Abstract
Caveolae are plasma membrane microdomains that have been implicated in the regulation of several intracellular signaling pathways. Previous studies suggest that caveolin-1, the main structural protein of caveolae, could function as a tumor suppressor. Caveolin-1 is highly expressed in terminally differentiated mesenchymal cells including adipocytes, endothelial cells, and smooth muscle cells. To study whether caveolin-1 is a possible tumor suppressor in human mesenchymal tumors, we have analyzed the expression using immunohistochemistry in normal mesenchymal tissues, 22 benign and 79 malignant mesenchymal tumors. Caveolin-1 was found to be expressed in fibromatoses, leiomyomas, hemangiomas, and lipomas at high levels comparable to normal mesenchymal tissues. The expression of caveolin-1 was slightly reduced in four of six well-differentiated liposarcomas and strongly reduced or lost in three of three fibrosarcomas, 17 of 20 leiomyosarcomas, 16 of 16 myxoid/round cell/pleomorphic liposarcomas, five of eight angiosarcomas, 15 of 18 malignant fibrous histiocytomas, and eight of eight synovial sarcomas. The immunohistochemical findings were confirmed by Western blot analysis in a number of tumors. High levels of both the 24-kd [α]- and the 21-kd [β]-isoform of caveolin-1 were detected in the nontumorigenic human fibroblast cell line IMR-90. In contrast, in HT-1080 human fibrosarcoma cells, caveolin-1 is strongly down-regulated. We show that the [α]-isoform of caveolin-1 is potently up-regulated in HT-1080 cells by inhibition of the mitogen-activated protein kinase-signaling pathway with the specific inhibitor PD 98059, whereas the specific inhibitor of DNA methylation 5-aza-2′-deoxycytidine only marginally up-regulates caveolin-1. In addition, re-expression of caveolin-1 in HT-1080 fibrosarcoma cells potently inhibited colony formation. From these we conclude that caveolin-1 is likely to act as a tumor suppressor gene in human sarcomas.
Caveolae are 50- to 100-nm ω-shaped invaginations of the plasma membrane that have been implicated in endocytosis and signal transduction. 1 Caveolae membranes are enriched with specific lipids (glycosphingolipids, sphingomyelin, and cholesterol) and different lipid-modified signaling molecules. 2 Caveolins, a family of highly conserved 20- to 25-kd integral membrane proteins, are the principal protein component of caveolae. Thus far the mammalian caveolin family consists of four proteins, caveolin-1[α], -1[β], -2, and -3 encoded by three genes. Caveolins-1 and -2 are co-expressed, whereas the expression of caveolin-3 is muscle-specific. 3
In general, caveolins bind to and inactivate signaling molecules including receptor tyrosine kinases, their downstream targets (eg, H-RAS, MEK1, and ERK2), serpentine receptors, G-proteins, and eNOS. 4 Direct interaction of caveolin-1 with signaling molecules leads to their inhibition, 5 therefore it has been suggested that caveolin-1 may possess transformation suppressor activity. Consistent with this hypothesis it was shown that 1) the caveolin-1 and -2 genes are co-localized to a suspected tumor suppressor locus in mice and humans [7q31.1/D7S522] 6 and, 2) the first and second exons of the caveolin-1 gene are embedded within CpG islands. 7 Therefore, it was proposed that regulation of caveolin-1 expression may be controlled, in part, by methylation of these regions. 3) Caveolin-1 expression is reduced or absent in NIH 3T3 cells transformed by activated oncogenes such as v-Abl, Bcr-Abl, or H-Ras [G12V], and caveolae are missing from these transformed cells. 8 4) In addition, activation of the Ras-p42/44 mitogen-activated protein (MAP) kinase and protein kinase A (PKA) signaling pathways transcriptionally down-regulate caveolin-1. 9 5) Most importantly, recombinant expression of caveolin-1 in transformed NIH 3T3 cells and breast cancer cells inhibits anchorage-independent growth. 10,11
Caveolin-1 is most abundantly expressed in terminally differentiated mesenchymal cells such as smooth muscle cells, adipocytes, and endothelial cells. 5 Tumors of the soft tissue and bone represent a heterogeneous group of mesenchymal lesions, which account for ∼1% of all malignancies. Impaired function or inactivation of several tumor suppressor gene products has been implicated in the development of soft tissue tumors, including p53, the retinoblastoma gene product, p16INK4A, p18, and p21WAF1/CIP1. 12 To investigate whether caveolin-1 is a candidate tumor suppressor in sarcomas, we have analyzed its expression in normal human mesenchymal tissues, benign mesenchymal tumors, and sarcomas. We demonstrate that caveolin-1 is expressed in benign mesenchymal tumors at levels comparable to normal mesenchymal cells, whereas the majority of sarcomas have a reduced expression. Our data suggests that caveolin-1 may act as a tumor suppressor in human sarcomas.
