Abstract
Matrix metalloproteinase-1 (MMP-1, collagenase-1), which degrades interstitial collagen, is expressed at high levels by some tumor cells and is thought to enhance their invasiveness and metastatic potential. We recently described a common single nucleotide insertion polymorphism (2G allele) at −1,607 bp in the promoter of the MMP-1 gene that creates a binding site for the ETS family of transcription factors, and that is associated with enhanced transcription of this gene and increased enzyme activity. Allelic loss at the MMP-1 locus on chromosome 11 occurs in many tumors including melanoma, an invasive and aggressive cancer. We hypothesized that although loss of either the 1G or 2G allele from 1G/2G heterozygotes is random, retention of the transcriptionally more active 2G allele would favor tumor invasion and metastasis. As a result, a higher proportion of metastases would contain the 2G genotype than the 1G genotype. We report here the development of quantitative methods for assessing allelic loss at the MMP-1 locus, and demonstrate that 83% of the metastatic melanomas with loss of heterozygosity at this locus retained the 2G allele. This supports the hypothesis that retention of the 2G allele favors tumor invasion and metastasis in melanoma.
Matrix metalloproteinase-1 (MMP-1, collagenase-1), a member of a family of MMPs that degrades most components of the extracellular matrix, breaks down the interstitial collagens types I, II, and III. 1-3 MMP-1 is ubiquitously expressed by most normal cell types, usually at low levels. Expression can be increased to high levels by numerous cytokines and growth factors, and this enzyme can mediate connective tissue destruction in diseases such as arthritis, 1,4 periodontitis, 5,6 and atherosclerosis. 7 In contrast, some tumor cells express high levels of MMP-1 constitutively, 8-11 which may contribute to the invasive and aggressive behavior of the tumor. Indeed, evidence suggests that expression of MMP-1 is linked to poor outcome in several cancers, including colon carcinoma, esophageal carcinoma, pancreatic carcinoma, and melanoma. 12-15
Recently we described a common single nucleotide polymorphism (SNP) in the promoter of MMP-1, in which one allele is associated with enhanced transcription of this gene. 8 The SNP is located at −1,607 bp and is represented by the presence or absence of a guanine (G) at this position. In the absence of the G, the sequence reads 5′-GAA-3′, whereas the presence of an extra G gives the sequence 5′-GGAA-3′, a consensus binding site for the ETS family of transcription factors. Indeed, MMP-1 genes with promoters carrying the 2G allele are transcribed at a significantly higher rate in both normal fibroblasts and in tumor cells than are MMP-1 genes with promoters carrying the 1G allele. 8 The frequencies of the 1G and 2G alleles appear to be approximately equal in normal populations, whereas in a small sample of tumor cell lines tested, the frequency of the 2G allele was increased to 60%. 8 In addition, there was a higher than expected proportion of apparent 2G homozygotes in these cell lines, suggesting that the 2G allele may be associated with aggressive and invasive cancers. 8 This concept is supported by a significant association of the 2G allele with ovarian 16 and endometrial cancer, 17 and with increased expression of MMP-1 protein in these diseases. 16,17 Although the findings in tumor cell lines may simply reflect a greater proportion of tumors derived from homozygous 2G hosts, it is also possible that these aggressive tumors result from the loss of the 1G allele in tumors from heterozygous hosts [loss of heterozygosity (LOH)].
LOH is a common event in many tumors and is often associated with advancing disease, presumably because of loss of tumor suppressor gene(s) at the deleted locus. 18-26 LOH occurs at several chromosomal loci, including the MMP-1 locus at 11q22.23. 27 We tested the hypothesis that if metastatic tumors were derived from cells that retained the 2G allele, they would have a selective invasive advantage and would, therefore, be overrepresented in heterozygous hosts compared to tumors that had retained the 1G allele. Accordingly, we genotyped normal and metastatic tumor tissues from patients with melanoma to determine the frequency of the 2G allele and the occurrence of LOH. To perform LOH studies with a relatively large number of patient samples, we developed a rapid method to screen samples for heterozygosity and precise quantitative techniques to objectively assess LOH.
Materials and Methods
Patient Specimens
Samples were available from a total of 61 patients: frozen tumor and peripheral blood leukocytes from 12 patients; formalin-fixed, paraffin-embedded normal and tumor tissue from 44 patients; and both frozen and paraffin-embedded samples from five patients. The frozen tissues had been stored for up to 3 years at −70OC; the paraffin blocks had been stored for up to 7 years at room temperature.
In addition to these paired normal tissue and tumor samples, control germline DNA samples from known heterozygotes were run with each experiment to provide additional data for calculation of interassay variability.
DNA Isolation
Frozen Tumor Tissue and Peripheral Blood Leukocytes
Genomic DNA was isolated from frozen peripheral blood lymphocytes or from frozen single-cell tumor suspensions using Gentra’s Puregene (Minneapolis, MN) kit according to the manufacturer’s instructions. These tumor samples had been collected for the purpose of vaccine production and had been trimmed, originally, to be free of normal tissue; however, this was not confirmed at the time of this study.
