Abstract
The principal enzyme responsible for the β-site cleavage of amyloid precursor protein (APP) in the brain is a membrane-bound aspartyl protease β-site APP cleaving enzyme (BACE). We examined human APP (hAPP) and BACE mRNA expression by in situ hybridization in young and old hAPP transgenic mice from two lines: Tg2576, hAPP KM670–671NL (hAPPSw) at 4 and 15 months; and PDAPP, hAPP V717F, at 4 and 11 months. In transgene-positive mice from both lines, hAPP expression was most prominent in cortical, cerebellar, and hippocampal neuronal populations. Cingulate, entorhinal, and hippocampal amyloid burden in transgene-positive 16-month Tg2576 mice was 4 to 8%, and in 12-month PDAPP mice, 2 to 4%; there was no cerebellar amyloid deposition. BACE expression in transgenic and nontransgenic mice was highest in the cerebellar granule cell layer and hippocampal neuronal layers, intermediate in cortex, lower in subcortical regions, and minimal or absent in white matter of the cerebellum. Emulsion-dipped sections confirmed a predominantly neuronal pattern of expression. The amount of hybridization signal did not differ between transgenic and nontransgenic mice, or young and old mice, within each line. Thus, hAPP and endogenous BACE expression in similar anatomical localizations allow for processing of hAPP and Aβ formation in hAPP transgenic mice, but these are modified by additional age-related and anatomical factors.
Alzheimer’s disease is characterized pathologically by amyloid β protein (Aβ) deposition, neurofibrillary tangle formation, and neuronal loss in specific neuroanatomical regions. Transgenic mice expressing mutant human amyloid precursor protein (hAPP) have been developed as animal models of Alzheimer’s disease. 1 Several lines of hAPP transgenic mice develop cerebral amyloid deposits with aging, 2-7 including the PDAPP mouse expressing an hAPPV717F minigene under the human platelet-derived growth factor b-chain (PDGFb) promoter, 2 and the Tg2576 mouse expressing the 695-amino acid isoform of hAPP with the KM670–671NL Swedish double mutation (hAPPSw) under the hamster prion protein (PrP) promoter. 3 Two remarkable features of both of these hAPP transgenic mice are (i) that Aβ deposits occur only in aged animals, and (ii) that the Aβ deposits occur in a restricted set of characteristic locations in the cortex and hippocampus. The sites of amyloid deposition do not reflect the regional expression of either the hAPPSw or hAPPV717F transgenes, which are widely expressed in neurons throughout the brain. 8,9 Of interest, however, is that the anatomical pattern parallels the pattern seen in human Alzheimer’s disease, where amyloid plaques occur in a stereotyped distribution in the neocortex and hippocampus, including the outer molecular layer of the dentate gyrus. These results imply that other factors, in addition to hAPP expression, impact the location and age dependency of Aβ generation and deposition in hAPP transgenic mice.
Aβ is produced from proteolytic processing of APP by the action of γ- and β-secretases. Presenilin-1 is essential for the γ-secretase cleavage of APP. 10 Presenilin-1 is widely expressed in the human and mouse brain, overlapping with APP, but with highest expression in areas that do not develop Aβ deposits, such as the cerebellum. 11 Moreover, presenilin-1 mRNA levels are highest in the embryo, then decline markedly to remain stable with increasing age. 12,13 Thus, presenilin-1 expression patterns do not correlate well with the anatomical pattern or age relationship of Aβ deposition.
Recently, the enzyme responsible for the β-site cleavage of APP in brain has been identified as BACE. BACE is a 501-amino acid membrane-bound aspartyl protease with an acidic pH optimum, widely expressed in the brain, pancreas, and other tissues, 14-17 localized in neuronal cell bodies and proximal dendrites, 17 and colocalizing with Golgi and endosomal markers. 14,15 A homologous protein, BACE2, 18 may also cleave APP, 19 but is expressed in very low levels in the adult human and rat brain. 19,20 Because BACE is the principal β-secretase in neural tissues, we assessed BACE mRNA expression by in situ hybridization in the hAPP transgenic mouse models described above. We asked if the age and region dependence of Aβ deposition could be explained by patterns of BACE expression with age, or in brain regions susceptible to amyloid deposition; we also examined whether BACE expression was altered by overexpression of its substrate, hAPP, in transgenic mice.