Materials and Methods
Tissue Samples
This study was conducted on formalin-fixed, paraffin-embedded specimens of benign and malignant mesenchymal tumors selected from the archives of the Institutes of Pathology, Universitätsklinikum Charité, Berlin, Germany. Histopathological diagnosis and grading of the tumors were performed using established diagnostic criteria. 13,14 The tissues analyzed included seven leiomyomas, six hemangiomas, five fibromatoses, four lipomas, three fibrosarcomas (two grade 1 and one grade 2), 20 leiomyosarcomas (four grade 1, eight grade 2, and eight grade 3), 22 liposarcomas (six well-differentiated, 14 myxoid/round cell, and two pleomorphic), eight angiosarcomas (two grade 1, one grade 2, and five grade 3), 18 malignant fibrous histiocytomas (six grade 2 and 12 grade 3), and eight synovial sarcomas (six grade 2 and two grade 3). Normal mesenchymal tissues surrounding the tumors were analyzed for each case. In addition, snap-frozen benign and malignant tissues of selected cases were used for Western blot analysis.
Immunohistochemistry
Paraffin sections were cut on to silane-coated slides. Antigen retrieval was performed on deparaffinized sections by boiling in 10 mmol/L sodium citrate buffer, pH 6.0, for 5 minutes. The mouse monoclonal anti-caveolin-1 antibody (clone 2297; Transduction Labs, Lexington, KY) was applied for 1 hour at a dilution of 1:500. To ensure consistency of staining intensity, normal mesenchymal tissue at the periphery of each tumor specimen and endothelial cells of capillaries within the tumors were evaluated as a positive control. In negative controls the primary antibody was omitted or replaced by an antibody with irrelevant specificity (mouse IgG1, X 0931; DAKO, Hamburg, Germany). Immunostaining was accomplished using a Vectastain ABC-AP kit (Vector Laboratories, Burlingame, CA) as recommended by the manufacturer. The sections were then counterstained with hematoxylin and mounted in Permount. Digital images were acquired with an Olympus DP-10 charge-coupled device camera. Adjustments in image contrast were performed identically on the images.
For caveolin-1 immunostaining, a semiquantitative estimation was made, using a composite score obtained by multiplying the values of the immunoreaction intensity and relative abundance of caveolin-1-positive cells slightly modified as described previously. 15 The intensity was graded as 1 (weakly positive), 2 (moderately positive), or 3 (intense stain equivalent to normal smooth muscle cells, adipocytes, or endothelial cells). The abundance of caveolin-1-positive cells was graded from 0 to 4 (0, <5% positive cells; 1, 5 to 25%; 2, 26 to 50%; 3, 51 to 75%; 4, 76 to 100%).
Cell Culture and Inhibitors
The fibrosarcoma cell line HT-1080 and the SV40-immortalized nontumorigenic fibroblast cell line IMR-90 were obtained from the American Type Culture Collection (Rockville, MD). Cell lines were maintained at 37°C in a humidified atmosphere of 5% CO2 and 95% air in Dulbecco’s modified Eagle’s medium containing 10% fetal calf serum and 2 mmol/L glutamine. The MAP kinase or ERK kinase (MEK) inhibitor PD 98059 (Calbiochem, San Diego, CA) was dissolved in dimethyl sulfoxide at a concentration of 50 mmol/L and used at a final concentration of 50 μmol/L for 48 hours. The inhibitor of DNA methylation, 5-aza-2′-deoxycytidine (Sigma, St. Louis, MO), was dissolved in phosphate buffer, pH 6.0, at a concentration of 1 mmol/L and applied at a concentration of 1 μmol/L for 48 hours.
Western Blot Analysis
To obtain total protein lysates, 2 × 10 6 cells plated on 10-cm dishes, were washed twice with cold phosphate-buffered saline and incubated on ice for 30 minutes in RIPA-buffer [150 mmol/L NaCl, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% sodium dodecyl sulfate (SDS), 50 mmol/L Tris-HCl, pH 8.0, and 2 μg/ml aprotinin]. Cells were scraped off the plates, lysates were mixed with 2× SDS sample buffer (120 mmol/L Tris-HCl, pH 6.8, 0.2 mol/L dithiothreitol, 4% SDS, 20% glycerol, 0.002% bromphenol blue), boiled for 5 minutes, and centrifuged for 5 minutes at 12,000 × g. The protein concentration of the supernatants was measured using the amido black method as described by Schaffner and Weissmann. 16 Equal amounts of protein (10 μg) were separated on 12% polyacrylamide gels by SDS-gel electrophoresis and transferred to polyvinylidene difluoride membranes (Hybond-P; Amersham, Freiburg, Germany). Immunodetection was performed using the caveolin-1-specific monoclonal antibody clone 2297 at a dilution of 1:1,000 followed by detection with the enhanced chemiluminescence system (Amersham). To ensure equal loading amounts, the blots were stripped in 200 mmol/L glycine, 1% Tween-20, 0.1% SDS, pH 2.2, for 2 hours at room temperature and reprobed using a monoclonal antibody to actin (clone C4; Roche, Mannheim, Germany) at a dilution of 1:5,000. Frozen samples of tumors and benign tissues were homogenized in RIPA buffer and processed as described above.
