in adult brain regions that exhibit synaptic plasticity supports the hypothesis that this protein is a member of the group of extracellular proteases and protease inhibitors that orchestrate brain development, function, and anatomical integrity. 12,13 As with many proteins expressed by the central nervous system, the identification of mutant forms of the protein, and diseases associated with these molecular defects, provides a new window to decipher the role of the protein in normal physiology. At the same time the metabolic basis of previously undefined disease is revealed. This is particularly true for the neurodegenerative diseases that are characterized by dementia. In the past, clinical and anatomical characterizations of these diseases have, by comparing similarities and dissimilarities, resulted in grouping together diseases that at the molecular level are distinct entities.
The PAS-positive reaction of the proband neuronal inclusions distinguishes them from Pick bodies, Lewy bodies, and Hirano bodies that are all PAS-negative (Table 1) ▶ . The morphological features and the PAS-positivity of the intraneuronal inclusions of the proband are analogous to other types of PAS-positive inclusions, including corpora amylacea, Lafora bodies, Bielschowsky bodies, the inclusions of type IV glycogenosis, and the polyglucosan bodies of adult polyglucosan body disease. 14-20 However, the proband neuronal inclusions differ from these PAS-positive inclusions in structure and composition (Table 1) ▶ . On electron microscopic examination, the proband inclusions showed primarily granular material with a limiting membrane, whereas the other PAS-positive inclusions are composed of fine filaments without a limiting membrane. Moreover, the proband inclusions were composed of a variant neuroserpin as indicated by protein sequencing in this study, and there was no evidence of polyglucosan deposition as seen in other inclusions mentioned above.
Table 1.
Characteristics of Inclusions in Central Nervous System
| Inclusion type | Diameter (μm) | Ultrastructure | Limiting membrane | PAS* | Biochemical content |
|---|---|---|---|---|---|
| Pick body | 2–20 | Filaments | − | − | Tau |
| Lewy body | 8–30 | Filaments | − | − | α-Synuclein |
| Hirano body | 8–30 | Filaments | − | − | Actin |
| Corpora amylacea | 2–20 | Filaments | − | + | Polyglucosan |
| Bielschowsky body | –40 | Filaments | − | + | Polyglucosan |
| Lafora body | 1–30 | Filaments | − | + | Polyglucosan |
| Inclusion of type IV glycogenosis | –20 | Filaments | − | + | Polyglucosan |
| Inclusion of adult polyglucosan body disease | 1–25 | Filaments | − | + | Polyglucosan |
| Inclusion of the proband in this study | 1–30 | Granular material | + | + | Neuroserpin |
* PAS: +, positive staining; −, negative staining.
The 410-residue neuroserpin deduced from chicken cDNA has a hydrophobic amino terminal sequence of 16 amino acid residues conforming to a consensus signal sequence. 11 The N-terminus of purified chicken neuroserpin protein started with residue 17 of the cDNA-encoded protein in agreement with the first 16 residues being the signal peptide. 11 Mouse and rat neuroserpin cDNAs have also been cloned and sequenced. 12,13,21 Both encode for a 410-residue protein, and the N-terminal 16 residues of both were proposed as signal peptides by analogy with chicken neuroserpin. Human neuroserpin cDNA was originally isolated from a fetal retina cDNA library. 7 From analysis of the N-terminal sequence of the encoded 410-residue protein, Schrimpf and colleagues 7 concluded that the Ala16-Thr17 and Ala 19-Thr20 peptide bonds had equal probability as signal peptide cleavage sites. By analogy to chicken neuroserpin, they assigned the N-terminal 16 residues as the signal peptide. Recently, Davis and colleagues 1,2 isolated a mutant neuroserpin from inclusion bodies in the brain of a patient with familial dementia. The Ser49-Pro mutant neuroserpin had an N-terminus starting with Thr20. The Ser52-Arg mutant neuroserpin characterized in the present study indicates a heterogeneous N-terminus with the majority starting at Thr20 and ∼30% at Ala19. Analysis of the human neuroserpin N-terminal sequence by von Heijne’s probability matrix indicates that whereas the Gly18-Ala19 peptide bond has less probability than the Ala16-Thr17 and Ala19-Thr20 peptide bonds as signal peptidase cleavage sites, it does have greater probability than other peptide bonds in the region. 22 This is supported by the finding that a PCR product encoding for residues 1 to 410 of human neuroserpin inserted into a baculovirus expression vector and expressed in Sf9 insect cells yielded a purified recombinant neuroserpin protein that had equal amounts beginning with Ala19 and Thr20. 23 These results show that the signal peptidase in this system cleaves the expressed normal neuroserpin after Gly18 and Ala19, similar to the results in our characterization of the Ser52-Arg mutant neuroserpin.