Procedure
Transgenic Mice and Tissue Preparation
Tg2576 mice were bred from lines described previously. 3,9 The transgene is expressed in C57B6/SJL F1 mice backcrossed to C57B6/SJL breeders. Age-matched nontransgenic littermates served as controls. Three to six heterozygote transgenic and six nontransgenic mice were studied at ages of 4 and 15 months for in situ hybridization (total of 3 male and 6 female transgenic and 8 male and 4 female nontransgenic). Four from each group were studied at 16 months for amyloid burden, as published previously. 9
Heterozygous PDAPP transgenic mice were bred from the previously established line PDAPP-109 over several generations on hybrid backgrounds representing combinations of C57BL/6, DBA, and Swiss-Webster strains. 2,21,22 Four heterozygous transgenic and four nontransgenic littermates were studied at 4 months and 11 months of age (total of 4 male and 4 female transgenic mice, 4 male and 4 female nontransgenic mice). Four from each group were studied at 12 months for amyloid burden, as published previously. 8
Mice were sacrificed under CO2 and brains were removed and snap-frozen in isopentane chilled with dry ice. Twelve-micron sagittal (for Tg2576 mice) or 14-μm coronal (for PDAPP mice) cryostat sections from one hemisphere were thaw-mounted onto polylysine-coated glass slides and stored at −70°C.
In Situ Hybridization
In situ hybridization was performed according to previously published protocols. 8,9,23 Sections were fixed for 5 minutes in ice-cold 4% paraformaldehyde and stored in 95% ethanol at 4°C. Sections were hybridized overnight with [35S]adenosine (DuPont/NEN, Boston, MA) end-labeled 45-mer oligonucleotide sense and antisense probes (10,000 cpm/μl) for BACE (GenBank AF190726, nucleotides 881 to 925) and hAPP695 24 at 42°C in sealed chambers humidified with 50% formamide/4× standard sodium citrate (SSC) water, then washed in 1× SSC at 55°C. Slides were exposed to Amersham β-max autoradiography film (Amersham Pharmacia Biotech, Piscataway, NJ) for 1 to 7 days, then Amersham Hypermax β emulsion for 3 to 6 weeks, and counterstained with thioflavin-S (thio-s) and thionin.
Autoradiographic images of coronal sections were captured using a Bio-Rad (Hercules, CA) GS-700 Imaging Densitometer under maximal resolution (1200 dpi, pixel depth 12) for relative optical density measurement using Molecular Analyst software (Bio-Rad). Mean relative optical density in measurement frames traced over the brain region of interest was corrected for background. Hybridization with sense probes yielded no detectable signal.
Statistical Assessment
We used a mixed between (genotype and age)- and within (brain region)-subject analysis of variance to analyze mRNA signal (mean relative optical density) in Tg2576 and PDAPP mice (using the SAS GLM procedure). 25 Data were pooled for male and female animals. In the case of sagittal sections from the Tg2576 mice, brain regions were dentate gyrus, CA1, CA3, frontal cortex, posterior cortex, striatum, thalamus, cerebellar granule layer, and cerebellar molecular layer. In the coronal sections from the PDAPP mice, they were dentate gyrus, CA1, CA3, entorhinal cortex, cingulate, and thalamus. Degrees of freedom were adjusted 26 to correct for violation of analysis of variance assumptions of independent errors.
Results
Expression of hAPP mRNA in Tg2576 and PDAPP Mice
Human APP expression in Tg2576 mice and PDAPP mice was prominent in neurons, with strongest signal in cortical, cerebellar, and hippocampal neuronal populations (Figure 1, A and B) ▶ . Cortical transgene expression in the PDAPP mouse was most prominent in cingulate, retrosplenial, neocortical, and superficial piriform/entorhinal regions, with less signal in deep piriform/entorhinal cortex. No signal was detected in nontransgenic mice. Emulsion-dipped sections demonstrated neuronal expression of the transgene. 8,9 This is consistent with the neuron-specific promoters for these mouse lines.