Caveolin-1 Expression Construct
A 631-bp cDNA fragment containing the complete coding region of human caveolin-1 was amplified by reverse transcriptase-polymerase chain reaction using the caveolin-1-specific forward primer 5′ CCTCCTCACAGTTTTCATCCA 3′ and the reverse primer 5′ ACTTGAAATTGGCACCAGGA 3′ and cloned into the HpaI site of the pLNHX expression vector (Clontech, Palo Alto, CA). The insert was sequenced to ensure for a correct caveolin-1 sequence.
Colony Formation Assay
HT-1080 cells (2 × 105) were plated onto 25 cm 2 dishes and transfected after 24 hours with 1.5 μg of caveolin-1 expression plasmid or the empty pLNHX vector as a control using the Fugene 6 reagent (Roche) according to the manufacturers instructions. Forty-eight hours after transfection, 850 μg/ml of geneticin (G418) was added to the culture medium and the colonies were stained and counted after 10 days of selection. In addition, several G418-resistant colonies with the caveolin-1 expression plasmid and the pLNHX vector were expanded and analyzed for caveolin-1 expression by Western blot as described above.
Statistical Analyses
Statistical analysis were made using Fisher’s exact test. Differences between two populations were considered significant when confidence intervals were >95% (P < 0.05).
Results
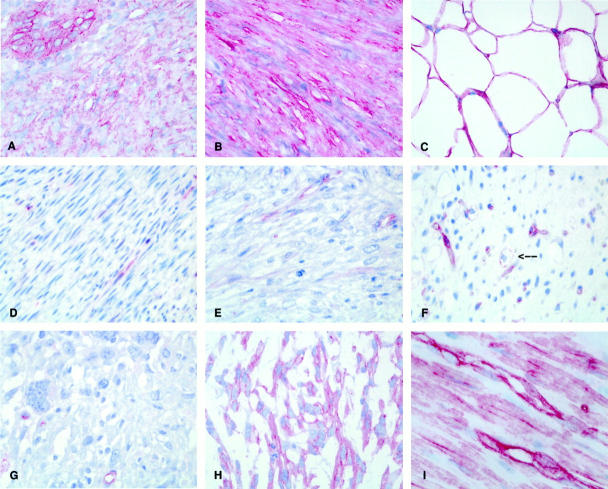
Caveolin-1 expression was detected using immunohistochemistry at high levels in fibroblasts, smooth muscle cells, adipocytes, and in endothelial cells with a fine granular membranous and a diffuse cytoplasmic staining pattern (Figure 1I) ▶ . The semiquantitative expression results for benign mesenchymal tumors and sarcomas are summarized in Table 1 ▶ . High levels of caveolin-1 expression, comparable to normal mesenchymal cells, were retained in all benign mesenchymal tumors, including five of five fibromatoses (Figure 1A) ▶ , seven of seven leiomyomas (Figure 1B) ▶ , four of four lipomas (Figure 1C) ▶ , and six of six hemangiomas. Caveolin-1 expression was found to be absent or strongly reduced in three of three fibrosarcomas (Figure 1D) ▶ , 17 of 20 leiomyosarcomas (Figure 1E) ▶ , five of eight angiosarcomas, 15 of 18 malignant fibrous histiocytomas (Figure 1G) ▶ , and eight of eight synovial sarcomas. Most of these sarcomas show a weak staining in distinct areas of the tumor.
Figure 1.
Expression of caveolin-1 in normal mesenchymal tissue, benign mesenchymal tumors and sarcomas detected by immunohistochemistry with the monoclonal caveolin-1-specific antibody 2297. A–C: High levels of caveolin-1 protein comparable to normal mesenchymal cells in benign mesenchymal tumors: fibromatosis (A), leiomyoma (B), and lipoma (C). D–G: Reduction of caveolin-1 in sarcomas: fibrosarcoma (D), leiomyosarcoma (E), myxoid liposarcoma (F), malignant fibrous histiocytoma (G). Note caveolin-1-positive endothelial cells within negative tumors (D–G) and single caveolin-1-positive lipoblasts in myxoid liposarcoma (arrow in F). H: High levels of caveolin-1 expression in a subset of sarcomas (malignant fibrous histiocytoma, case 1 in Table 2 ▶ ). I: High levels of caveolin-1 expression in normal mesenchymal tissue (smooth muscle and endothelial cells from stomach). Original magnifications: ×400 (A–G), ×600 (I).
Table 1.
