Abstract
No objective parameters have been found so far that can predict the biological behavior of early stages of prostatic cancer, which are encountered frequently nowadays due to surveillance and screening programs. We have applied comparative genomic hybridization to routinely processed, paraffin-embedded radical prostatectomy specimens derived from patients who participated in the European Randomized Study of Screening for Prostate Cancer. We defined a panel consisting of 36 early cancer specimens: 13 small (total tumor volume (Tv) < 0.5 ml) carcinomas and 23 intermediate (Tv between 0.5–1.0 ml) tumors. These samples were compared with a set of 16 locally advanced, large (Tv > 2.0 ml) tumor samples, not derived from the European Randomized Study of Screening for Prostate Cancer. Chromosome arms that frequently (ie, ≥15%) showed loss in the small tumors included 13q (31%), 6q (23%), and Y (15%), whereas frequent (ie, ≥15%) gain was seen of 20q (15%). In the intermediate cancers, loss was detected of 8p (35%), 16q (30%), 5q (26%), Y (22%), 6q, and 18q (both 17%). No consistent gains were found in this group. In the large tumors, loss was seen of 13q (69%), 8p (50%), 5q, 6q (both 31%), and Y (15%). Gains were observed of 8q (37%), 3q (25%), 7p, 7q, 9q, and Xq (all 19%). Comparison of these early, localized tumors with large adenocarcinomas showed a significant increase in the number of aberrant chromosomes per case (Rs = 0.36, P = 0.009). The same was true for the number of lost or gained chromosomes per case (Rs = 0.27, P = 0.05; Rs = 0.48, respectively; P < 0.001). Interestingly, chromosomal alterations that were found in previous studies to be potential biomarkers for tumor aggressiveness, ie, gain of 7pq and/or 8q, were already distinguished in the small and intermediate cancers. In conclusion, our data show that chromosomal losses, more specifically of 6q and 13q, are early events in prostatic tumorigenesis, whereas chromosomal gains, especially of 8q, appear to be late events in prostatic tumor development. Finally, early localized tumors, as detected by screening programs, harbor cancers with aggressive genetic characteristics.
The incidence of prostate cancer has dramatically increased during the last two decades. It is now the most commonly diagnosed noncutaneous malignancy in men in Western countries with a high socio-economic standard, and its mortality is surpassed only by that of lung cancer. 1 The exponential increase in incidence, which peaked in the early 1990s, 2 has been largely attributed to the increased use of prostate-specific antigen (PSA) in prostate carcinoma detection and population-based screening programs, rather than considered a true increase in incidence. 3,4 These surveillance and early detection programs have led to an increasing number of patients being diagnosed with early, clinically localized prostate carcinoma. 2,4,5 On the one hand, patients with these tumors likely benefit from curative treatment and mostly have a good prognosis after therapy. 6,7 On the other hand, with increased detection comes the risk of finding small, organ-confined, well differentiated tumors, which may pose relatively little threat to the patient. 6-10 The latter category of cancers might best be left untreated, especially in older patients, 7,11 if they could be clearly identified on a pretreatment basis. However, the available diagnostic tools fail to provide consistent predictive information for firm clinical decisionmaking in individual cases. 7 The analysis of molecular (cyto)genetic changes associated with the initiation and progression of prostate cancer may enable us to establish accurate methods of prognostication when the disease is encountered in its earliest stages of development.
Conventional cytogenetic studies of prostatic adenocarcinoma have consistently revealed loss of the Y chromosome, trisomy of chromosome 7, del(7)(q22), del(8)(p21), and del(10)(q24). 12 Loss of heterozygosity (LOH) analyses have shown frequent loss on chromosome arms 3p, 6q, 7q, 8p, 9p, 10pq, 13q, 16q, 17q, and 18q. 13-24 Furthermore, comparative genomic hybridization (CGH) applied to primary tumors revealed losses of 5q, 6q, 8p, and 13q, as well as gain of 8q in over 30% of cases. 25,26 CGH studies of advanced stages of prostate cancer detected frequent (≥50%) loss of 5q, 6q, 8p, 10q, 13q, 16q, and 17p, as well as gain of 1q, 3q, 7pq, 8q, 11p, 17q, and Xpq sequences. 25-28 Fluorescent in situ hybridization studies revealed numerical alterations of chromosomes 7, 8, 10, 16, 17, 18, X, and Y, 12,29 as well as deletions and amplifications of specific chromosomal regions, eg, loss of 8p22 30 and gain of MYC on 8q24. 31 Furthermore, we 25,32 and others 33,34 have identified alterations of chromosomes 7 and/or 8 as potential tumor progression markers. Altogether, these molecular (cyto)- genetic studies have identified multiple, non-random genetic alterations in prostate cancer. However, at present, knowledge concerning the initial stages of prostate cancer is very limited.
In the present study we have applied CGH to a unique panel of archival tumor material obtained from patients who participated in the European Randomized Study of Screening for Prostate Cancer (ERSPC). 35 To the best of our knowledge, this is the first study to present molecular cytogenetic data concerning tumors derived from a population-based screening program. Our goals were (i) to obtain an overview of chromosomal alterations occurring in these early, localized prostate cancers; (ii) to compare the molecular cytogenetic characteristics of these tumors with those of larger, clinically apparent prostate cancers; and (iii) to see whether early cancers, as detected by screening programs, harbor tumors with aggressive genetic features.
Materials and Methods
Patient Material
The original screening algorithm of the screening arm of the Rotterdam section of the ERSPC called for a biopsy in all men who had at least one of the following results: a suspicious digital rectal examination (DRE), a suspicious transrectal ultrasound (TRUS), or a PSA level ≥4.0 ng/ml. In March 1997, a major protocol change was implemented within the ERSPC, when the study group decided to perform a biopsy on all men with PSA levels ≥3.0 ng/ml, irrespective of findings on DRE or TRUS. DRE and TRUS were omitted as a screening tool if an individual’s PSA level was <3.0 ng/ml. 35 Approximately half of the patients with a positive biopsy were treated by radical prostatectomy. We collected formalin-fixed, paraffin-embedded materials, obtained between 1994 and 1998, derived from radical prostatectomies of 36 patients participating in the first round of the ERSPC. These samples were compared with a set of 16 archival, clinically apparent tumors, obtained between 1990 and 1992 and not derived from the ERSPC study.