One distinguishing characteristic of the neuroserpin inclusions is that they contain carbohydrate as indicated by their positive staining with PAS reagent. 4 Human, mouse, and rat neuroserpins have three potential carbohydrate attachment sites at Asn residues 157, 321, and 401. 7,12,13 Chicken neuroserpin has only two sites, as Asp is at position 321 instead of Asn. 11 All species have carbohydrate attached at least to one site because the size of the protein on SDS-PAGE analysis is greater than its molecular weight calculated from its amino acid sequence. 2,11-13 Sequence analysis of the Ser52-Arg mutant neuroserpin is consistent with glycosylation of all three Asns. The tertiary structure model, based on the primary structure, positions each of these Asn residues on the surface of the molecule and, therefore, indicates that a carbohydrate moiety can be readily accommodated at all three sites. 24
The neuroserpin inclusion bodies in the brain bear similarity to inclusion bodies in the liver composed of α1-antitrypsin, another member of the serpin superfamily. Although >70 α1-antitrypsin variants are known, only three are associated with liver abnormalities. 25 Like the neuroserpin mutants (Ser49Pro and Ser52Arg), two α1-antitrypsins, Malton (ΔPhe51) and Siiyama (Ser53Phe) have mutations in the shutter region of the molecule. 26,27 The third, α1-antitrypsin Z (Glu342Lys), involves loss of a salt bridge. 28-30 Although inclusion bodies from homozygous variant α1-antitrypsin patients have been characterized, we are not aware of any studies on inclusion bodies from heterozygous patients investigating if only the variant protein is present as we found with the neuroserpin inclusion bodies. Variant α1-antitrypsin isolated from plasma and liver inclusion bodies has altered carbohydrate compared to the normal protein with no sialic acid and high mannose content. 31-34 The incomplete carbohydrate maturation results in its accumulation in the liver endoplasmic reticulum and reduced secretion into the blood stream. Although mutant neuroserpin is glycosylated, its carbohydrate structure is unknown. Elucidation and comparison of the carbohydrate structures on mutant and normal neuroserpin should provide insight on the role of glycosylation in neuroserpin inclusion body formation.
These mutations in α1-antitrypsin and presumably in neuroserpin result in two effects leading to intracellular inclusion body formation. First, the mutation alters the molecule’s structure sufficiently to impair carbohydrate maturation that results in reduced secretion and increased intracellular concentration. Second, the mutation alters the protein’s structure or stability to allow aggregation by the insertion of the reactive loop of one mutant molecule into the β-sheet of a second mutant molecule at the increased intracellular concentration. This aggregation phenomenon is common in the serpin superfamily as certain mutants of antithrombin, C1-inhibitor, and α1-antichymotrypsin also form inclusion bodies and/or multimers. 35-37
The fact that individuals heterozygous for variant neuroserpins have normal central nervous system development suggests that function of the mutant protein is intact, the expression of one normal allele during fetal development is sufficient, or that neuroserpin is not essential for brain development. The finding of only mutant neuroserpin in the inclusions indicates that synthesis and processing of normal neuroserpin is unaltered. It has been postulated that neuroserpin is part of the protease and protease inhibitor complex that determines neuronal growth and synaptic plasticity processes that occur extracellularly. 12,13 In the case of mutant neuroserpin, the protein accumulates within neurons of the cerebral cortex, brain stem nuclei, and dorsal root ganglia. 2,4 The mutant protein is secreted past the ribosomal membrane with cleavage of the signal peptide, it is posttranscriptionally modified by glycosylation, but is not excreted. If the mutant does retain its function of serine protease inhibition, it is not transported to the location where this function is required. Intracellular accumulation of the abnormally aggregating protein is, therefore, the most likely cause of neuronal dysfunction.