Figure 1.
hAPP695 mRNA expression (A and B) and Aβ immunoreactivity (C and D) in Tg2576 (A and C) and PDAPP (B and D) transgenic mice. In situ hybridization in sagittal Tg2576 mouse sections (A) and coronal PDAPP mouse sections (B) demonstrate a neuronal pattern of transgene expression, especially in cortical and hippocampal neuron layers. Bi-3D6 Aβ immunoreactivity in 16-month Tg2576 (C) and 12-month PDAPP (D) mice demonstrate cortical (ctx) and hippocampal Aβ deposits, especially in dentate gyrus (dg) and CA1. Scale bars, 2 mm (A and B) and 500 μm (C and D).
Aβ Deposition in Tg2576 and PDAPP Mice
Amyloid deposition in heterozygous 16-month Tg2576 mice showed a characteristic anatomical pattern (Figure 1, C and D) ▶ . Amyloid burden (the percentage of surface area of an anatomical region covered by Aβ immunoreactivity) using the antibody bi-3D6 was 4% to 8% in cingulate cortex, entorhinal cortex, and hippocampus, and absent in thalamus, basal ganglia, and cerebellum. There were no amyloid deposits in 4-month Tg2576 mice. 9
Amyloid deposition in 12-month PDAPP mice showed essentially the same pattern. Amyloid burden was 2% to 4% in cingulate and entorhinal cortex and hippocampus. There were no amyloid deposits in thalamus, basal ganglia, and cerebellum. Four-month mice did not have any amyloid deposits. 8
Expression of BACE mRNA in Tg2576 and PDAPP Mice
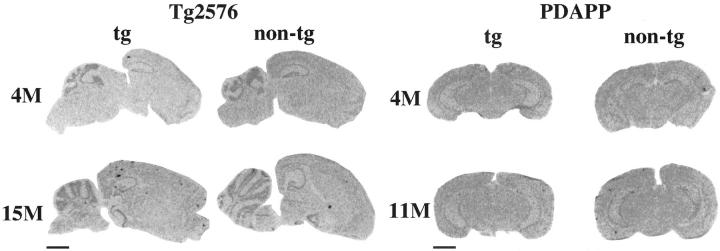
In transgenic and nontransgenic mice, the pattern of BACE expression was highest in the cerebellar granule cell layer and hippocampal neuronal layers (dentate gyrus granule cells, CA1 and CA3 pyramidal cell layer), with intermediate expression in the cortex, lower levels in the subcortical regions, and lowest in the white matter of the cerebellar molecular layer (Figures 2–4) ▶ ▶ ▶ . The intensity of BACE signal corresponded to neuronal density, with the highest signals in the highly packed pyramidal and granule cell layers of hippocampus and cerebellum. Emulsion-dipped sections confirmed the neuronal expression of BACE, with absent or minimal glial expression (Figure 3) ▶ . Cored plaques labeled by thio-S in the Tg2576 mice and PDAPP mice also bound the BACE mRNA probe. However, most of these plaques were not associated with any thionin-labeled cellular structures, apart from nearby neurons, suggesting nonspecific binding of the mRNA probe to compact plaques, although we cannot exclude specific binding to neuronal BACE mRNA trapped in plaques. 27
Figure 2.
BACE mRNA expression in young and old Tg2576 and PDAPP transgenic and nontransgenic mice demonstrates cortical, subcortical, and hippocampal BACE expression. Cored plaques also bind the probe (Scale bars, 2 mm).
Figure 3.
Emulsion-dipped BACE in situ hybridization with thionin counterstain demonstrates neuronal expression of BACE mRNA in 15-month transgenic Tg2576 mouse cortex (A) and 11-month transgenic PDAPP mouse CA3 (B). Scale bar, 10 μm.