After fixation, each specimen was step-sectioned at 4-mm intervals and totally embedded in paraffin blocks as described previously. 36 From each paraffin block, standard hematoxylin-eosin-stained slides were prepared for routine histopathological examination, including determination of pathological TNM stage 37 and Gleason score 38 (see Table 1 ▶ ). Tumor volumes were measured as reported before. 36 Briefly, after histological examination, all areas containing cancer were outlined on the slides. Digital morphometric analysis (Kontron Imaging System, model KS 400; Kontron Elektronik GmbH, Eching, Germany; ERSPC cancers) and/or a microscopic grid method (non-ERSPC cancers) was performed to measure each tumor area . The two methods correlated excellently (Rs = 1.00; P < 0.001). We determined the total tumor volume (Tv) by adding all measured tumor areas (in mm2) and multiplying them by 4 (ie, the thickness, in millimeters, of the original slices). On the basis of their total tumor volume, our set of 52 tumors (all peripheral zone adenocarcinomas) was arbitrarily divided into three categories: 13 small (Tv < 0.5 ml) carcinomas, 23 intermediate (Tv between 0.5–1.0 ml) tumors, and 16 large (Tv > 2.0 ml) tumor samples (Table 1) ▶ . The subdivision of the early cancers in small and intermediate tumors was based partially on data from literature, in which it was reported that localized (pT2) tumors with a Tv <0.5 ml and lacking a Gleason growth pattern of 4 or 5 may be considered insignificant and/or minimal 9,36 or clinically unimportant. 10
Table 1.
Clinicopathological Data and Results of CGH Analysis
| Case | Age (years) | Pre- operative PSA (ng/ml) | Pre- operative clinical stage T* | pT stage | Tumor volume (ml) | Gleason score | CGH loss | CGH gain |
|---|---|---|---|---|---|---|---|---|
| 1 | 57 | 2.2 | T2a | pT2a | 0.04 | 6 | 8p11.2-p22 | |
| 2 | 57 | 2.1 | T2a | pT2b | 0.12 | 6 | ||
| 3 | 62 | 2.4 | T2b | pT2b | 0.18 | 5 | ||
| 4 | 56 | 5.6 | T2a | pT2a | 0.20 | 7 | ||
| 5 | 69 | 6.5 | T3a | pT2b | 0.21 | 6 | ||
| 6 | 61 | 1.2 | T2a | pT2b | 0.23 | 6 | 4q27, 6p21.3-p23, 6q14-q21, 7p13-p15, 7q11.2, 13q12-q22, Y | |
| 7 | 58 | 1.2 | T2a | pT2b | 0.32 | 5 | 13q14-q31 | |
| 8 | 56 | 1.3 | T2x | pT2a | 0.35 | 6 | 5q11.2, 5q15-q22, 6q12-q21, Y | 3q13.2-q22, 4p15.3-pter, 8q23-qter, 20q13.1-q13.2 |
| 9 | 66 | 13.5 | T2a | pT2b | 0.36 | 6 | 6q21-q22, 13q14-q21 | |
| 10 | 65 | 4.1 | T1c | pT2a | 0.38 | 6 | ||
| 11 | 62 | 3.6 | T2x | pT2b | 0.38 | 6 | 9q31-qter, 17q23-qter, 20q12 | |
| 12 | 61 | 0.4 | T2a | pT2b | 0.43 | 6 | ||
| 13 | 57 | 0.9 | T2a | pT2a | 0.46 | 6 | 3q13.1-q21, 13q21-q22 | 16p11.2-pter |
| 14 | 62 | 3.0 | T2x | pT2b | 0.50 | 7 | 3p24-pter, 5q14-q23, 5q31, 6q15-q21, 8p11.2-p22, 18q21-q22 | |
| 15 | 66 | 9.1 | T2a | pT2b | 0.51 | 6 | 5p13-pter, 5q12-q13 | |
| 16 | 59 | 7.2 | T1c | pT2a | 0.52 | 6 | 6q22-q25, 16q13-q23 | |
| 17 | 62 | 8.0 | n.a. | pT2b | 0.53 | 7 | 9p21-p23, 11p13-p14 | |
| 18 | 64 | 3.9 | T3x | pT2b | 0.55 | 7 | 2q21-q31, 8p11.2-p21, 11q14-q22, 12p11.2, Y | 5p12-pter |
| 19 | 58 | 4.7 | T1c | pT2a | 0.55 | 6 | 6q15-q22, 16q12.1-q23, 18q21, Xq21-q26, Y | 8q11.2-qter |
| 20 | 64 | 1.6 | T1c† | pT2b | 0.58 | 6 | 4p12-p15.3, 4q12-q21, 16q22-qter | |
| 21 | 70 | 6.4 | T2a | pT2b | 0.59 | 7 | ||
| 22‡ | 64 | 3.9 | n.a. | pT2b | 0.61 | 7 | 16q12.2-qter | |
| 23 | 62 | 1.5 | T2b | pT2b | 0.61 | 5 | 5q12-q21, 9p13-p22, 12p11.2-p12, 13q21 | |
| 24 | 66 | 2.9 | T2a | pT2b | 0.63 | 7 | 5q14-q21, 6q16-q21, 8p12-p22, 18q21, Y | #7 |
| 25 | 65 | 13.0 | T1c | pT2b | 0.63 | 7 | 8p11.2-p23, 16p12-p13.1, 18q11.2-qter, Y | 8q21.1-q22, 12p12 |
| 26 | 60 | 3.7 | T2a | pT2b | 0.64 | 7 | 5p15.1, 5q14-q23, 11q14-q24, 14q24-q31, 16q21-qter | 3q13.3-q21 |
| 27 | 70 | 2.8 | T1c§ | pT2b | 0.69 | 7 | 3p13-p21, 8p12-p21, 16q22-q23 | 3q24, 4q24-q26, 4q32, 7p13-p14, 9q21 |
| 28 | 69 | 4.4 | T2a | pT2b | 0.70 | 6 | 12p13, 17p11.2-pter | |
| 29‡ | 60 | 4.8 | T1c | pT2b | 0.77 | 5 | 10p11.2-p13 | |
| 30 | 66 | 10.6 | T3a | pT2a | 0.78 | 7 | 2q13-q22, 8p11.2-p21 | 7q11.2-q21 |
| 31‡ | 71 | 4.7 | T2a | pT2b | 0.83 | 7 | 5q23-q31, 8p21-pter, 16q23 | 8q11.2-qter |
| 32 | 63 | 3.6 | T2a | pT2b | 0.84 | 7 | 2q14.2-q21, 13q14-q21, Y | |
| 33 | 57 | 4.1 | T2a | pT2b | 0.84 | 6 | Xp11.2-p22.1 | |
| 34 | 73 | 8.3 | T2a | pT2b | 0.85 | 7 | ||
| 35‡ | 70 | 5.8 | T1c | pT2b | 0.85 | 5 | 8p11.2-p22 | |
| 36 | 57 | 4.6 | T1c | pT2b | 0.90 | 5 | ||
| 37 | 59 | 17.2 | T3x | pT2b | 2.1 | 7 | 8p11.2-pter, 12p11.2-pter | |
| 38 | 51 | 16.1 | T2a | pT2a | 2.2 | 7 | 13q21 | |