Paraffin-Embedded Tissues
One 5-μm section and three adjacent 10-μm serial sections of formalin-fixed, paraffin-embedded tumor and normal tissue were mounted on glass slides. The 5-μm section was stained with hematoxylin and eosin. Under a dissecting microscope, normal and tumor tissue were identified by a pathologist on unstained sections by comparing these to the adjacent stained section, and were scraped separately with a sterile pin into a microfuge tube. The pathologist performing the dissections estimated that no more than 20% of the cells dissected from tumor were contaminating normal cells. Digestion buffer, consisting of 25 μl of 50 mmol/L Tris-HCl, pH 8.5, 2% Laureth-12, 400 μg/ml proteinase K, was added, and the samples were incubated overnight in a 55°C oven to minimize evaporation. An additional 25 μl of digestion buffer was added and the samples were incubated at 55°C for an additional 4 to 5 hours. The proteinase K was inactivated by heating to 95°C for 10 minutes. The samples were spun at 13,000 × g in a microcentrifuge for 2 minutes and the supernatant was used for polymerase chain reaction (PCR). DNA from some tumor samples could not be amplified using this lysis method, perhaps because of high melanin concentrations that may have interfered with the PCR reaction. 28 Those samples were recut and DNA was isolated using the Puregene kit.
SNP Determination by Restriction Fragment Length Polymorphism (RFLP) Analysis
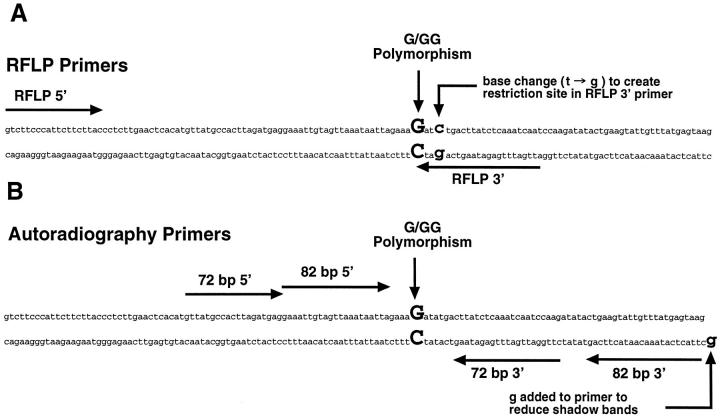
An RFLP method to distinguish the SNP was developed to screen samples rapidly and to identify heterozygotes for LOH studies. Restriction enzyme sites were introduced into the PCR amplicon at the SNP locus by making a single base change in the downstream primer, such that the 1G allele digested with BglII and the 2G allele digested with Alw I (Figure 1A) ▶ . Purified DNA (100 ng) or lysate (5 μl) from the paraffin-embedded samples was amplified using 50 pmol of each primer (RFLP forward: GTC TTC CCA TTC TTC TTA CC; RFLP reverse: ATT GAT TTG AGA TAA GTC AGA TC) in 1× Qiagen (Valencia, CA) PCR buffer (Tris-Cl, pH 8.7, at 20°C, KCl, (NH4)2SO4, 1.5 mmol/L MgCl2), 200 μmol/L each dNTP, 1.5 mmol/L additional MgCl2, for a total of 3 mmol/L MgCl2, 2.5 U HotStarTaq (Qiagen) in a 100 μl reaction. Cycling conditions were 95°C for 15 minutes followed by 35 cycles of 95°C for 30 seconds, 50°C for 30 seconds, and 72°C for 30 seconds with a final 72°C extension for 2 minutes. Samples (20 μl) were digested using 10 U of Alw I or BglII (New England Biolabs, Beverly, MA) at 37°C for 2 to 3 hours. The digests were electrophoresed on 3% SFR Agarose (Amresco, Solon, OH) and visualized with ethidium bromide.
Figure 1.