Expression of Caveolin-1 Protein in Human Mesenchymal Tumors
| Histology | Histological grade | Number of samples | Mean staining intensity | % of stained cells | Mean score ± SEM |
|---|---|---|---|---|---|
| Fibroblastic tumors | |||||
| Fibromatoses | 5 | 3.0 | 83 | 11.4 ± 0.6 | |
| Fibrosarcoma | Grade 1 | 2 | 0.5 | 10 | 0.5 |
| Grade 2 | 1 | 0.0 | 0 | 0.0 | |
| Smooth muscle tissue tumors | |||||
| Leiomyoma | 7 | 2.7 | 88 | 11.1 ± 0.9 | |
| Leiomyosarcoma | Grade 1 | 4 | 2.0 | 45 | 5.3 ± 1.9 |
| Grade 2 | 8 | 1.0 | 21 | 2.4 ± 1.1 | |
| Grade 3 | 8 | 1.3 | 25 | 2.4 ± 1.1 | |
| Lipogenic tumors | |||||
| Lipoma | 4 | 3.0 | 92 | 12.0 | |
| Well-differentiated liposarcoma | 6 | 2.3 | 55 | 8.0 | |
| Myxoid/round cell liposarcoma | 14 | 1.2 | 11 | 0.9 | |
| Pleomorphic liposarcoma | 2 | 1.5 | 15 | 1.5 | |
| Vascular tumors | |||||
| Hemangioma | 6 | 3.0 | 100 | 12.0 | |
| Angiosarcoma | Grade 1 | 2 | 3.0 | 60 | 9.0 |
| Grade 2 | 1 | 3.0 | 50 | 6.0 | |
| Grade 3 | 5 | 1.8 | 54 | 4.0 ± 1.6 | |
| Malignant fibrous histiocytoma | Grade 2 | 6 | 2.2 | 45 | 5.0 ± 1.5 |
| Grade 3 | 12 | 1.4 | 22 | 2.7 ± 1.1 | |
| Synovial sarcoma | Grade 2 | 6 | 0.5 | 15 | 1.3 ± 1.0 |
| Grade 3 | 2 | 0.0 | 0 | 0.0 |
Score, obtained by multiplying the values of the immunoreaction intensity and relative abundance of caveolin-1-positive cells as described in Materials and Methods.
In three malignant fibrous histiocytomas (Figure 1H) ▶ , three leiomyosarcomas, and three angiosarcomas high levels of caveolin-1 protein expression (immunoreactivity scores ranging from 8 to 12) were detected. Although expression of caveolin-1 in leiomyosarcomas, angiosarcomas, and malignant fibrous histiocytomas displayed some correlation with the histological grade, these differences were not significant.
Regarding malignant lipogenic tumors, a marginally reduced expression of caveolin-1 was observed in four of six well-differentiated liposarcomas, whereas in myxoid/round cell and pleomorphic liposarcomas the caveolin-1 expression was strongly reduced or absent in all 16 tumors. In these forms of liposarcomas, only some scattered multivacuolar and univacuolar lipoblasts stained positive for caveolin-1 (Figure 1F ▶ , arrow).
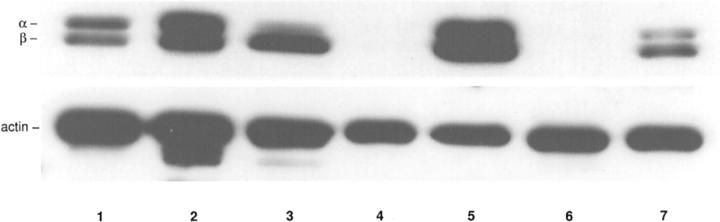
The immunohistochemical findings were confirmed by Western blot analysis of normal mesenchymal tissues and tumors with high and low expression of caveolin-1, where snap-frozen tissue was available (Table 2 ▶ and Figure 2 ▶ ). In two samples derived from normal mesenchymal tissue and three tumors with a high expression of caveolin-1 in immunohistochemistry (Table 2 ▶ ; cases 1, 3, and 5), the 24-kd [α]- and the 21-kd [β]-isoforms of caveolin-1 were detectable by Western blot analysis (Figure 2 ▶ ; lanes 1, 2, 3, 5, and 7). In two tumors with a reduced expression of caveolin-1 in immunohistochemistry (Table 2 ▶ , cases 2 and 4) no caveolin-1 protein was detected by Western blot analysis (Figure 2 ▶ , lanes 4 and 6).
Table 2.
Tumors Selected for Western Blot Analysis of Caveolin-1 Expression
| Case no. | Histology | Staining intensity | Percentage of positive cells | Score |
|---|---|---|---|---|
| 1 | MFH | 3+ | 70 | 9 |
| 2 | MFH | 2+ | 30 | 4 |
| 3 | MFH | 3+ | 90 | 12 |
| 4 | Leiomyosarcoma | 1+ | 50 | 2 |
| 5 | Leiomyosarcoma | 3+ | 70 | 9 |
Score, obtained by multiplying the values of the immunoreaction intensity and relative abundance of caveolin-1-positive cells as described in Materials and Methods.
MFH, malignant fibrous histiocytoma.
Figure 2.