| 39 | 66 | 5.0 | T2b | pT3a | 2.3 | 7 | 13q14-q21 | 3p12-pter, 3q13.3-qter |
| 40 | 47 | 11.2 | T3x | pT3a | 2.3 | 7 | 5q12-q33, 8p21-pter, 9p13-pter, 13q21-qter, Y | 3q21 |
| 41 | 63 | 4.5 | T3x | pT3a | 2.4 | 5 | 2q21-q22, 4q22-q27, 6q13-q22, 13q13-q31 | 5q34-qter, 9q32-qter |
| 42 | 66 | 5.8 | T2a | pT2b | 2.4 | 5 | 8p11.2-p22, 16q22-q23 | |
| 43 | 53 | 16.5 | T3x | pT3a | 4.2 | 7 | 6q12-q25, 13q13-q21 | # X |
| 44 | 60 | 32.9 | T3x | pT3x | 6.0 | 10 | 6q12-q16, 8p11.2-pter, 13q12-qter, 16q12.1-qter, 18q11.2-qter, 22q11.2 | 1q21-qter, 1p31-pter, 1p13, 3p24-pter, 3p13-p21, 7q11.2-q32, 8q21-qter, 9p23-p24, 9q13-qter, 11q12-qter |
| 45 | 63 | 18.7 | T3x | pT4 | 6.9 | 7 | 5q15-q23, 6q13-q22, 8p11.2-pter, 11q14-q23, 12q15-q21, 13q13-q21 | 3q12-qter, 7p11.2-pter, 7q11.2-q31, 8q24.1-qter |
| 46 | 55 | 13.2 | T2a | pT4 | 7.0 | 7 | 5q11.2, 13q21, Y | 2q35-qter, 8q24.1-qter, 9q32-qter, Xq27-qter |
| 47 | 51 | 2.8 | T2a | pT4 | 7.2 | 6 | 1p13-p31, 3q24-q25, 4q23-q28, 5q14-q21, 13q14-q31 | 6p21.1-p21.3, 7p21-pter, 14q24.3-qter, 20q11.2-qter |
| 48 | 57 | 12.4 | T2a | pT4 | 9.8 | 7 | 4q12-q13, # X | |
| 49 | 65 | 181.4 | T2a | pT4 | 12.5 | 9 | ||
| 50 | 58 | 108 | T2b | pT4 | 14.8 | 5 | 8p11.2-p22, 13q13-q21, Y | 8q22-qter |
| 51 | 64 | 5.9 | T4x | pT4 | 15.7 | 9 | 6q11.2-q24, 8p11.2-pter, 13q14-qter | 2q32-qter, 6p11.2-pter, 8q23-qter, 17q23-qter |
| 52 | 67 | 32 | T3x | pT4 | 21.6 | 9 | 5q14-q21, 8p11.2-pter, 12p11.2-pter | 1q23-q31, 3q26.1-q27, 7p11.2-pter, 7q21, 7q32-qter, 8q12-qter |
* Staging of localized prostatic adenocarcinoma (all N0M0) according to the clinical and pathological (pT) TNM classification 1997 36 T1c, clinically inapparent tumor, not palpable nor visible by imaging, identified by needle biopsy (e.g. because of elevated PSA); T2, tumor confined to prostate, involving 1 lobe (T2a), or both lobes (T2b); T3 tumor extends through prostatic capsule (T3a) or tumor invades seminal vesicle(s) (T3b); T4, tumor is fixed or invades adjacent structures other than seminal vesicles, including microscopic invasion of bladder wall; Tx primary tumor cannot be assessed.
† PSA at time of biopsy 3.1 ng/ml.
‡ These patients showed biochemical progression in follow-up.
§ PSA at time of biopsy 5.5 ng/ml. n.a., not available.
CGH of Archival Material
Isolation of DNA from the formalin-fixed, paraffin-embedded tumor material was performed as described by Alers et al. 32 Briefly, the tissue blocks were counterstained in 4′,6-diamidino-2-phenylindole (DAPI) and placed under a fluorescence microscope, enabling a precise selection of the tumor area. Microdissection of the tumor areas was performed using a hollow bore coupled to the microscope. In cases of very small tumor areas, manual microdissection of the selected areas was performed by scraping successive hematoxylin-stained 10-μm tissue sections using a hollow needle under a stereo microscope. Lower boundaries were checked for the presence of tumor on 4-μm hematoxylin-eosin-stained tissue sections. Isolation of DNA from the formalin-fixed, paraffin-embedded material was performed using the Puregene DNA isolation kit (Gentra Systems, Minneapolis, MN). Tumor DNA with a fragment size <1 kb was chemically labeled with biotin-universal linkage system (Kreatech Diagnostics, Amsterdam, The Netherlands). 39 Tumor DNA with larger DNA fragment sizes was labeled with biotin by nick translation (Nick Translation System, Gibco BRL, Gaithersburg, MD). Likewise, male reference DNA (Promega, Madison, WI) was labeled by nick translation with digoxigenin (Boehringer Mannheim, Indianapolis, IN). The reaction time and the amount of DNase were adjusted to obtain a matching probe size for reference and tumor DNAs. The labeled DNAs were hybridized onto normal male metaphase chromosomes (Vysis Inc., Downers Grove, IL), as described previously. 25,32,40 CGH analysis was accomplished with Quips XL software from Vysis (version 3.1.1). Loss of DNA sequences was defined as chromosomal regions where the mean green:red ratio was below 0.85, whereas gain was defined as chromosomal regions where the ratio was above 1.15. These threshold values were based on series of normal controls. Some chromosomal alterations, such as simultaneous gains of chromosome 1pter, 9q34, 11q13, #19, and 22q, were disregarded and excluded from analysis, since these areas are known to present variation in normal controls. 41 In contrast to some other studies, we have included genomic imbalances of the Y chromosome as we have confirmed CGH data by in situ hybridization results with a chromosome Y-specific DNA probe.