PCR primers for MMP-1 SNP analysis. The 1G/2G polymorphism in the promoter is indicated in bold upper case. A: Primers for RFLP analysis of the SNP. Note the base change (t->g) in the 3′ primer that creates a BglII site in the 1G allele and an Alw I site in the 2G allele. B: Primers used for quantitative autoradiographic analysis of LOH at the MMP-1 SNP locus. The 3′ primer for the 82-bp amplicon has an added 5′ terminal “g”. This additional base eliminated shadow bands that interfered with accurate quantification of the alleles. 29
Quantitative Analysis of LOH at the SNP Locus: High-Resolution Electrophoresis of 32P-Labeled Amplicons
Tumor and normal tissue samples from heterozygotes were analyzed by two quantitative PCR assays, using sets of nonoverlapping primer pairs, that generated either a 72-bp amplicon or an 82-bp amplicon. This dual amplification with no overlap between the primer sets controlled for the possibility that a point mutation was present under either of the primer binding sites in the tumor DNA (that might alter the relative amplification of the 1G and 2G alleles), thereby giving a false indication of LOH. The primers for the 72-bp product were: forward: GTT ATG CCA CTT AGA TGA GG, end-labeled with γ-32P ATP (3,000 Ci/mmol); reverse: CTT GGA TTG ATT TGA GAT AAG (Figure 1B) ▶ . The primers for the 82-bp product were: forward: GAA ATT GTA GTT AAA TAA TTA G, end-labeled with γ-32P ATP; reverse: GCT TAC TCA TAA ACA ATA CTT CAG (Figure 1B) ▶ . The reverse primer of the 82-bp set was modified by the addition of a 5′ terminal G that, along with the cycling conditions, helped drive the PCR reaction to allele + A. 29 This modification eliminated shadow bands seen in previous studies using a different primer set. 8,30
For both PCR reactions 100 ng of purified DNA or 5 μl lysate from the paraffin-embedded samples was amplified using 50 pmol of each primer in 1× Qiagen PCR buffer (Tris-Cl, pH 8.7, at 20°C, KCl, (NH4)2SO4, 1.5 mmol/L MgCl2), 200 μmol/L each dNTP, 1.5 mmol/L additional MgCl2, for a total of 3 mmol/L MgCl2, 2.5 U HotStarTaq (Qiagen) in a 100 μl reaction. Samples containing known ratios of 1G and 2G DNA were prepared for a standard curve (see Results) using purified peripheral blood DNA from 1G and 2G homozygotes. For the 72-bp product, cycling conditions were 95°C for 15 minutes, followed by 30 cycles of 95°C for 30 seconds, 50°C for 30 seconds, and 72°C for 30 seconds with a final 72°C extension for 2 minutes. For the 82-bp product, cycling conditions were 94°C for 15 minutes, followed by 10 cycles of 94°C for 15 seconds, 50°C for 30 seconds, and 72°C for 60 seconds, then 20 cycles of 89°C for 15 seconds, 50°C for 30 seconds, and 72°C for 60 seconds with a final 72°C extension for 30 minutes.
Five μl of PCR product from each reaction was then mixed with 5 μl of formamide loading buffer and denatured at 95°C for 5 minutes. Five μl aliquots of the sample mixtures were loaded on an 8% denaturing acrylamide gel and run at 55 W constant power until the xylene cyanol was ∼6 cm from the bottom of the gel. Under these conditions there was excellent resolution of the 1G and 2G alleles for both the 72/73-bp and 82/83-bp products (see Results). Gels were exposed to a phosphorimager screen (Molecular Dynamics, Sunnyvale, CA) for 15 to 60 minutes and to film for 30 minutes to several hours.
For quantitative analysis of allele intensity, the phosphorimager screens were scanned and the band intensities quantified using ImageQuant v. 1.2 (Molecular Dynamics) by drawing a 2 by 1 matrix around each lane, placing the 2G allele in the upper box and the 1G allele in the lower box. The percent 2G was calculated by dividing the signal in the upper box by the total signal in both boxes and multiplying by 100. The signals were not corrected for background.
Statistical Testing Methods
Under the null hypothesis, assuming that allele retention is a random event, we would expect the 1G or 2G allele to be retained in an equal number of tumors (ie, a genotype ratio of 1:1). We calculated an exact P value using a binomial distribution 31 for the likelihood that the observed genotype ratio in the 12 tumors with LOH was a chance finding.
Results
SNP Detection by RFLP Analysis
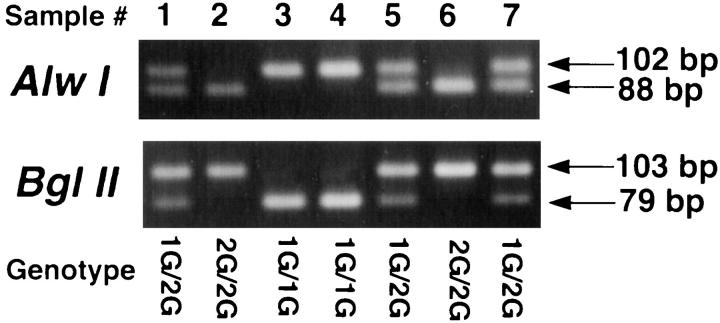
Normal DNA from 61 patients was genotyped using the RFLP method, yielding 102- or 103bp products. There were 31 1G/2G heterozygotes, 16 1G/1G homozygotes, and 14 2G/2G homozygotes (allele frequencies: 1G = 0.52, 2G = 0.48). Figure 2 ▶ demonstrates the RFLP pattern after digestion with BglII (the 102-bp 1G allele is cleaved) and after digestion with Alw I (the 103-bp 2G allele is cleaved). This method is rapid and does not require radioactivity, and the use of separate restriction enzymes to recognize each of the two alleles serves as an internal control.
Figure 2.