Top: Western blot analysis of caveolin-1 expression in normal mesenchymal tissues and sarcomas. In normal mesenchymal tissues (lanes 1 and 2) and three tumors (lanes 3, 5, and 7) with a high expression of caveolin-1 in immunohistochemistry (Table 2 ▶ ; cases 1, 3, and 5) the 24-kd [α]- and the 21-kd [β]-isoforms of caveolin-1 were detectable. In two tumors (lanes 4 and 6) with a reduced expression of caveolin-1 in immunohistochemistry (Table 2 ▶ , cases 2 and 4) no caveolin-1 protein was detected. Bottom: To ensure equal loading amounts, the blots were stripped and reprobed with an antibody to actin.
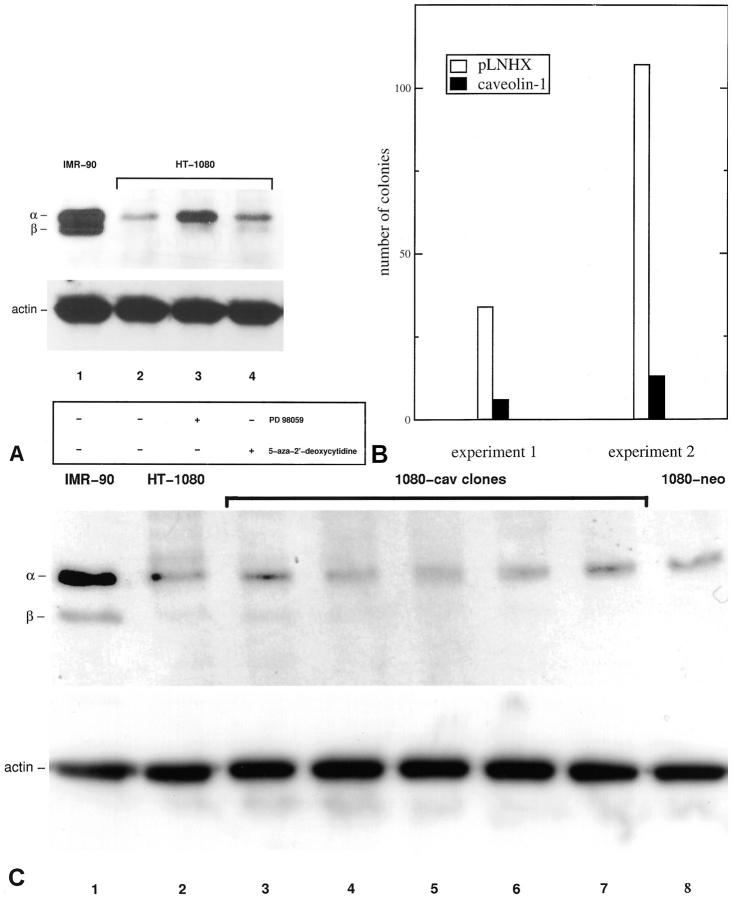
Next, we examined the expression of caveolin-1 in the nontumorigenic human fibroblast cell line IMR-90 and in the human fibrosarcoma cell line HT-1080. As shown in Figure 3A ▶ , IMR-90 cells express high levels of the 24-kd [α]- and the 21-kd [β]-isoform of caveolin-1. In contrast, caveolin-1 expression is strongly down-regulated in HT-1080 fibrosarcoma cells (Figure 3A ▶ , lanes 1 and 2). Treatment of HT-1080 cells with the well-characterized MEK inhibitor PD 98059 (50 μmol/L) potently up-regulates the expression of the 24-kd [α]-isoform of caveolin-1, whereas treatment with the inhibitor of DNA methylation, 5-aza-2′-deoxycytidine (1 μmol/L), only marginally up-regulates caveolin-1 (Figure 3A ▶ , lanes 3 and 4).
Figure 3.
A: Western blot analysis of caveolin-1 expression in the nontumorigenic human fibroblast cell line IMR-90 and in the human fibrosarcoma cell line HT-1080 (top). Lane 1: IMR-90 cells express high levels of the 24-kd [α]- and the 21-kd [β]-isoform of caveolin-1. Lane 2: Caveolin-1 expression is strongly down-regulated in HT-1080 fibrosarcoma cells. Lane 3: Inhibition of the MAP kinase pathway by treatment of HT-1080 cells with the MEK inhibitor PD 98059 (50 μmol/L) for 48 hours potently up-regulates the expression of the 24-kd [α]-isoform of caveolin-1. Lane 4: Treatment with the inhibitor of DNA methylation, 5-aza-2′-deoxycytidine (1 μmol/L) for 48 hours, only marginally up-regulates caveolin-1. To ensure equal loading amounts, the blots were stripped and re probed with an antibody to actin (bottom). B: Colony formation assay demonstrating growth suppression of HT-1080 fibrosarcoma cells by expression of caveolin-1. Two independent experiments are shown. HT-1080 cells were transfected with either the caveolin-1 expression plasmid or the empty pLNHX vector as a control. Cells were selected in medium containing 850 μg/ml of G418 for 10 days, fixed, stained, and the colonies were counted. C: Western blot analysis of caveolin-1 expression of expanded G418-resistant colonies transfected with the caveolin-1 plasmid (top). No stable overexpression is observed in HT-1080 clones with the caveolin-1 plasmid. Lane 1: IMR-90-positive control; lane 2, parental HT-1080 cells; lanes 3–7, individual G418-resistant HT-1080 clones transfected with the caveolin-1 plasmid; lane 8, neo control (pLNHX). Bottom: actin control.