Statistical Analysis
Percentages between groups were compared using Fisher’s exact test or the χ 2 test for trend if indicated. Comparison between percentages of chromosomal alterations between groups were only performed if the alteration occurred in >10% of total cases. Comparison of the average number of aberrations and clinical stage was performed using the Mann-Whitney U test. Correlation coefficients (Rs) given are Spearman’s. Multivariate analysis using multiple regression was performed to investigate which of the clinicopathological parameters, ie, tumor volume, pathological tumor stage, and pathological grade, played a dominant role regarding the genetic changes found. In this analysis, Tv and the dependent variable, ie, the number of aberrant, lost, or gained chromosomes per case, had to be transformed logarithmically to reduce skewness of the distributions. P = 0.05 (two-sided) was considered the limit of significance.
Results
Overview of Genetic Changes
Clinicopathological data and results of CGH analysis of patients with different tumor volumes, ie, 13 small tumors (Tv < 0.5 ml), 23 intermediate tumors (Tv 0.5–1.0 ml; all 36 cases derived from ERSPC), and 16 large (Tv > 2.0 ml tumors; not derived from ERSPC), are summarized in Table 1 ▶ and Figure 1 ▶ .
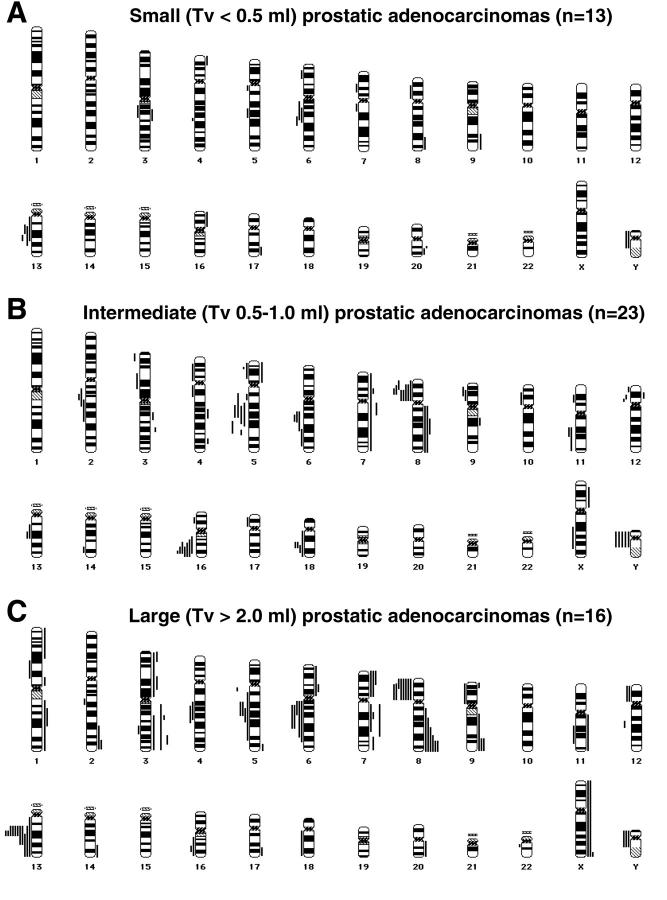
Figure 1.
Chromosomal ideograms showing the summary of DNA copy number changes, detected by CGH, in tumors of 52 patients with different tumor volumes. Losses are displayed on the left of the ideogram, gains are shown on the right. A: Small cancers (Tv < 0.5 ml; n = 13), showing loss of 6q, 13q, and Y, as well as gain of 20q. B: Intermediate cancers (Tv between 0.5–1.0 ml; n = 23), displaying loss of 5q, 6q, 8p, 16q, 18q, and Y. C: Large cancers (Tv > 2.0 ml; n = 16), revealing recurrent loss of 5q, 6q, 8p, 13q, and Y and frequent gain of 3q, 7pq, 8q, 9q, and Xq sequences.
In small carcinomas, chromosomal losses were seen repetitively of 13q (31%), 6q (23%), and Y (15%), whereas gain of 20q was observed in 15% of cases (Figure 1A) ▶ . In the intermediate tumors losses were most often detected of 8p (35%), 16q (30%), 5q (26%), Y (22%), 6q, and 18q sequences (both 17%), whereas no recurrent gains were seen (Figure 1B) ▶ . However, gain of 7pq and/or gain of 8q sequences, potential biomarkers for tumor aggressiveness identified in previous studies, 25,32-34 were already encountered in 19% of the early (ie, small and intermediate combined) cancers. In the large cancers, frequent loss was observed of 13q (69%), 8p (50%), 5q, 6q (both 31%), and Y (15%; Figure 1C ▶ ). Gains most often involved 8q (37%), 3q (25%), 7p, 7q, 9q, and Xq (all 19%; Figure 1C ▶ ).
Genetic Changes and Tumor Volume
The results of CGH analysis of the three different tumor volume groups are presented in Figure 2 ▶ , whereas statistical correlations between different clinicopathological parameters and chromosomal alterations, as detected by CGH, are depicted in Table 2 ▶ .
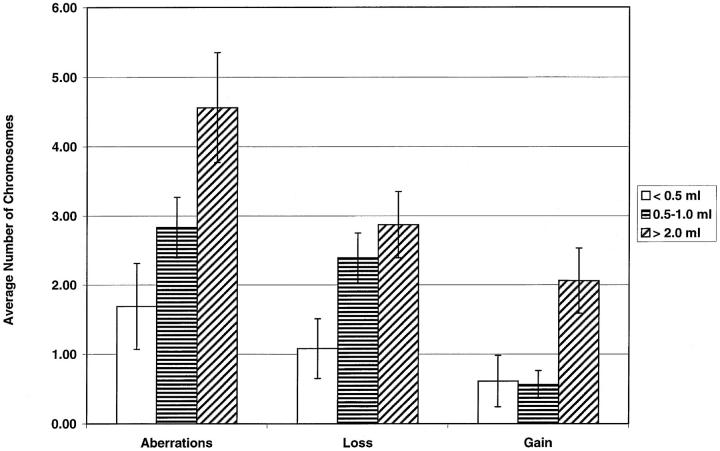
Figure 2.