RFLP analysis of MMP-1 SNP. Alw I digests the 103-bp 2G amplicon (88 bp + 15 bp), leaving intact the 1G amplicon (102 bp); BglII digests the 102-bp 1G amplicon (79 bp + 23 bp), leaving intact the 2G amplicon (103 bp). Samples 3 and 4 are 1G/1G homozygotes. Samples 1, 5, and 7 are 1G/2G heterozygotes. Samples 2 and 6 are 2G/2G homozygotes.
Quantitative Analysis of LOH at the SNP Locus: High-Resolution Electrophoresis of 32P-Labeled Amplicons
Of the 31 heterozygous patients identified by RFLP analysis, one patient had three metastases available for study, one patient had two metastases available for study (both of which were refractory to DNA amplification), and 29 had one metastasis available for study (one of which was refractory to amplification). The three samples refractory to DNA amplification were from paraffin blocks. In summary, 31 tumors and normal tissue from 29 patients were tested successfully for LOH at the SNP locus with the quantitative 72-bp and 82-bp amplicon assays.
The most sensitive means of detecting LOH was by visual inspection of exposed film, the unaided eye readily seeing small deviations from the pattern of equal allele intensity seen with normal DNA from heterozygotes. However, at the outset of the study it was decided that an objective, quantitative method of assessing allelic imbalance was preferable to visual inspection to eliminate any possibility of observer bias. Accordingly, each experiment included seven standards, composed of various proportions of DNA from 1G and 2G homozygotes (0/100, 25/75, 40/60, 50/50, 60/40, 75/25, 100/0) expressed as “percent 2G” (Figure 3) ▶ . A standard curve (x axis = actual percent 2G, y axis = observed percent 2G) was plotted and analyzed by linear regression, and the regression equation was used to calculate the actual percent 2G in the unknown samples. Because the phosphorimager data used to determine observed percent 2G were not corrected for background (see Materials and Methods), the regression line has a positive intercept on the y axis. The standard curves from successive experiments with each primer pair were remarkably reproducible and linear (Figure 4) ▶ .
Figure 3.
Autoradiographic detection of the MMP-1 SNP and quantification of LOH in tumor tissue. Data from the 72-bp amplicon assay (A) and the 82-bp amplicon assay (B) are shown. Normal tissue/tumor pairs from three patients are at the left; standards, composed of known mixtures of DNA from 1G and 2G homozygous controls, are at the right. The “%2G” shown for the tumor samples was determined by quantitative phosphorimager analysis and calculated from a standard curve (see Figure 4 ▶ ) constructed from standards run on the same gel. Tumors from patients 65 and 34 have lost the 1G allele. The tumor from patient 22 has lost the 2G allele.
Figure 4.
Standard curves for the 72-bp amplicon assay (A) and the 82-bp amplicon assay (B). Standards were composed of mixtures of DNA from 1G and 2G homozygous controls that were analyzed concurrently with patient samples. The actual composition of the standards is plotted on the x axis as “Actual %2G.” Data from quantitative phosphorimager analysis of the standards (see text) is plotted on the y axis as “Observed %2G.” Data from multiple experiments are shown, plotted as mean ± SD. Standard curves were used to convert “Observed %2G” of tumor samples to “Actual %2G.”
The interassay variability of the percent 2G for heterozygous samples in the 72-bp amplicon assay was calculated from data obtained in multiple experiments from 94 normal tissue samples and control blood samples; 33 samples provided similar data for the 82-bp amplicon assay. The calculated percent 2G of heterozygous samples using the 72-bp amplicon assay was 50.5 ± 6.7 (mean, ±2 SD); for the 82-bp amplicon assay it was 49.9 ± 4.3 (mean, ±2 SD). These ranges are indicated as dashed lines in Figure 5 ▶ .
Figure 5.
Summary of MMP-1 LOH data from all tumors where LOH was detected, using the 72-bp amplicon assay (A) and the 82-bp amplicon assay (B). The “%2G” for each tumor was determined as described in Figures 4 and 5 ▶ ▶ , and is represented here as a bar. Replicate assays are indicated by individual bars. Fifty percent 2G represents no allelic loss, characteristic of normal tissues. The hatched lines represent the imprecision (2 SD) of the estimate of “%2G” in normal heterozygous tissue samples and control blood samples. Tumor samples whose “%2G” was at or exceeded the imprecision limits in both assays were classified as having undergone LOH. Ten tumors lost the 1G allele; two tumors lost the 2G allele.
Tumor samples were classified as having undergone LOH if the calculated percent 2G exceeded the 2 SD range for heterozygotes in each of the two assays (Figure 5) ▶ . If the deviation from heterozygosity was large, single concordant results from each assay were accepted as sufficient. If the deviation from heterozygosity was small or borderline, consistent reproducible findings with both primer pairs were required to classify the sample as having undergone LOH. In general, differences between replicate samples were small and the results of the two assays were in close agreement (Figure 5) ▶ .