To determine, whether caveolin-1 affects growth of human sarcoma cells, we transfected HT-1080 cells with a caveolin-1 expression vector. Colony formation, as measured after G418 selection, was reduced to 18 and 12% of the vector control in two independent experiments (Figure 3B) ▶ . Additionally, G418-resistant colonies transfected with the caveolin-1 expression vector and the empty vector were expanded and caveolin-1 expression was analyzed by Western blot analysis (Figure 3C) ▶ . In these experiments, however, it was not possible to obtain HT-1080 clones stably overexpressing caveolin-1.
Discussion
Our data presented here shows that caveolin-1 is expressed at high levels in normal mesenchymal tissues and in benign mesenchymal tumors. In contrast, the expression of caveolin-1 is strongly reduced in the majority of sarcomas of various histological types. In support of these in vivo findings, the nontumorigenic fibroblast cell line IMR-90 shows high levels of caveolin-1 expression, whereas in contrast the fibrosarcoma cell line HT-1080 has a strong down-regulation of caveolin-1.
Caveolin-1 was first identified as a major v-Src substrate in Rous sarcoma virus-transformed cells. It was proposed that caveolin-1 may represent a critical target during cell transformation. 17 In support of this notion, caveolin-1 expression is reduced in NIH 3T3 cells transformed by a variety of activated oncogenes, such as v-Abl, Bcr-Abl, and H-Ras [G12V]. 8 Targeted down-regulation of caveolin-1 in NIH 3T3 cells by expression of an antisense RNA induces cell transformation and activation of the p42/44 MAP kinase cascade. 18 Most importantly, heterologous expression of caveolin-1 in transformed NIH 3T3 cells and breast cancer cells inhibits their anchorage-independent growth capabilities. 10,11
The genes encoding caveolin-1 and -2 were previously mapped to the D7S522 locus (7q31.1), a known fragile site that shows deletions using loss of heterozygosity analysis in a variety of human malignancies such as cancers of the breast, 19 prostate, 20 kidney, 21 and colon. 22 Therefore it was suggested that caveolin-1 may represent a tumor suppressor gene at the D7S522 (7q31.1) locus. 6 However, in a variety of human tumor cell lines, including ovarian, breast and prostate cancer, osteosarcoma and HT-1080 fibrosarcoma cell lines, no mutations of the caveolin-1 gene were detected and some of these cell lines express caveolin-1. 23
The exons 1 and 2 of the caveolin-1 gene are embedded within CpG islands 7 and it was proposed that the expression of caveolin-1 may be down-regulated by methylation of these CpG regions. It was recently shown that in two human breast cancer cell lines, that fail to express the protein, the 5′ promoter region of the caveolin-1 gene is methylated, 7 however, others did not detect methylation of the caveolin-1 gene promoter in a variety of human tumor cell lines and human ovarian cancers. 23 We did observe only a marginal up-regulation of caveolin-1 protein expression in HT-1080 fibrosarcoma cells after treatment with the DNA methylation inhibitor 5-aza-2′-deoxycytidine. This indicates that in HT-1080 cells, suppression of caveolin-1 by methylation does not play a significant role, which complements with previous findings for this cell line. 23
It was shown that activation of the Ras-p42/44 MAP kinase and PKA cascades transcriptionally down-regulate caveolin-1 promoter activity in NIH 3T3 fibroblasts. 9 The transformed phenotype of HT-1080 fibrosarcoma cells is controlled by an activating mutation of the N-RAS gene 24 and we observed a re-expression of caveolin-1 by inhibition of the p42/44 MAP kinase pathway by treatment with the MEK inhibitor PD 98059 in this cell line. This effect was selective for the 24-kd [α]-isoform of caveolin-1, indicating a different regulation of the caveolin-1 isoforms. Caveolin-1 isoforms are encoded by distinct mRNAs generated by alternative transcription initiation. 25 We found both [α]- and [β]-isoforms co-expressed in mesenchymal tissues and IMR-90 fibroblasts. It was recently shown that re-expression of the [β]-isoform but not the [α]-isoform of caveolin-1 up-regulates volume-regulated anion channels in colon and breast cancer cell lines. 26 Therefore it would be of interest to analyze whether separate re-expression of the caveolin-1 isoforms exerts different biological effects in mesenchymal tumor cells.
To summarize, our expression data obtained from benign and malignant tissues and cells and from colony formation experiments, suggests that caveolin-1 may act as a class II tumor suppressor in human sarcomas. Class II tumor suppressor genes, 27 as defined by their reversible down-regulation, may be useful because their re-expression may be inducible by drugs and may improve treatment of human tumors.