Comparison of the chromosomal aberrations between small, intermediate, and large prostate cancers, showing the average number (± SEM) of altered chromosomes per patient. The average number of aberrant chromosomes per patient significantly increased with increasing exact tumor volume (Rs = 0.36, P = 0.009). The same was true for the number of chromosomes showing loss or, especially, gain per case (Rs = 0.27, P = 0.05; Rs = 0.48, respectively; P < 0.001).
Table 2.
Statistical Correlations between Clinicopathological Parameters and Genetic Changes as Detected by CGH
| Tumor volume | pT stage | Pathological grade* | |
|---|---|---|---|
| Number of cases with aberrations | Ptrend = 0.009 | Ptrend = 0.3, n.s. | Ptrend = 0.6, n.s. |
| Number of cases with loss | Ptrend = 0.01 | Ptrend = 0.6, n.s. | Ptrend = 0.6, n.s. |
| Number of cases with gain | Ptrend = 0.005 | Ptrend < 0.001 | Ptrend = 0.02 |
| Average number of aberrant chromosomes per case | Rs = 0.36, P = 0.009 | Rs = 0.41, P = 0.002 | Rs = 0.28, P = 0.04 |
| Average number of chromosomes with loss per case | Rs = 0.27, P = 0.05 | Rs = 0.27, P = 0.05 | Rs = 0.23, P = 0.1, n.s. |
| Average number of chromosomes with gain per case | Rs = 0.48, P < 0.001 | Rs = 0.59, P < 0.001 | Rs = 0.36, P = 0.008 |
| Number of cases with 8p loss | Ptrend = 0.02 | Ptrend = 0.2, n.s. | Ptrend = 0.03 |
| Number of cases with 8q gain | Ptrend = 0.04 | Ptrend = 0.001 | Ptrend = 0.03 |
| Number of cases with gain of 7pq and/or 8q | Ptrend = 0.03 | Ptrend = 0.002 | Ptrend = 0.02 |
n.s., not statistically significant.
*Gleason score.
The number of cases showing a chromosomal aberration significantly increased, from 54% in small cancers to 87% in intermediate tumors to 94% in large cancers (Ptrend = 0.009). Also, the number of cases showing loss or gain was higher in larger tumors (Ptrend = 0.01 and Ptrend = 0.005, respectively; Table 2 ▶ ). Furthermore, the average number of aberrant chromosomes per case increased along with the exact tumor volume (Table 2 ▶ , Figure 2 ▶ ; Rs = 0.36, P = 0.009). The same was true for the number of chromosomes showing loss or gain per case (Table 2 ▶ , Figure 2 ▶ ; Rs = 0.27, P = 0.05; Rs = 0.48, respectively; P < 0.001).
A gradual increase was seen of cases with loss of 8p, going from 8% in small cancers to 35% in intermediate tumors to 50% in large carcinomas (Table 2 ▶ ; Ptrend = 0.02). Likewise, a significant rise in cases with gain of 8q sequences was observed with increasing tumor volume (Table 2 ▶ ; Ptrend = 0.04). Potential biomarkers for tumor aggressiveness, ie, gain of 7pq and/or 8q, were less frequently discriminated in the small (8%) and intermediate cancers (26%) than in the large tumor samples (44%; Table 2 ▶ ; Ptrend = 0.03).
Genetic Changes and Clinicopathological Parameters
As shown in Table 2 ▶ , the average number of chromosomes with aberrations, especially gains, per case increased with tumor stage and grade. More specifically, an increase in the number of cases with gain of 8q sequences was found in high stage, high grade tumors. For corresponding P values, see Table 2 ▶ . Further, no differences were found for the molecular cytogenetic parameters described here between preoperative clinical stage T1c (nonpalpable, invisible) versus T2/T3 (palpable) tumors (all P values >0.5; Table 1 ▶ ). In addition, it appeared that the clinicopathological parameters of tumor volume, pathological tumor stage, and pathological grade (Gleason score) were closely related (tumor volume versus stage Rs = 0.75, P < 0.001; tumor volume versus grade Rs = 0.37, P = 0.006; stage versus grade Rs = 0.35, P = 0.01). Multiple regression analysis of these features showed that for most of the genetic parameters described here, no significant additional predictive value on top of tumor volume was observed for Gleason score and stage. Due to the strong correlation between tumor volume and tumor stage, however, the effects of these parameters were difficult to separate. Only for the number of chromosomal gains tumor stage was a better predictor (P = 0.04) than tumor volume (P = 0.55).
Statistically significant correlations were found between preoperative PSA and tumor volume (Rs = 0.61, P < 0.001), preoperative PSA and tumor stage (Rs = 0.51, P < 0.001), and preoperative PSA and grade (Rs = 0.45, P = 0.001), as has been described by others. 42 Importantly, no direct correlations were observed between preoperative PSA levels and the molecular cytogenetic parameters mentioned here.