Acknowledgments
We thank D. Dian Martin for assistance in preparation of the manuscript.
Footnotes
Address reprint requests to Merrill D. Benson, M.D., Pathology and Laboratory Medicine, Indiana University School of Medicine, 975 West Walnut St., #IB-503 Indianapolis, IN 46202-5121. E-mail: mdbenson@iupui.edu.
Supported in part by the Public Health Service (grants AG10133, DK42111, and RR-00750), the Veteran Affairs Medical Research, the Marion E. Jacobson Fund, and the Machado Family Research Fund. M. Yazaki is a postdoctoral fellow from Shinshu University and is supported by Japanese Gift Foundation, Boshi-Aiiku-Kai. M. Takao is a postdoctoral fellow from Keio University and is supported by the Sasakawa Health Science Foundation.
References
- 1.Davis RL, Shrimpton AE, Holohan PD, Bradshaw C, Feiglin D, Collins GH, Sonderegger P, Kinter J, Becker LM, Lacbawan F, Krasnewich D, Muenke M, Lawrence DA, Yerby MS, Shaw C-M, Gooptu B, Elliott PR, Finch JT, Carrell RW, Lomas DA: Familial dementia caused by polymerization of mutant neuroserpin. Nature 1999, 401:376-379 [DOI] [PubMed] [Google Scholar]
- 2.Davis RL, Holohan PD, Shrimpton AE, Tatum AH, Daucher J, Collins GH, Todd R, Bradshaw C, Kent P, Feiglin D, Rosenbaum A, Yerby MS, Shaw C-M, Lacbawan F, Lawrence DA: Familial encephalopathy with neuroserpin inclusion bodies. Am J Pathol 1999, 155:1901-1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yerby MS, Shaw C-M, Watson JMD: Progressive dementia and epilepsy in a young adult: unusual intraneuronal inclusions. Neurology 1986, 36:68-71 [DOI] [PubMed] [Google Scholar]
- 4.Takao M, Benson MD, Murrell JR, Yazaki M, Piccardo P, Unverzagt F, Davis RL, Holohan PD, Lawrence DA, Richardson R, Farlow MR, Ghetti B: Neuroserpin mutation S52R causes neuroserpin accumulation in neurons and is associated with progressive myoclonus epilepsy. J Neuropathol Exp Neurol 2000, 59:1069-1085 [DOI] [PubMed] [Google Scholar]
- 5.Nichols WC, Gregg RE, Brewer HB, Benson MD: A mutation in apolipoprotein A-I in the Iowa type of familial amyloidotic polyneuropathy. Genomics 1990, 8:318-323 [DOI] [PubMed] [Google Scholar]
- 6.Zeldenrust SR, Benson MD: A new test for detection of the His58 variant transthyretin allele in hereditary amyloidosis; creation of diagnostic restriction endonuclease recognition sites by PCR based mutagenesis. Amyloid Int J Exp Clin Invest 1994, 1:154-159 [Google Scholar]
- 7.Schrimpf SP, Bleiker AJ, Brecevic L, Kozlov SV, Berger P, Osterwalder T, Krueger SR, Schinzel A, Sonderegger P: Human neuroserpin (PI12): cDNA cloning and chromosomal localization to 3q26. Genomics 1997, 40:55-62 [DOI] [PubMed] [Google Scholar]
- 8.Marshall RD, Neuberger A: Aspects of the structure and metabolism of glycoproteins. Tipson RS Horton D eds. Advances in Carbohydrate Chemistry and Biochemistry, 1970, vol 25.:pp 407-478 Academic Press, New York and London [Google Scholar]
- 9.Orpiszewski J, Schormann N, Kluve-Beckerman B, Liepnieks JJ, Benson MD: Protein aging hypothesis of Alzheimer disease. FASEB J 2000, 14:1255-1263 [DOI] [PubMed] [Google Scholar]
- 10.Stoeckli ET, Lemkin PF, Kuhn TB, Ruegg MA, Heller M, Sonderegger P: Identification of proteins secreted from axons of embryonic dorsal-root-ganglia neurons. Eur J Biochem 1989, 180:249-258 [DOI] [PubMed] [Google Scholar]