Twelve of the 31 tumors (39%) showed marked or consistent deviation from the 1G/2G ratio characteristic of normal heterozygous tissue and were classified as having undergone LOH. Each of these tumors was from a different patient. Ten (83%) of the tumors underwent LOH with retention of the 2G allele, and two (17%) tumors underwent LOH with retention of the 1G allele (Figure 5) ▶ . This departure from the expected 1:1 ratio was statistically significant (P = 0.04).
Table 1 ▶ lists some of the clinical characteristics of the patients whose tumors had undergone LOH, the tissue source of the metastatic tumors that were analyzed, and the likely mode of spread (lymphatic or hematogenous) of each of the metastases.
Table 1.
Summary of Heterozygous Patients with Metastases Demonstrating LOH at the MMP-1 Locus
| Patient no. | Age | Sex | Location of metastasis | Mechanism of spread* | ||||
|---|---|---|---|---|---|---|---|---|
| Loss of 1G allele in metastasis | ||||||||
| 8 | 54 | F | Soft tissue | Lymphatic | ||||
| 14 | 64 | F | Lymph node | Lymphatic | ||||
| 34 | 75 | M | Lung | Hematogenous | ||||
| 38 | 42 | F | Lung | Hematogenous | ||||
| 4 | 75 | F | Lymph node | Unknown† | ||||
| 48 | 71 | M | Lymph node | Lymphatic | ||||
| 53 | 39 | M | Lymph node | Lymphatic | ||||
| 54 | 77 | F | Soft tissue | Lymphatic | ||||
| 61 | 67 | M | Subcutaneous | Lymphatic | ||||
| 65 | 64 | M | Lymph node | Hematogenous | ||||
| Loss of 2G allele in metastasis | ||||||||
| 1 | 38 | M | Lymph node | Lymphatic | ||||
| 22 | 53 | M | Soft tissue | Hematogenous | ||||
*Mechanism of spread was scored as lymphatic if the metastasis was in a lymph node or in soft tissue in the path of lymphatic drainage of the primary tumor; other metastases were scored as hematogenous.
†This was scored as unknown because the location of the primary tumor in this patient was unknown.
Some of the tumors displayed marked allelic imbalance, suggesting that the tissue samples that were analyzed consisted almost entirely of tumor cells, all of which had undergone LOH, and few nonneoplastic stromal or inflammatory cells. Other tumors had a lesser degree of allelic imbalance, suggesting the presence of a larger fraction of nonneoplastic cells in the sampled tissue or heterogeneity within the tumor cells, with only a fraction having undergone LOH. In the simplest model of LOH, loss of one allele with no other confounding genetic alterations, this study was capable of detecting LOH if at least 20 to 25% of the total cells in the sampled tissue had undergone allelic loss.
Discussion
In this report we describe methods for the rapid detection of a SNP in the MMP-1 (collagenase 1) promoter that can increase transcription of this gene, and thus enhance the destruction of the extracellular matrix necessary for tumor invasion and metastasis. In addition, we developed rapid and quantitative techniques for assessing LOH at this locus. We chose metastatic melanoma for this study because expression of collagenase 1 is associated with a poor prognosis in this highly invasive and aggressive disease. 15 We tested 31 metastatic melanomas from heterozygous patients and detected LOH in 12 cases: 83% retained the 2G allele and 17% retained the 1G allele. This disproportionate representation of the 2G allele in these metastases supports our hypothesis that the 2G genotype of the MMP-1 promoter may provide some tumors with a selective and/or invasive advantage, which is manifested by the development of metastatic lesions that preferentially contain the 2G genotype. Our previous studies showing that the 2G allele was overrepresented in melanoma and breast cancer cell lines are consistent with this hypothesis. 8 Studies of patients with ovarian 16 and endometrial cancer 17 also bear indirectly on our findings. In these studies the proportion of patients who were either heterozygotes or homozygotes for the 2G allele was significantly higher than that observed in individuals without cancer, suggesting that the risk of developing these diseases was increased in individuals who carried a 2G allele. Furthermore, the levels of MMP-1 protein in cancer tissues from patients with one or more copies of the 2G allele were higher than protein levels in 1G homozygotes, consistent with a higher level of transcription from the 2G allele. A high level of MMP-1 expression has been associated previously with poor patient outcome in a number of cancers. 12-15