Discussion
This study reports for the first time a genome-wide survey of the DNA copy number changes occurring in early localized prostatic tumors derived from patients participating in a population-based screening study. Furthermore, the observed molecular cytogenetic changes were correlated with tumor volume and different clinicopathological parameters. Our data show not only that larger cancers have more frequent chromosomal alterations, but also that more chromosomes are affected. This may potentially be a result of an increased genetic instability in larger tumors. 43 Chromosomal losses, especially loss of 6q and 13q, appeared to be relatively early changes in prostatic tumor development, since they were already frequently encountered in the small tumors. This suggests that one or more tumor suppressor genes located on these chromosomes may be important for prostatic tumorigenesis. Loss of 6q sequences has been reported to be both an early 12 and a late event in prostatic tumorigenesis. 14,26 In one study, it occurred in about one-third of primary tumors and 73% of distant metastases. 25 Loss of 13q is a recurrent finding in LOH 13,14,21 and CGH 25-28 studies of prostate cancer, with frequencies ranging from 22% 13 to as high as 91% in advanced cases. 21 Allelic loss of 6q and 13q was found in 18% and 8%, respectively, of prostatic intraepithelial neoplasia (PIN) lesions, a putative precursor lesion of prostate cancer, illustrating their early appearance in prostatic tumor development. 14 A gradual increase in the frequency of loss of 8p sequences was found with increasing tumor volume. In contrast, Vocke et al 18 did not find a correlation between 8p loss and tumor stage or grade. Loss of 8p is one of the most common genetic alterations in prostate cancer, with frequencies of 8p LOH as high as 86% in a large panel of primary prostate cancers. 18 LOH at 8p was also described in PIN lesions, 44,45 thereby being a possible initial event in prostatic tumorigenesis. In the group of early cancers, we found loss of 8p in nine cases (25%), again revealing 8p loss as an early event. This percentage of 8p loss is somewhat lower than reported in LOH studies, which may be due to the fact that the resolution of CGH for detecting loss is approximately 10 Mb. 46 Another recurrent finding, especially in the group of intermediate cancers, was the loss of 16q sequences. This alteration was less prominent in the large cancers (12%), which we attribute to sample size effects. Loss of 16q is also one of the most consistent genetic alterations in prostate cancer, 13,14,22,25-28 with CGH analyses reporting frequencies ranging from 19% in primary tumors to 56% and 55% in recurrent cancers and metastases, respectively. 26,27 Loss of 5q appeared a relatively frequent finding in our panel of both intermediate and large cancers. This alteration has been predominantly reported in advanced tumors. 14,25-27 Y chromosome loss was also an early finding, occurring already in the small and intermediate cancers. We have seen loss of Y throughout the spectrum of prostatic tumorigenesis, ranging from PIN lesions 47 to distant metastases. 32
Chromosomal gains were found to be relatively late events in prostatic tumor development, as can be seen by a sharp increase in the average number of chromosomes with gains going from intermediate tumors to large cancers (Figure 2) ▶ . It suggests involvement of oncogenes in later stages of prostatic tumorigenesis, as opposed to the more gradual increase of loss of tumor suppressor gene sites (Figure 2) ▶ . Gains appeared to be adverse prognostic indicators after radical prostatectomy. 25 In this study, a gradual increase was seen of the number of cases with 8q gain with increasing tumor volume. In several cases (Table 1) ▶ , gain of (part of) 8q was accompanied by 8p loss. This may be suggestive for i(8q) formation. 32,48,49 In addition, gain of 8q has been reported in advanced stages of prostate cancer, 25-28 and appeared to be associated with short progression-free survival. 25,34,48 Interestingly, a gradual increase was found for the number of cases with gain of 7pq and/or 8q along with increasing tumor volume. Gains of chromosome 7 and/or 8 are regarded as potential biomarkers for tumor aggressiveness. 25,32-34 These alterations were predominantly found in distant metastases and in primary tumors that showed progression after radical prostatectomy. 25,32 Noteworthy, a subset of the tumors derived from the ERSPC showed gain on chromosome 7pq and/or 8q. Furthermore, other chromosomal alterations that are reported in advanced stages of disease only, such as chromosomal gains in general and loss of 5q and 16q sequences, were also seen in the early cancers. These tumors may, therefore, be regarded as potentially aggressive, which may have therapeutic implications.
Although the period of follow-up of these ERSPC patients is short at present (mean, 37.5 months; range, 6–62 months), preliminary data show that of the 36 patients described in this study, four patients with tumor volumes between 0.5 and 1.0 ml have shown biochemical progression of the disease. All these cases displayed chromosomal alterations, including one with 8q gain combined with 8p loss (Table 1) ▶ . This is in line with the observation that the majority of nonpalpable, invisible (T1c) tumors derived from screening programs, as judged by their pathological characteristics, are clinically significant tumors. 2,9,10,36 Further follow-up studies of patients participating in population-based screening programs, such as ERSPC, will answer the question whether the detection and treatment of early, localized tumors is justified. Noteworthily, no direct correlations were observed between preoperative PSA levels and the genetic parameters mentioned here. Therefore, in our opinion, an important role might be reserved for genetic biomarkers that can predict the biological behavior of the tumor in the earliest stages of clinical decisionmaking. Importantly, this genetic test can be performed on archival prostatic biopsies (Alers JC, unpublished results). Currently, we are conducting a CGH study to examine whether the pattern of genetic changes in pre-operative needle biopsies accurately represents the tumor in the corresponding radical prostatectomy specimens. Finally, we are further defining the 7pq-8q biomarker in a large cohort of patients.
Footnotes
Address reprint requests to Dr. Janneke C. Alers, Department of Pathology, Josephine Nefkens Institute, Erasmus University Rotterdam, P.O. Box 1738, 3000 DR Rotterdam, The Netherlands. E-mail: alers@path.fgg.eur.nl.
Supported by the Dutch Cancer Society Grant EUR 97–1404 and the Sascha Swarttouw-Hijmans Foundation.