- 11.Osterwalder T, Contartese J, Stoeckli ET, Kuhn TB, Sonderegger P: Neuroserpin, an axonally secreted serine protease inhibitor. EMBO J 1996, 15:2944-2953 [PMC free article] [PubMed] [Google Scholar]
- 12.Hill RM, Parmar PK, Coates LC, Mezey E, Pearson JF, Birch NP: Neuroserpin is expressed in the pituitary and adrenal glands and induces the extension of neurite-like processes in AtT-20 cells. Biochem J 2000, 345:595-601 [PMC free article] [PubMed] [Google Scholar]
- 13.Krueger SR, Ghisu G-P, Cinelli P, Gschwend TP, Osterwalder T, Wolfer DP, Sonderegger P: Expression of neuroserpin, an inhibitor of tissue plasminogen activator, in the developing and adult nervous system of the mouse. J Neurosci 1997, 17:8984-8996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yokoi S, Austin T, Witmer F, Sakai M: Studies in myoclonus epilepsy (Lafora body form). 1. Isolation and preliminary characterization of Lafora bodies in two cases. Arch Neurol 1968, 19:15-33 [DOI] [PubMed] [Google Scholar]
- 15.Sakai M, Austin J, Witmer F, Trueb L: Studies of corpora amylacea. 1. Isolation and preliminary characterization by chemical and histochemical techniques. Arch Neurol 1969, 21:526-544 [DOI] [PubMed] [Google Scholar]
- 16.Sakai M, Austin J, Witmer F, Trueb L: Studies in myoclonus epilepsy (Lafora body form). II. Polyglucosans in the systemic deposits of myoclonus epilepsy and in corpora amylacea. Neurology 1970, 20:160-176 [DOI] [PubMed] [Google Scholar]
- 17.Collins GH, Cowden RR, Nevis AH: Myoclonus epilepsy with Lafora bodies: an ultrastructural and cytochemical study. Arch Pathol 1968, 86:239-254 [PubMed] [Google Scholar]
- 18.Schochet SS, McCormick WF, Zellweger H: Type IV glycogenosis (amylopeptinosis): light and electron microscopic observations. Arch Pathol 1970, 90:354-363 [PubMed] [Google Scholar]
- 19.Robitaille Y, Carpenter S, Karpati G, Dimauro S: A distinct form of adult polyglucosan body disease with massive involvement of central and peripheral neuronal processes and astrocytes: a report of four cases and a review of the occurrence of polyglucosan bodies in other conditions such as Lafora disease and normal aging. Brain 1980, 103:315-336 [DOI] [PubMed] [Google Scholar]
- 20.Cavanagh JB: Corpora-amylacea and the family of polyglucosan disease. Brain Res Rev 1999, 29:265-295 [DOI] [PubMed] [Google Scholar]
- 21.Berger P, Kozlov SV, Krueger SR, Sonderegger P: Structure of the mouse gene for the serine protease inhibitor neuroserpin (PI12). Gene 1998, 214:25-33 [DOI] [PubMed] [Google Scholar]
- 22.Von Heijne G: Patterns of amino acids near signal-sequence cleavage sites. Eur J Biochem 1983, 133:17-21 [DOI] [PubMed] [Google Scholar]
- 23.Hastings GA, Coleman TA, Haudenschild CC, Stefansson S, Smith EP, Barthlow R, Cherry S, Sandkvist M, Lawrence DA: Neuroserpin, a brain-associated inhibitor of tissue plasminogen activator is localized primarily in neurons: implications for the regulation of motor learning and neuronal survival. J Biol Chem 1997, 272:33062-33067 [DOI] [PubMed] [Google Scholar]