In contrast to these findings, Thompson and co-workers 28 recently reported elevated caveolin-1 levels in prostate cancer and they show that caveolin-1 suppresses c-myc-induced apoptosis in prostate cancer cell lines. 29 We have analyzed the expression of caveolin-1 in human carcinomas of various sites and found that caveolin-1 is down-regulated in several human cancers but elevated in some other tumors, including prostate cancer (Wiechen K, Diatchenko L, Agoulnik A, Zhumabayeva B, Desai S, Atun S, Scharff KM, Hydes K, Siebert PD, Dietel M, Schafer R, Sers C, submitted for publication). Therefore, caveolin-1 is likely to act as a tumor suppressor in some human malignancies such as sarcomas and may have other functions in prostate cancer.
Further studies are necessary to characterize the mechanism of down-regulation of caveolin-1 in sarcomas. It will be of interest to analyze whether the expression of caveolin-1 in distinct tumor areas of leiomyosarcomas, angiosarcomas, and malignant fibrous histiocytomas and single lipoblasts in myxoid/round cell and pleomorphic liposarcomas reflects a higher differentiation. In addition, it will be important to examine the possible relationship between loss or preservation of caveolin-1 expression and clinical outcome in patients with sarcomas.
Acknowledgments
We thank Anke Sager, Ina Wendler, and Susanne Metzkow for excellent technical assistance and Gabriele Höppner for excellent editorial help.
Footnotes
Address reprint requests to Dr. Kai Wiechen, Institute of Pathology, Universitätsklinikum Charité, Medizinische Fakultät der Humboldt-Universität Berlin, Schumannstr. 20/21, 10117 Berlin, Germany. E-mail: kai.wiechen@charite.de.
The research was performed at the Institute of Pathology, Universitätsklinikum Charité, Medizinische Fakultät der Humboldt-Universität Berlin, 10117 Berlin, Germany and at the Clinic of Surgery and Surgical Oncology, Robert-Rössle Klinik, 13122 Berlin-Buch, Germany.
References
- 1.Severs NJ: Caveolae static inpocketings of the plasma membrane, dynamic vesicles or plain artifact? J Cell Sci 1988, 90:341-348 [DOI] [PubMed] [Google Scholar]
- 2.Schlegel A, Volonte D, Engelman JA, Galbiati F, Mehta P, Zhang XL, Scherer PE, Lisanti MP: Crowded little caves: structure and function of caveolae. Cell Signal 1998, 10:457-463 [DOI] [PubMed] [Google Scholar]
- 3.Parton RG: Caveolae and caveolins. Curr Opin Cell Biol 1996, 8:542-548 [DOI] [PubMed] [Google Scholar]
- 4.Okamoto T, Schlegel A, Scherer PE, Lisanti MP: Caveolins, a family of scaffolding proteins for organizing “preassembled signaling complexes” at the plasma membrane. J Biol Chem 1998, 273:5419-5422 [DOI] [PubMed] [Google Scholar]
- 5.Lisanti MP, Scherer PE, Vidugiriene J, Tang Z, Hermanowski-Vosatka A, Tu YH, Cook RF, Sargiacomo M: Characterization of caveolin-rich membrane domains isolated from an endothelial-rich source: implications for human disease. J Cell Biol 1994, 126:111-126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Engelman JA, Zhang XL, Lisanti MP: Genes encoding human caveolin-1 and -2 are co-localized to the D7S522 locus (7q31.1), a known fragile site (FRA7G) that is frequently deleted in human cancers. FEBS Lett 1998, 436:403-410 [DOI] [PubMed] [Google Scholar]
- 7.Engelman JA, Zhang XL, Lisanti MP: Sequence and detailed organization of the human caveolin-1 and -2 genes located near the D7S522 locus (7q31.1). Methylation of a CpG island in the 5′ promoter region of the caveolin-1 gene in human breast cancer cell lines. FEBS Lett 1999, 448:221-230 [DOI] [PubMed] [Google Scholar]
- 8.Koleske AJ, Baltimore D, Lisanti MP: Reduction of caveolin and caveolae in oncogenically transformed cells. Proc Natl Acad Sci USA 1995, 92:1381-1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engelman JA, Zhang XL, Razani B, Pestell RG, Lisanti MP: p42/44 MAP kinase-dependent and -independent signaling pathways regulate caveolin-1 gene expression. Activation of Ras-MAP kinase and protein kinase a signaling cascades transcriptionally down-regulates caveolin-1 promoter activity. J Biol Chem 1999, 274:32333-32341 [DOI] [PubMed] [Google Scholar]
- 10.Lee SW, Reimer CL, Oh P, Campbell DB, Schnitzer JE: Tumor cell growth inhibition by caveolin re-expression in human breast cancer cells. Oncogene 1998, 16:1391-1397 [DOI] [PubMed] [Google Scholar]