References
- 1.Landis SH, Murray T, Bolden S, Wingo PA: Cancer statistics, 1999. CA Cancer J Clin 1999, 4:8-31 [DOI] [PubMed] [Google Scholar]
- 2.Stamey AT, Donaldson AN, Yemoto CE, McNeal JE, Sözen S, Gill H: Histological and clinical findings in 896 consecutive prostates treated only with radical retropubic prostatectomy: epidemiologic significance of annual changes. J Urol 1998, 160:2412-2417 [DOI] [PubMed] [Google Scholar]
- 3.Jacobsen SJ, Katusic SK, Bergstralh EJ, Oesterling JE, Ohrt D, Klee GG, Chute CG, Lieber MM: Incidence of prostate cancer diagnosis in the eras before and after serum prostate-specific antigen testing. JAMA 1995, 274:1445-1449 [PubMed] [Google Scholar]
- 4.Hankey BF, Feuer EJ, Clegg LX, Hayes RB, Legler JM, Prorok PC, Ries LA, Merrill RM, Kaplan RS: Cancer surveillance series: interpreting trend in prostate cancer-part 1: Evidence of the effects of screening in recent prostate cancer incidence, mortality, and survival rates. J Natl Cancer Inst 1999, 91:1017-1024 [DOI] [PubMed] [Google Scholar]
- 5.Catalona WJ, Smith DS, Ratliff TL, Basler JW: Detection of organ-confined prostate cancer is increased through prostate-specific antigen-based screening. JAMA 1993, 270:948-954 [PubMed] [Google Scholar]
- 6.Scardino PT, Weaver R, Hudson MA: Early detection of prostate cancer. Hum Pathol 1992, 23:211-222 [DOI] [PubMed] [Google Scholar]
- 7.Frydenberg M, Stricker PD, Kaye KW: Prostate cancer diagnosis and management. Lancet 1997, 349:1681-1687 [DOI] [PubMed] [Google Scholar]
- 8.Kirkels WJ, Rietbergen JBW: Screening for prostate cancer. Urol Res 1997, 25:S53-S56 [DOI] [PubMed] [Google Scholar]
- 9.Epstein JI, Walsh PC, Carmichael M, Brendier CB: Pathologic and clinical findings to predict tumor extent of nonpalpable (stage T1c) prostate cancer. JAMA 1994, 271:368-374 [PubMed] [Google Scholar]
- 10.Ohori M, Wheeler TM, Dunn JK, Stamey TA, Scardino PT: The pathological features and prognosis of prostate cancer detectable with current diagnostic tests. J Urol 1994, 152:1714-1720 [DOI] [PubMed] [Google Scholar]
- 11.Albertsen PC, Fryback DG, Storer BE, Kolon TF, Fine J: Long-term survival among men with conservatively treated localized prostate cancer. JAMA 1995, 274:626-631 [PubMed] [Google Scholar]
- 12.Brothman AR, Maxwell TM, Cui J, Deubler DA, Zhu XL: Chromosomal clues to the development of prostate tumors. Prostate 1999, 38:303-312 [DOI] [PubMed] [Google Scholar]
- 13.Cunningham JM, Shan A, Wick MJ, McDonnell SK, Schaid DJ, Tester DJ, Qian J, Takahashi S, Jenkins RB, Bostwick DG, Thibodeau SN: Allelic imbalance and microsatellite instability in prostatic adenocarcinoma. Cancer Res 1996, 56:4475-4482 [PubMed] [Google Scholar]
- 14.Saric T, Brkanac Z, Troyer DA, Padalecki SS, Sarosdy M, Williams K, Abadesco L, Leach RJ, O’Connell P: Genetic pattern of prostate cancer progression. Int J Cancer 1999, 81:219-224 [DOI] [PubMed] [Google Scholar]
- 15.Dahiya R, McCarville J, Hu W, Lee C, Chui RM, Kaur G, Deng G: Chromosome 3p24–26 and 3p22–12 loss in human prostatic adenocarcinoma. Int J Cancer 1997, 71:20-25 [DOI] [PubMed] [Google Scholar]
- 16.Cooney KA, Wetzel JC, Consolino CM, Wojno KJ: Identification and characterization of proximal 6q deletions in prostate cancer. Cancer Res 1996, 56:4150-4153 [PubMed] [Google Scholar]
- 17.Takahashi S, Shan AL, Ritland SR, Delacey KA, Bostwick DG, Lieber MM, Thibodeau SN, Jenkins RB: Frequent loss of heterozygosity at 7q31.1 in primary prostate cancer is associated with tumor aggressiveness and progression. Cancer Res 1995, 55:5115-5119 [PubMed] [Google Scholar]
- 18.Vocke CD, Pozzatti RO, Bostwick DG, Florence CD, Jennings SB, Strup SE, Duray PH, Lioatta LA, Emmert-Buck MR, Linehan WM: Analysis of 99 microdissected prostate carcinomas reveals a high frequency of allelic loss on chromosome 8p12–p21. Cancer Res 1996, 56:2411-2416 [PubMed] [Google Scholar]
- 19.Perinchery G, Bukurov N, Nakajima K, Chang J, Li LC, Dahiya R: High frequency of deletion on chromosome 9p21 may harbor several tumor-suppressor genes in human prostate cancer. Int J Cancer 1999, 83:610-614 [DOI] [PubMed] [Google Scholar]
- 20.Gray IC, Philips SMA, Lee SJ, Neoptolemos JP, Weissenbach J, Spurr NK: Loss of chromosomal region 10q23–q25 in prostate cancer. Cancer Res 1995, 55:4800-4803 [PubMed] [Google Scholar]
- 21.Hyytinen ER, Frierson HF, Boyd JC, Chung LWK, Dong JT: Three distinct regions of allelic loss at 13q14, 13q21–22, and 13q33 in prostate cancer. Genes Chromosomes Cancer 1999, 25:108-114 [PubMed] [Google Scholar]
- 22.Li C, Berx G, Larsson C, Auer G, Aspenblad U, Pan Y, Sundelin B, Ekman P, Nordenskjöld M, van Roy F, Bergerheim USR: Distinct deleted regions on chromosome segment 16q23–24 associated with metastases in prostate cancer. Genes Chromosomes Cancer 1999, 24:175-182 [PubMed] [Google Scholar]
- 23.Gao X, Zacharek A, Grignon DJ, Sakr W, Powell IJ, Porter AT, Honn KV: Localization of potential tumor suppressor loci to a <2 Mb region on chromosome 17q in human prostate cancer. Oncogene 1995, 11:1241-1247 [PubMed] [Google Scholar]
- 24.Ueda T, Komiya A, Emi M, Suzuki H, Shiraishi T, Yatani R, Masai M, Yasuda K, Ito H: Allelic loss on 18q21 are associated with progression and metastasis in human prostate cancer. Genes Chromosomes Cancer 1997, 20:140-147 [DOI] [PubMed] [Google Scholar]
- 25.Alers JC, Rochat J, Krijtenburg PJ, Hop WCJ, Kranse R, Rosenberg C, Tanke HJ, Schröder FH, van Dekken H: Identification of genetic markers for prostatic cancer progression. Lab Invest 2000, 80:931-942 [DOI] [PubMed] [Google Scholar]
- 26.Visakorpi T, Kallioniemi AH, Syvänen AC, Hyytinen ER, Karhu R, Tammela T, Isola JJ, Kallioniemi OP: Genetic changes in primary and recurrent prostate cancer by comparative genomic hybridization. Cancer Res 1995, 55:342-347 [PubMed] [Google Scholar]