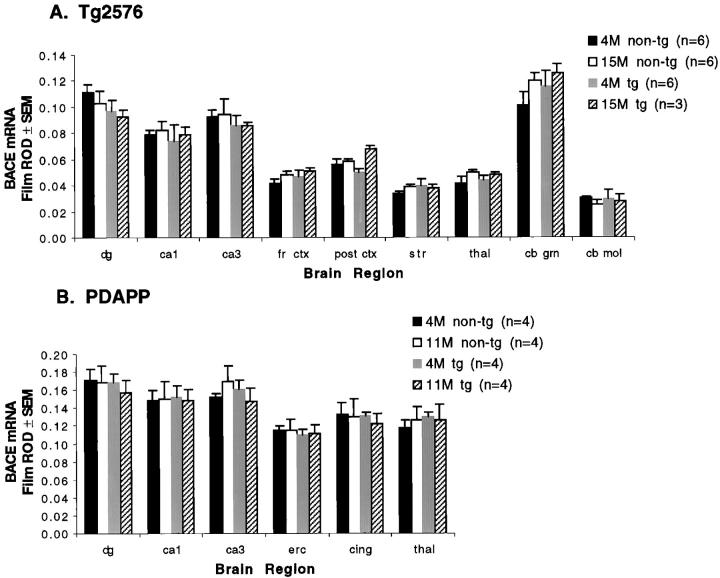
Figure 4.
Quantitation of regional BACE mRNA expression (film relative optical density (ROD) above background ± SEM) by in situ hybridization in Tg2576 (A) and PDAPP (B) mice. n = 3 to 6 in each group, as shown in legend. dg, dentate gyrus; fr ctx, frontal cortex; post ctx, posterior occipital cortex; str, striatum; thal, thalamus; cb grn, cerebellar granule cell layer; cb mol, cerebellar molecular layer; cing, cingulate cortex.
We compared the BACE pattern of expression to that seen for the hAPP transgene, to test the hypothesis that endogenous BACE acts on hAPP to generate Aβ. hAPP695 mRNA is more robustly expressed. The regional distribution of hAPP matches closely that of BACE, suggesting that hAPP and BACE are expressed in overlapping brain regions. Comparing this pattern of overlap to the pattern of Aβ deposition, however, suggests that although coexpression of hAPP and BACE is necessary, it is not sufficient for Aβ deposits. Coexpression of hAPP and BACE mRNA in hippocampus and cortex is associated with amyloid deposition in these regions, but prominent coexpression in the cerebellar granule cell layer does not result in cerebellar amyloid plaques.
The transgenic Tg2576 mice do not develop amyloid deposits until 9 to 11 months of age; the PDAPP mice do not develop them until 6 to 9 months of age. To determine whether alterations in BACE expression with hAPP expression or aging could contribute to the development of amyloid deposition in these mice, we compared BACE mRNA levels in transgenic and nontransgenic mice before amyloid deposition (4-month Tg2576 and PDAPP) and after amyloid deposition (15-month Tg2576 and 11-month PDAPP) (Figure 4) ▶ . In the case of analysis of variance for the Tg2576 mice as well as that for the PDAPP mice, the only significant effect found was a main effect for brain region (P < 0.0001), reflecting expected differences in BACE mRNA levels among regions. No significant genotype or age effects, or interactions involving them, were found. Thus, BACE hybridization signal did not differ between transgenic mice and nontransgenic mice, or with aging, and overexpression of hAPP did not alter BACE expression.
Discussion
The pattern of BACE expression closely matches the regional expression of the hAPP transgenes, demonstrating that both the hAPP and endogenous BACE are expressed in overlapping brain regions, and that the hAPP may therefore be susceptible to processing by endogenous BACE. Although BACE and hAPP expression are high in cortical and limbic areas that develop Aβ deposition, high expression is also seen in the cerebellum, which does not develop significant amyloid deposition, indicating that other regionally specific factors are necessary. We tested the hypothesis that the age and anatomical specificity of Aβ deposition in hAPP transgenic mice is related to overlap of hAPP and its processing enzymes. Our data show that this is not necessarily the case. The coexpression of APP and BACE supports a role for BACE in Aβ generation; however, it cannot account for the anatomical specificity and age dependence of Aβ deposition. Although it is difficult to extrapolate from mRNA levels to protein levels and enzyme functional activity, these results support previous studies looking at the β-cleaved metabolite of APP (APP-β) produced by BACE, which does not vary with age in the hippocampus, cortex, and cerebellum of transgenic PDAPP mice. 22 The BACE homologue, BACE2, may process APP, but it is only present in low levels in brain, and is responsible for a small proportion of β-secretase activity by cell culture relative to BACE. 18 Thus, hAPP and endogenous BACE expression in similar anatomical localizations allow for processing of APP and Aβ formation in these hAPP transgenic mice, but the amount, form, and location of Aβ deposition are modified by age and anatomical factors, as well as additional proteins such as presenilin and apolipoprotein E. 28-31
Acknowledgments
We thank Karen K. Hsiao-Ashe, M.D., Ph.D. (University of Minnesota) for kindly providing Tg2576 breeders, and Dora Games, Ph.D., and Dale Schenk, Ph.D. (Elan Pharmaceuticals) for generously providing PDAPP brain tissue.