The varying degrees of LOH observed in our study most likely reflect either the presence of contaminating stromal cells and/or tumor heterogeneity, with only a portion of the sampled tumor having undergone allelic loss. An alternative explanation is that there was amplification of one MMP-1 allele, not allelic loss. Indeed, there are examples of amplification at 11q13 associated with aggressive breast carcinoma 32 and rare examples of MLL amplification at 11q23 associated with acute myelogenous leukemia. 33 Although we did not investigate this possibility, it seems to be an unlikely explanation for our findings, because a large body of literature has documented frequent LOH at this locus, not gene amplification, in many tumors including melanoma. Furthermore, we have preliminary data from analysis of satellite markers flanking the MMP-1 locus that large regions of LOH occurred in five metastatic melanomas that were tested, in some cases extending at least over the region 11q21-q24 (data not shown). Finally, a recent report describes allelic loss at 11q23 in regional lymph node metastases in melanoma, providing further support for our findings. 34
Loss of regions of chromosomes 1, 3, 6, 7, 8, 9, 10, and 18 has been documented in many tumors, 18-26 suggesting that genes in several chromosomal locations may be involved in tumor formation and/or progression. LOH is often thought to indicate the loss of one copy of a tumor suppressor gene at that locus, the other copy previously having undergone loss or inactivation by another mechanism, such as point mutation. LOH at 11q22-q23 has been described in numerous cancers, including melanoma, breast, ovarian, lung, and oral squamous cell carcinomas. 18-26 The region involved includes a known tumor suppressor gene, ATM, 19 as well as several candidate tumor suppressor genes, eg, IGSF4, 35 VACM-1, 36 and PPP2R1B. 20
It is likely that deletion of one or more of these or other genes at this locus is a major determinant of aggressive behavior and metastasis in melanoma, independent of the patient’s MMP-1 genotype. Because deletions at this locus may be large, it is also likely that the deleted region will often include the MMP-1 gene. Furthermore, because it is reasonable to assume that LOH is a random event, there is an equal probability of retaining either of the two MMP-1 alleles. Consequently, if a patient is a 1G/2G MMP-1 heterozygote, and deletion occurs on the chromosome that carries the 1G allele, leaving behind an active 2G allele, that tumor cell may have an additional growth advantage over a cell that undergoes a similar deletion of 11q on the chromosome carrying the 2G allele.
A model of how a tumor suppressor gene and MMP-1 may cooperate to enhance tumor growth and invasion is suggested by a recent study of PPP2R1B, a candidate tumor suppressor gene located in an interval that flanks the MMP-1 locus. 20 PPP2R1B encodes an isoform of the A subunit of the serine/threonine protein phosphatase 2A. Deletion/mutation of this gene generates a truncated protein that is unable to bind to the catalytic subunit of the PP2A holoenzyme, suggesting that the PP2R1B gene product may suppress tumor development through its role in cell cycle regulation and control of cell growth. If loss of this tumor suppressor gene coincides with LOH at the MMP-1 locus, tumor growth may be enhanced through de-regulation of cell growth, and the invasive potential of the tumor is maximized if the 2G allele is retained.
Increasing evidence suggests that elevated MMP-1 expression is associated with tumor invasiveness and with a poor prognosis in a variety of cancers, including colorectal, esophageal, breast, ovarian and endometrial carcinomas, and melanoma. 12-17 Genotyping for the MMP-1 SNP, and rapid quantitative analysis of LOH in tumors may, therefore, serve as an additional prognostic indicator.
Acknowledgments
We thank Dr. Daniel S. Longnecker for instruction in tissue microdissection and for reviewing the manuscript; Dr. Barbara Schaeffer for organizing the logistics of sample procurement and distribution; and Christine Hodorowski for organizing the preparation of the histological sections.
Footnotes
Address reprint requests to Constance E. Brinckerhoff, Ph.D., Dartmouth Medical School, Depts. of Medicine and Biochemistry, HB 7200, Hanover, NH 03755. E-mail: c.e.brinckerhoff@dartmouth.edu.
Supported by National Institute of Health grants CA-77267 (to C. E. B.), ST32-CA09658 (to J. L. R.), CA-23108 to the Norris Cotton Cancer Center from the National Cancer Institute, and a grant from the Ronya and Gregory Kozmetsky Foundation (to C. E. B.).
Present address of Joni L. Rutter, Ph.D.: National Cancer Institute, NIH, DCEG, Lab of Population Genetics, 41 Library Dr./Bldg. 41/Room D701, Bethesda, MD 20892 MSC 5060.