- 24.Huntington JA, Pannu NS, Hazes B, Read RJ, Lomas DA, Carrell RW: A 2.6 Å structure of a serpin polymer and implications for conformational disease. J Mol Biol 1999, 293:449-455 [DOI] [PubMed] [Google Scholar]
- 25.Cox DW: α1-Antitrypsin deficiency. The Metabolic and Molecular Bases of Inherited Disease, Connective Tissues, ed 7, vol III, chap 138, part 18. Edited by CR Scriver, AL Beaudet, WS Sly, D Valle. New York, McGraw Hill, 1995, pp 4125–4158
- 26.Seyama K, Nukiwa T, Takabe K, Takahashi H, Miyake K, Kira S: Siiyama (Serine 53 (TCC) to Phenylalanine 53 (TTC)): a new α1-antitrypsin-deficient variant with mutation on a predicted conserved residue of the serpin backbone. J Biol Chem 1991, 266:12627-12632 [PubMed] [Google Scholar]
- 27.Curiel DT, Holmes MD, Okayama H, Brantly ML, Vogelmeier C, Travis WD, Stier LE, Perks WH, Drystal RG: Molecular basis of the liver and lung disease associated with the α1-antitrypsin deficiency allele Mmalton. J Biol Chem 1989, 264:13938-13945 [PubMed] [Google Scholar]
- 28.Jeppsson J-O: Amino acid substitution Glu→Lys in α1-antitrypsin PiZ. FEBS Lett 1976, 65:195-197 [DOI] [PubMed] [Google Scholar]
- 29.Yoshida A, Lieberman J, Gaidulis L, Ewing C: Molecular abnormality of human alpha1-antitrypsin variant (Pi-ZZ) associated with plasma activity deficiency. Proc Natl Acad Sci USA 1976, 73:1324-1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carrell RW, Jeppsson J-O, Laurell C-B, Brennan SO, Owen MC, Vaughan L, Boswell DR: Structure and variation of human α1-antitrypsin. Nature 1982, 298:329-334 [DOI] [PubMed] [Google Scholar]
- 31.Bell OF, Carrell RW: Basis of the defect in α-1-antitrypsin deficiency. Nature 1973, 243:410-411 [DOI] [PubMed] [Google Scholar]
- 32.Eriksson S, Larsson C: Purification and partial characterization of PAS-positive inclusion bodies from the liver in alpha1-antitrypsin deficiency. N Engl J Med 1975, 292:176-180 [DOI] [PubMed] [Google Scholar]
- 33.Jeppsson J-O, Larsson C, Eriksson S: Characterization of α1-antitrypsin in the inclusion bodies from the liver in α1-antitrypsin deficiency. N Engl J Med 1975, 293:576-579 [DOI] [PubMed] [Google Scholar]
- 34.Bathurst IC, Travis J, George PM, Carrell RW: Structural and functional characterization of the abnormal Z α1-antitrypsin isolated from human liver. FEBS Lett 1984, 177:179-183 [DOI] [PubMed] [Google Scholar]
- 35.Lindo VS, Kakkar VV, Learmonth M, Melissari E, Zappacosta F, Panico M, Morris HR: Antithrombin-TRI (Ala382 to Thr) causing severe thromboembolic tendency undergoes the S-to-R transition and is associated with a plasma-inactive high-molecular-weight complex of aggregated antithrombin. Br J Haematol 1995, 89:589-601 [DOI] [PubMed] [Google Scholar]
- 36.Aulak KS, Eldering E, Hack CE, Lubbers YPT, Harrison RA, Mast A, Cicardi M, Davis AE: A hinge region mutation in C1-inhibitor (Ala436⇒Thr) results in nonsubstrate-like behavior and in polymerization of the molecule. J Biol Chem 1993, 268:18088-18094 [PubMed] [Google Scholar]
- 37.Faber J-P, Poller W, Olek K, Baumann U, Carlson J, Lindmark B, Eriksson S: The molecular basis of α1-antichymotrypsin deficiency in a heterozygote with liver and lung disease. J Hepatol 1993, 18:313-321 [DOI] [PubMed] [Google Scholar]