- 27.Cher ML, Bova GS, Moore DH, Small EJ, Carroll PR, Pin SS, Epstein JI, Isaacs WB, Jensen RH: Genetic alterations in untreated metastases and androgen-independent prostate cancer detected by comparative genomic hybridization. Cancer Res 1996, 56:3091-3102 [PubMed] [Google Scholar]
- 28.Nupponen NN, Kakkola L, Koivisto P, Visakorpi T: Genetic alterations in hormone-refractory recurrent prostate carcinomas. Am J Pathol 1998, 153:141-148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alers JC, van Dekken H: Interphase cytogenetic analysis of solid tumors by non-isotopic DNA in situ hybridization. Prog Histochem Cytochem 1996, 31:1-133 [DOI] [PubMed] [Google Scholar]
- 30.Huang SF, Xiao S, Renshaw AA, Loughlin KR, Hudson TJ, Fletcher JA: Fluorescence in situ hybridization evaluation of chromosome deletion patterns in prostate cancer. Am J Pathol 1996, 149:1565-1573 [PMC free article] [PubMed] [Google Scholar]
- 31.Bubendorf L, Kononen J, Koivisto P, Schraml P, Moch H, Gasser TC, Willi N, Mihatsch MJ, Sauter G, Kallioniemi OP: Survey of gene amplifications during prostate cancer progression by high-throughput fluorescence in situ hybridization on tissue microarrays. Cancer Res 1999, 59:803-806 [PubMed] [Google Scholar]
- 32.Alers JC, Krijtenburg PJ, Rosenberg C, Hop WCJ, Verkerk AM, Schröder FH, van der Kwast ThH, Bosman FT, van Dekken H: Interphase cytogenetics of prostatic tumor progression: Specific chromosomal abnormalities are involved in metastasis to the bone. Lab Invest 1997, 77:437-448 [PubMed] [Google Scholar]
- 33.Alcaraz A, Takahashi S, Brown JA, Herath JF, Bergstralh EJ, Larson-Keller JJ, Lieber MM, Jenkins RB: Aneuploidy and aneusomy of chromosome 7 detected by fluorescence in situ hybridization are markers for poor prognosis in prostate cancer. Cancer Res 1994, 54:3998-4002 [PubMed] [Google Scholar]
- 34.Sato K, Qian J, Slezak JM, Lieber MM, Bostwick DG, Bergstralh EJ, Jenkins RB: Clinical significance of alterations of chromosome 8 in high-grade, advanced, nonmetastatic prostate carcinoma. J Natl Cancer Inst 1999, 91:1574-1580 [DOI] [PubMed] [Google Scholar]
- 35.Schröder FH, van der Maas P, Beemsterboer P, Boeken Kruger A, Hoedemaeker R, Rietbergen J, Kranse R: Evaluation of the digital rectal examination as a screening test for prostate cancer: Rotterdam section of the European Randomized Study of Screening for Prostate Cancer. J Natl Cancer Inst 1998, 90:1817–1823 [DOI] [PubMed]
- 36.Hoedemaeker RF, Rietbergen JBW, Kranse R, van der Kwast ThH, Schröder FH: Comparison of pathologic characteristics of T1c and non-T1c cancers detected in a population-based screening study, the European Randomized Study of Screening for Prostate Cancer. World J Urol 1997, 15:339-345 [DOI] [PubMed] [Google Scholar]
- 37.Hermanek P, Hutter RVP, Sobin LH, Wagner G, Wittekind Ch: TNM Atlas: Illustrated Guide to the TNM/pTNM Classification of Malignant Tumors, 4th ed. 1997, :pp 272-280 Springer Verlag, Berlin [Google Scholar]
- 38.Gleason DF: Histologic grading of prostate cancer. Hum Pathol 1992, 23:273-279 [DOI] [PubMed] [Google Scholar]
- 39.Alers JC, Rochat J, Krijtenburg PJ, van Dekken H, Raap AK, Rosenberg C: Universal linkage system (ULS): An improved method for labeling archival DNA for comparative genomic hybridization. Genes Chromosomes Cancer 1999, 25:301-305 [DOI] [PubMed] [Google Scholar]
- 40.Kallioniemi A, Kallioniemi OP, Sudar D, Rutovitz D, Gray JW, Waldman F, Pinkel D: Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science 1992, 258:818-821 [DOI] [PubMed] [Google Scholar]
- 41.Kirchhoff M, Gerdes T, Rose H, Maahr J, Ottesen AM, Lundsteen C: Detection of chromosomal gains and losses in comparative genomic hybridization analysis bases on standard reference intervals. Cytometry 1998, 31:163-173 [PubMed] [Google Scholar]
- 42.Stamey TA, Yemoto CM, McNeal JE, Sigal BM, Johnstone IM: Prostate cancer is highly predictable: A prognostic equation based on all morphological variables in radical prostatectomy specimens. J Urol 2000, 163:1155-1160 [DOI] [PubMed] [Google Scholar]
- 43.Nowell PC: Mechanisms of tumor progression. Cancer Res 1986, 46:2203-2207 [PubMed] [Google Scholar]
- 44.Emmert-Buck MR, Vocke CD, Pozzatti RO, Duray PH, Jennings SB, Florence CD, Zhuang Z, Bostwick DG, Lioatta LA, Linehan WM: Allelic loss on chromosome 8p12–p21 in microdissected prostatic intraepithelial neoplasia. Cancer Res 1995, 55:2959-2962 [PubMed] [Google Scholar]
- 45.Sakr WA, Macoska JA, Benson P, Grignon DJ, Wolman SR, Pontes JE, Crissman JD: Allelic loss in locally metastatic, multisampled prostate cancer. Cancer Res 1994, 54:3273-3277 [PubMed] [Google Scholar]
- 46.Bentz M, Plesch A, Stilgenbauer S, Döhner H, Lichter P: Minimal sizes of deletions detected by comparative genomic hybridization. Genes Chromosomes Cancer 1998, 21:172-175 [PubMed] [Google Scholar]
- 47.Alers JC, Krijtenburg PJ, Vissers KJ, Bosman FT, van der Kwast ThH, van Dekken H: Interphase cytogenetics of prostatic adenocarcinomas and precursor lesions: analysis of 25 radical prostatectomies and 17 adjacent prostatic intraepithelial neoplasias. Genes Chromosomes Cancer 1995, 12:241-250 [DOI] [PubMed] [Google Scholar]
- 48.Macoska JA, Trybus TM, Wojno KJ: 8p22 loss concurrent with 8c gain is associated with poor outcome in prostate cancer. Urology 2000, 55:776-782 [DOI] [PubMed] [Google Scholar]
- 49.Virgin JB, Hurley PM, Nahhas FA, Bebchuk KG, Mohamed AN, Sakr WA, Bright RK, Cher ML: Isochromosome 8q formation is associated with 8p loss of heterozygosity in a prostate cancer cell line. Prostate 1999, 41:49-57 [DOI] [PubMed] [Google Scholar]