References
- 1.Vincenti MP, White LA, Schroen DJ, Benbow U, Brinckerhoff CE: Regulating expression of the gene for matrix metalloproteinase-1 (collagenase): mechanisms that control enzyme activity, transcription and mRNA stability. Crit Rev Eucaryotic Gene Express 1996, 6:391-411 [DOI] [PubMed] [Google Scholar]
- 2.Borden P, Heller RA: Transcriptional control of matrix metalloproteinases and the tissue inhibitors of matrix metalloproteinases. Crit Rev Eukaryotic Gene Express 1997, 7:159-178 [DOI] [PubMed] [Google Scholar]
- 3.Nagase H, Woessner JF, Jr: Matrix metalloproteinases. J Biol Chem 1999, 274:21491-21492 [DOI] [PubMed] [Google Scholar]
- 4.Yoshihara Y, Nakamura H, Obata K, Yamada H, Hayakawa T, Fujikawa K, Okada Y: Matrix metalloproteinases and tissue inhibitor of metalloproteinases in synovial fluids from patients with rheumatoid arthritis or osteoarthritis. Ann Rheum Dis 2000, 59:455-461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nomura T, Ishii A, Oishi Y, Kohma H, Hara K: Tissue inhibitor of metalloproteinases level and collagenase activity in gingival crevicular fluid: the relevance to periodontal disease. Oral Dis 1998, 4:231-240 [DOI] [PubMed] [Google Scholar]
- 6.Wassenaar A, Verschoor T, Kievits F, Den Hartog MT, Kapsenberg ML, Everts V, Snijders A: CD40 engagement modulates the production of matrix metalloproteinases by gingival fibroblasts. Clin Exp Immunol 1999, 115:161-167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galis ZS, Sukhova GK, Lark M, Libby P: Increased expression of matrix metalloproteinases and matrix degrading activity in vulnerable regions of human atherosclerotic plaques. J Clin Invest 1994, 94:2493-2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rutter JL, Mitchell TI, Buttice G, Meyers J, Gusella JF, Ozelius LJ, Brinckerhoff CE: A single nucleotide polymorphism in the matrix melloproteinase-1 promoter creates an Ets binding site and augments transcription. Cancer Res 1998, 58:5321-5325 [PubMed] [Google Scholar]
- 9.Giambernardi TA, Grant GM, Taylor GP, Hay RJ, Maher VM, McCormick JJ, Klebe RJ: Overview of matrix metalloproteinase expression in cultured human cells. Matrix Biol 1999, 16:483-496 [DOI] [PubMed] [Google Scholar]
- 10.Durko M, Navab R, Shibata HR, Brodt P: Suppression of basement membrane type IV collagen degradation and cell invasion in human melanoma cells expressing an antisense RNA for MMP-1. Biochem Biophys Acta 1997, 1356:271-280 [DOI] [PubMed] [Google Scholar]
- 11.Benbow U, Schoenermark MP, Orndorff KA, Given AL, Brinckerhoff CE: Human breast cancer cells activate procollagenase-1 and invade type I collagen: invasion is inhibited by all-trans retinoic acid. Clin Exp Metastasis 1999, 17:213-238 [DOI] [PubMed] [Google Scholar]
- 12.Murray GI, Duncan ME, O’Neil P, Melvin WT, Fothergill JE: Matrix metalloproteinase-1 is associated with poor prognosis in colorectal cancer. Nat Med 1996, 2:461-462 [DOI] [PubMed] [Google Scholar]
- 13.Murray GI, Duncan ME, O’Neil P, McKay JA, Melvin WT, Fothergill JE: Matrix metalloproteinase-1 is associated with poor prognosis in oesophageal cancer. J Pathol 1998, 185:256-261 [DOI] [PubMed] [Google Scholar]
- 14.Ito T, Ito M, Shiozawa J, Naito S, Kanematsu T, Sekine I: Expression of the MMP-1 in human pancreatic carcinoma: relationship with prognostic factor. Mod Pathol 1999, 12:669-674 [PubMed] [Google Scholar]
- 15.Airola K, Karonen T, Vaalamo M, Lehti K, Lohi J, Kariniemi AL, Keski-Oja J, Saarialho-Kee UK: Expression of collagenases-1 and -3 and their inhibitors TIMP-1 and -3 correlates with the level of invasion in malignant melanomas. Br J Cancer 1999, 80:733-743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanamori Y, Matsushima M, Minaguchi T, Kobayashi K, Sagae S, Kudo R, Terakawa N, Nakamura Y: Correlation between expression of the matrix metalloproteinase-1 gene in ovarian cancers and an insertion/deletion polymorphism in its promoter region. Cancer Res 1999, 59:4225-4227 [PubMed] [Google Scholar]
- 17.Nishioka Y, Kobayashi K, Sagae S, Ishioka S, Nishikawa A, Matsushima M, Kanamori Y, Minaguchi T, Nakamura Y, Tokino T, Kudo R: A single nucleotide polymorphism in the matrix metalloproteinase-1 promoter in endometrial carcinomas. Jpn J Cancer Res 2000, 91:612-615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Driouch K, Briffod M, Bieche I, Champeme M-H, Lidereau R: Location of several putative genes possibly involved in human breast cancer progression. Cancer Res 1998, 58:2081-2086 [PubMed] [Google Scholar]