Footnotes
Address reprint requests to Dr. Bradley T. Hyman, Alzheimer Disease Research Unit, Massachusetts General Hospital, East, 149 13th Street, Charlestown, MA 02129. E-mail: b_hyman@helix.mgh.harvard.edu.
Supported by National Institutes of Health grants AG00793 and AG05134 (to Massachusetts Alzheimer Disease Research Center).
References
- 1.Emilien G, Maloteaux JM, Beyreuther K, Masters CL: Alzheimer disease: mouse models pave the way for therapeutic opportunities. Arch Neurol 2000, 57:176-181 [DOI] [PubMed] [Google Scholar]
- 2.Games D, Adams D, Alessandrini R, Barbour R, Berthelette P, Blackwell C, Carr T, Clemens J, Donaldson T, Gillespie F, Guido T, Hagoplan S, Johnson-Wood K, Khan K, Lee M, Leibowitz P, Lieberburg I, Little S, Masliah E, McConlogue L, Montoya-Zavala M, Mucke L, Paganini L, Penniman E, Power M, Schenk D, Seubert P, Snyder B, Soriano F, Tan H, Vitale J, Wadsworth S, Wolozin B, Zhao J: Alzheimer-type neuropathology in transgenic mice overexpressing V717F β-amyloid precursor protein. Nature 1995, 373:523-527 [DOI] [PubMed] [Google Scholar]
- 3.Hsiao K, Chapman P, Nilsen S, Eckman C, Harigaya Y, Younkin S, Yang F, Cole G: Correlative memory deficits, Aβ elevation and amyloid plaques in transgenic mice. Science 1996, 274:99-102 [DOI] [PubMed] [Google Scholar]
- 4.Sturchler-Pierrat C, Abramowski D, Duke M, Wiederhold KH, Mistl C, Rothacher S, Ledermann B, Burki K, Frey P, Paganetti PA, Waridel C, Calhoun ME, Jucker M, Probst A, Staufenbiel M, Sommer B: Two amyloid precursor protein transgenic mouse models with Alzheimer disease-like pathology. Proc Natl Acad Sci USA 1997, 94:13287-13292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borchelt DR, Ratovitski T, van Lare J, Lee MK, Gonzales V, Jenkins NA, Copeland NG, Price DL, Sisodia SS: Accelerated amyloid deposition in the brains of transgenic mice coexpressing mutant presenilin 1 and amyloid precursor proteins. Neuron 1997, 19:939-945 [DOI] [PubMed] [Google Scholar]
- 6.Moechars D, Dewachter I, Lorent K, Reverse D, Baekelandt V, Naidu A, Tesseur I, Spittaels K, Haute CV, Checler F, Godaux E, Cordell B, Van Leuven F: Early phenotypic changes in transgenic mice that overexpress different mutants of amyloid precursor protein in brain. J Biol Chem 1999, 274:6483-6492 [DOI] [PubMed] [Google Scholar]
- 7.Lamb BA, Bardel KA, Kulnane LS, Anderson JJ, Holtz G, Wagner SL, Sisodia SS, Hoeger EJ: Amyloid production and deposition in mutant amyloid precursor protein and presenilin-1 yeast artificial chromosome transgenic mice. Nat Neurosci 1999, 2:695-697 [DOI] [PubMed] [Google Scholar]
- 8.Irizarry MC, Soriano F, McNamara M, Page KJ, Schenk D, Games D, Hyman BT: Aβ deposition is associated with neuropil changes, but not with overt neuronal loss in the PDAPP transgenic mice. J Neurosci 1997, 17:7053-7059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Irizarry MC, McNamara M, Fedorchak K, Hsiao K, Hyman BT: APP-Sw transgenic mice develop age related amyloid deposits and neuropil abnormalities, but no neuronal loss in CA1. J Neuropathol Exp Neurol 1997, 56:965-973 [DOI] [PubMed] [Google Scholar]