- 11.Engelman JA, Wykoff CC, Yasuhara S, Song KS, Okamoto T, Lisanti MP: Recombinant expression of caveolin-1 in oncogenically transformed cells abrogates anchorage-independent growth. J Biol Chem 1997, 272:16374-16381 [DOI] [PubMed] [Google Scholar]
- 12.Slominski A, Wortsman J, Carlson A, Mihm M, Nickoloff B, McClatchey K: Molecular pathology of soft tissue and bone tumors: a review. Arch Pathol Lab Med 1999, 123:1246-1259 [DOI] [PubMed] [Google Scholar]
- 13.Weiss SW: Histological typing of soft tissue tumours. World Health Organization, International Histological Classification of Tumours. 1994, Springer, Berlin
- 14.Enzinger FM, Weiss SW: Soft Tissue Tumors, ed 3 1994, Mosby, London
- 15.Gamallo C, Palacios J, Moreno G, Calvo de Mora J, Suarez A, Armas A: Beta-catenin expression pattern in stage I and II ovarian carcinomas: relationship with beta-catenin gene mutations, clinicopathological features, and clinical outcome. Am J Pathol 1999, 155:527-536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schaffner W, Weissmann C: A rapid, sensitive, and specific method for the determination of protein in dilute solution. Anal Biochem 1973, 56:502-514 [DOI] [PubMed] [Google Scholar]
- 17.Glenney JR: Tyrosine phosphorylation of a 22-kDa protein is correlated with transformation by Rous sarcoma virus. J Biol Chem 1989, 264:20163-20166 [PubMed] [Google Scholar]
- 18.Galbiati F, Volonte D, Engelman JA, Watanabe G, Burk R, Pestell RG, Lisanti MP: Targeted downregulation of caveolin-1 is sufficient to drive cell transformation and hyperactivate the p42/44 MAP kinase cascade. EMBO J 1998, 17:6633-6648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zenklusen JC, Bieche I, Lidereau R, Conti CJ: (C-A)n microsatellite repeat D7S522 is the most commonly deleted region in human primary breast cancer. Proc Natl Acad Sci USA 1994, 91:12155-12158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jenkins RB, Qian J, Lee HK, Huang H, Hirasawa K, Bostwick DG, Proffitt J, Wilber K, Lieber MM, Liu W, Smith DI: A molecular cytogenetic analysis of 7q31 in prostate cancer. Cancer Res 1998, 58:759-766 [PubMed] [Google Scholar]
- 21.Shridhar V, Sun QC, Miller OJ, Kalemkerian GP, Petros J, Smith DI: Loss of heterozygosity on the long arm of human chromosome 7 in sporadic renal cell carcinomas. Oncogene 1997, 15:2727-2733 [DOI] [PubMed] [Google Scholar]
- 22.Zenklusen JC, Thompson JC, Klein-Szanto AJ, Conti CJ: Frequent loss of heterozygosity in human primary squamous cell and colon carcinomas at 7q31.1: evidence for a broad range tumor suppressor gene. Cancer Res 1995, 55:1347-1350 [PubMed] [Google Scholar]
- 23.Hurlstone AF, Reid G, Reeves JR, Fraser J, Strathdee G, Rahilly M, Parkinson EK, Black DM: Analysis of the CAVEOLIN-1 gene at human chromosome 7q31.1 in primary tumours and tumour-derived cell lines. Oncogene 1999, 18:1881-1890 [DOI] [PubMed] [Google Scholar]
- 24.Paterson H, Reeves B, Brown R, Hall A, Furth M, Bos J, Jones P, Marshall C: Activated N-ras controls the transformed phenotype of HT1080 human fibrosarcoma cells. Cell 1987, 51:803-812 [DOI] [PubMed] [Google Scholar]
- 25.Kogo H, Fujimoto T: Caveolin-1 isoforms are encoded by distinct mRNAs. Identification of mouse caveolin-1 mRNA variants caused by alternative transcription initiation and splicing. FEBS Lett 2000, 465:119-123 [DOI] [PubMed] [Google Scholar]
- 26.Trouet D, Nilius B, Jacobs A, Remacle C, Droogmans G, Eggermont J: Caveolin-1 modulates the activity of the volume-regulated chloride channel. J Physiol 1999, 520:113-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee SW, Tomasetto C, Sager R: Positive selection of candidate tumor-suppressor genes by subtractive hybridization. Proc Natl Acad Sci USA 1991, 88:2825-2829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang G, Truong LD, Wheeler TM, Thompson TC: Caveolin-1 expression in clinically confined human prostate cancer: a novel prognostic marker. Cancer Res 1999, 59:5719-5723 [PubMed] [Google Scholar]
- 29.Timme TL, Goltsov A, Tahir S, Li L, Wang J, Ren C, Johnston RN, Thompson TC: Caveolin-1 is regulated by c-myc and suppresses c-myc-induced apoptosis. Oncogene 2000, 19:3256-3265 [DOI] [PubMed] [Google Scholar]