- 19.Laake K, Launonen B, Niederacher D, Gudlaugsdottir S, Seitz S, Rio P, Champeme M-H, Bieche I, Birnbaum D, White G, Sztan M, Sever N, Plummer S, Osorio A, Broeks A, Huusko P, Spurr N, Borg A, Cleton-Jansen A-M, van’t Veer L, Benitez J, Casey G, Peterlin B, Olah E, Varley J, Bignon Y-J, Scherneck S, Sigurdardottir V, Lidereau R, Eyfjord J, Beckmann MW, Winqvist R, Skovlund E, Borresen-Dale A-L, Breast Cancer Somatic Genetics Consortium: Loss of Heterozygosity at 11q23.1 and survival in breast cancer: results of a large European study. Genes, Chromosom Cancer 1999, 25:212–221 [PubMed]
- 20.Wang SS, Esplin ED, Li JL, H L, G A, Minna J, Evans GA: Alterations of the PPP2R1B gene in human lung and colon cancer. Science 1998, 282:284–287 [DOI] [PubMed]
- 21.Herbst RA, Larson A, Weiss J, Cavenee WK, Hampton GM, Arden KC: A defined region of loss of heterozygosity at 11q23 in cutaneous malignant melanoma. Cancer Res 1995, 55:2494-2496 [PubMed] [Google Scholar]
- 22.Herbst RA, Gutzmer R, Matiaske F, Mommert S, Casper U, Kapp A, Weiss J: Identification of two distinct deletion targets at 11q23 in cutaneous malignant melanoma. Int J Cancer 1999, 80:205-209 [DOI] [PubMed] [Google Scholar]
- 23.Launonen V, Stenback F, Puistola U, Bloigu R, Huuski P, Kytola K, Kauppila A, Winqvist R: Chromosome 11q22.3-q25 LOH in ovarian cancer: association with a more aggressive disease course and involved subregions. Gynecol Oncol 1998, 71:299-304 [DOI] [PubMed] [Google Scholar]
- 24.Lazar AD, Winter MR, Nogueira CP, Larson PS, Finnemore EM, Dolan RW, Fuleihan N, Chakravarti A, Zietman A, Rosenberg CL: Loss of heterozygosity at 11q23 in squamous cell carcinoma of the head and neck is associated with recurrent disease. Clin Cancer Res 1998, 4:2787-2793 [PubMed] [Google Scholar]
- 25.Morita R, Fujimoto A, Hatta N, Takehara K, Takata M: Comparison of genetic profiles between primary melanoma and their metastases reveals genetic alterations and clonal evolution during progression. J Invest Dermatol 1998, 111:919-924 [DOI] [PubMed] [Google Scholar]
- 26.Phillips KK, Welch DR, Miele ME, Lee J-H, Wei LL, Weissman BE: Suppression of MDA-MB-435 breast carcinoma cell metastasis following the introduction of human chromosome 11. Cancer Res 1996, 56:1222-1227 [PubMed] [Google Scholar]
- 27.Pendas AM, Balbin M, Llano E, Jimenez MG, Lopez-Otin C: Structural analysis and promoter characterization of the human collagenase-3 gene (MMP-13). Genomics 1997, 40:222-233 [DOI] [PubMed] [Google Scholar]
- 28.Price K, Linge C: The presence of melanin in genomic DNA isolated from pigmented cell lines interferes with successful polymerase chain reaction: a solution. Melanoma Res 1999, 9:5-9 [DOI] [PubMed] [Google Scholar]
- 29.Magnuson VL, Ally DS, Nylund SJ, Rayman JB, Knapp JI, Lowe AL, Ghosh S, Collins FS: Substrate nucleotide-determined non-templated addition of adenine by Taq DNA polymerase: implications for PCR-based genotyping and cloning. Biotechniques 1996, 21:700-709 [DOI] [PubMed] [Google Scholar]
- 30.Dunleavey L, Seyyare B, Ye S: Rapid genotype analysis of the matrix metalloproteinase-1 gene 1G/2G polymorphism that is associated with risk of cancer. Matrix Biol 2000, 19:175-177 [DOI] [PubMed] [Google Scholar]
- 31.Conover WJ: Practical Nonparametric Statistics, 2nd ed. 1980, Wiley, New York
- 32.Schuuring E: The involvement of the chromosome 11q23 region in human malignancies: cyclin D1 and EMS1 are two new candidate oncogenes—-a review. Gene 1995, 159:83-96 [DOI] [PubMed] [Google Scholar]
- 33.Park JP, Ladd SL, Ely P, Weiner NJ, Wojiski SA, Hawk AB, Noll WW, Mohandas TK: Amplification of the MLL region in acute myeloid leukemia. Cancer Genet Cytogenet 2000, 121:198-205 [DOI] [PubMed] [Google Scholar]
- 34.Herbst RA, Mommert S, Caper U, Podewski EK, Kiehl P, Kapp A, Weiss J: 11q23 allelic loss is associated with regional lymph node metastasis in melanoma. Clin Cancer Res 2000, 6:3222-3227 [PubMed] [Google Scholar]
- 35.Gomyo H, Arai Y, Tanigami A, Murakami Y, Hattori M, Hosoda F, Arai K, Aikawa Y, Tsuda H, Hirohashi S, Asakawa S, Shimizu N, Soeda E, Sakaki Y, Ohki M: A 2-Mb sequence-ready contig map and a novel immunoglobulin superfamily gene IGSF4 in the LOH region of chromosome 11q23.2. Genomics 1999, 62:139-146 [DOI] [PubMed] [Google Scholar]
- 36.Byrd PJ, Stankovic T, McConville CM, Smith AD, Cooper PR, Taylor AM: Identification and analysis of expression of human VACM-1, a cullin gene family member located on chromosome 11q22–23. Genome Res 1997, 7:71-75 [DOI] [PubMed] [Google Scholar]