- 10.Wolfe MS, Xia W, Ostaszewski BL, Diehl TS, Kimberly WT, Selkoe DJ: Two transmembrane aspartates in presenilin-1 required for presenilin endoproteolysis and gamma-secretase activity. Nature 1999, 398:513-517 [DOI] [PubMed] [Google Scholar]
- 11.Page K, Hollister R, Tanzi RE, Hyman BT: In situ hybridization analysis of presenilin 1 mRNA in Alzheimer’s disease and in lesioned rat brain. Proc Natl Acad Sci USA 1996, 93:14020-14024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berezovska O, Xia MQ, Page K, Wasco W, Tanzi RE, Hyman BT: Developmental regulation of presenilin mRNA expression parallels notch expression. J Neuropathol Exp Neurol 1997, 56:40-44 [DOI] [PubMed] [Google Scholar]
- 13.Flood FM, Cowburn RF, Johnston JA: Presenilin-1, amyloid precursor protein and amyloid precursor-like protein 2 mRNA levels in human superior frontal cortex during aging. Neurosci Lett 1997, 235:17-20 [DOI] [PubMed] [Google Scholar]
- 14.Vassar R, Bennett BD, Babu-Khan S, Kahn S, Mendiaz EA, Denis P, Teplow DB, Ross S, Amarante P, Loeloff R, Luo Y, Fisher S, Fuller J, Edenson S, Lile J, Jarosinski MA, Biere AL, Curran E, Burgess T, Louis JC, Collins F, Treanor J, Rogers G, Citron M: Beta-secretase cleavage of Alzheimer’s amyloid precursor protein by the transmembrane aspartic protease BACE. Science 1999, 286:735-741 [DOI] [PubMed] [Google Scholar]
- 15.Yan R, Bienkowski MJ, Shuck ME, Miao H, Tory MC, Pauley AM, Brashier JR, Stratman NC, Mathews WR, Buhl AE, Carter DB, Tomasselli AG, Parodi LA, Heinrikson RL, Gurney ME: Membrane-anchored aspartyl protease with Alzheimer’s disease beta-secretase activity. Nature 1999, 402:533-537 [DOI] [PubMed] [Google Scholar]
- 16.Sinha S, Anderson JP, Barbour R, Basi GS, Caccavello R, Davis D, Doan M, Dovey HF, Frigon N, Hong J, Jacobson-Croak K, Jewett N, Keim P, Knops J, Lieberburg I, Power M, Tan H, Tatsuno G, Tung J, Schenk D, Seubert P, Suomensaari SM, Wang S, Walker D, John V: Purification and cloning of amyloid precursor protein beta-secretase from human brain. Nature 1999, 402:537-540 [DOI] [PubMed] [Google Scholar]
- 17.Hussain I, Powell D, Howlett DR, Tew DG, Meek TD, Chapman C, Gloger IS, Murphy KE, Southan CD, Ryan DM, Smith TS, Simmons DL, Walsh FS, Dingwall C, Christie G: Identification of a novel aspartic protease (Asp 2) as beta-secretase. Mol Cell Neurosci 1999, 14:419-427 [DOI] [PubMed] [Google Scholar]
- 18.Saunders AJ, Kim T-W, Tanzi RE, Fan W, Bennet BD, Babu-Kahn S, Luo Y, Louis J-C, McCaleb M, Citron M, Vassar R, Richards WG: BACE maps to chromosome 11 and a BACE homolog, BACE2, reside in the obligate Down syndrome region of chromosome 21. Science 1999, 286:1255 [Google Scholar]
- 19.Farzan M, Schnitzler CE, Vasilieva N, Leung D, Choe H: BACE2, a β-secretase homologue, cleaves at the β site and within the amyloid-β region of the amyloid-β precursor protein. Proc Nat Acad Sci USA 2000, 97:9712-9717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bennett BD, Babu-Khan S, Loeloff R, Louis J-C, Curran E, Citron M, Vassar R: Expression analysis of BACE2 in brain and peripheral tissue. J Biol Chem 2000, 275:20647-20651 [DOI] [PubMed] [Google Scholar]
- 21.Masliah E, Sisk A, Mallory M, Mucke L, Schenk D, Games D: Comparison of neurodegenerative pathology in transgenic mice overexpressing V717F β-amyloid precursor protein and Alzheimer’s disease. J Neurosci 1996, 16:5795-5811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson-Wood K, Lee M, Motter R, Hu R, Gordon G, Barbour R, Khan K, Gordon M, Tan H, Games D, Lieberberg I, Schenk D, Seubert P, McConologue L: Amyloid precursor protein processing and Aβ42 deposition in a transgenic mouse model of Alzheimer’s disease. Proc Natl Acad Sci USA 1997, 94:1550-1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sirinathsinghji DJS, Dunnett SB: Imaging gene expression in neural grafts. Sharif NA eds. Molecular Imaging in Neuroscience: A Practical Approach. 1993, :pp 43-70 Oxford University Press, New York [Google Scholar]
- 24.Sola C, Mengod G, Probst A, Palacios JM: Differential regional and cellular distribution of beta-amyloid precursor protein messenger RNAs containing and lacking the Kunitz protease inhibitor domain in the brain of human, rat and mouse. Neuroscience 1993, 53:267-295 [DOI] [PubMed] [Google Scholar]
- 25.The GLM procedure. SAS/STAT User’s Guide, Volume 2. Version 6, ed 4. Cary, NC, SAS Institute Inc., 1990, pp 891–996
- 26.Greenhouse SW, Geisser S: On methods in the analysis of profile data. Psychometrika 1959, 32:95-112 [Google Scholar]
- 27.Ginsberg SD, Crino PB, Hemby SE, Weingarten JA, Lee VM, Eberwine JH, Trojanowski JQ: Predominance of neuronal mRNAs in individual Alzheimer’s disease senile plaques. Ann Neurol 1999, 45:174-181 [PubMed] [Google Scholar]
- 28.Bales K, Verina T, Dodel RC, Du Y, Altstiel L, Bender M, Hyslop P, Johnstone EM, Little SP, Cummins DJ, Piccardo P, Ghetti B, Paul SM: Lack of apolipoprotein E dramatically reduces amyloid β-protein deposition. Nat Genet 1997, 17:263-265 [DOI] [PubMed] [Google Scholar]
- 29.Holcomb L, Gordon MN, McGowan E, Yu X, Benkovic S, Jantzen P, Wright K, Saad I, Mueller R, Morgan D, Sanders S, Zehr C, O’Campo K, Hardy J, Prada CM, Eckman C, Younkin S, Hsiao K, Duff K: Accelerated Alzheimer-type phenotype in transgenic mice carrying both mutant amyloid precursor protein and presenilin 1 transgenes. Nat Med 1998, 4:97-100 [DOI] [PubMed] [Google Scholar]
- 30.Holtzman DM, Fagan AM, Mackey B, Tenkova T, Sartorius L, Paul S, Bales K, Hsiao-Ashe KK, Irizarry MC, Hyman BT: ApoE is required for the development of neuritic and cerebrovascular plaques in the APPsw model of Alzheimer’s disease. Ann Neurol 2000, 47:739-747 [PubMed] [Google Scholar]
- 31.Irizarry MC, Cheung BS, Rebeck GW, Paul SM, Bales KR, Hyman BT: Apolipoprotein E affects the amount, form, and anatomical distribution of Aβ deposition in homozygous APPV717F transgenic mice. Acta Neuropathol 2000, 100:451-458 [DOI] [PubMed] [Google Scholar]