Abstract
The interaction of proteins with ubiquitin receptors is key to solving the mystery that surrounds the functional role ubiquitin chains play in directing traffic. The specificity of these interactions is largely mediated by UbL/UBA domains. Sequestosome 1/p62 is a protein that is gaining attention as it is intimately involved in cell signaling, receptor internalization, and protein turnover. Herein we review recent advances in the field.
Keywords: Polyubiquitin, p62, scaffold, atypical protein kinase C
1. Overview
The UbL/UBA family of proteins is a rapidly expanding group involved in a diverse set of cellular functions. Members of this family have been shown to transport ubiquitinated cargo for degradation, and protect other proteins from degradation by sequestration of polyubiquitin chains. In S. cerevisiae, Rad23, Dsk2, and Ddi1 shuttle ubiquitinated proteins to the 26S proteasome for degradation. This is accomplished by interaction of their UBA domains with ubiquitinated substrates and binding of their N-terminal UbL domains with the proteasome [1]. Conversely, the Rad23 UBA2 domain sequesters K48 polyubiquitin chains preventing their elongation or de-ubiquitination, which blocks substrate degradation [2]. UBA domains have also been implicated in protecting UbL/UBA family members from their own degradation via the proteasome. Rad23 and Ddi1 heterodimerize by interaction of their respective UbL and UBA domains this interaction results in enhanced binding of polyubiquitin [3].
2. Functional Properties
2.1 Self Association – PB1 domain
Sequestosome (SQSTM) 1/p62, a co-interacting protein of the atypical PKC isoforms zeta/iota [4,5], has a UBA domain at its C-terminal end, which binds non-covalently to polyubiquitin chains. Several other protein interaction domains exist within p62 suggesting a possible role in the formation of multimeric signaling complexes [6]. At its N-terminus, p62 has a PB1 domain [7], which shares considerable structural homology with the UbL domain [8] and has been shown to interact with the 26S proteasome subunit Rpt1 [9]. Thus, p62 can serve as a shuttling factor that directs polyubiquitinated proteins interacting with the UBA domain for turnover [9]. The PB1 (Phox and Bem1p) domain is a protein-protein interaction domain present in 29 human proteins, including p40phox, p67phox, aPKC λ/ι and ζ, Par6, MEK5 and SQSTM1/p62 [10]. Proteins containing PB1 domains can form specific heterodimers between family members. Critical residues involved in this protein-protein interaction have been identified. A “basic cluster” of conserved Lys and Arg residues in one PB1 domain interacts with the highly conserved acidic OPCA motif of another in a front-to-back interaction topology [7]. Based on their domain architecture, three broad classes of PB1 domains have been identified (Fig. 1A) [10]. Type A PB1 domain proteins have the canonical OPCA motif but lack the basic cluster. By contrast, Type B family members possess the basic cluster but lack the OPCA motif. Type A and Type B PB1 domains can interact to form heterodimers; however, not all B-type domains interact with all A-type domains or vice versa. It is possible that nonconserved PB1-PB1 interactions confer the high specificity of heterodimerization reported between family members. A small group of PB1 domain proteins, aPKC, Par6 and p62, contain both the basic cluster and the OPCA motif [10]. The basic cluster of p62 interacts with the OPCA motif of aPKC [7]. In this configuration, both the OPCA motif of p62 and the basic cluster of aPKC are free and may interact with other PB1 domains, including that of p62 itself. In fact, p62 oligomerization has been observed where the basic cluster of one p62 molecule binds to the OPCA motif of another in a front-to-back arrangement [10]. p62 has been shown to sequester polyubiquitin into sequestosomes or aggresomes [11], therein, interactions between p62 molecules may enhance the size of the aggresomes. In this regard, p62’s UBA domain is necessary for recruitment of polyubiquitin and aggresome formation [9]. Alternatively, p62 may be required either directly for formation of aggresomes or for a process whose inhibition indirectly disrupts the formation of aggresomes. The physiological relevance of aggresomes is still a matter of debate. However, they are a common pathological feature in diseases that affect brain, muscle, and liver [12,13].
Fig. 1.
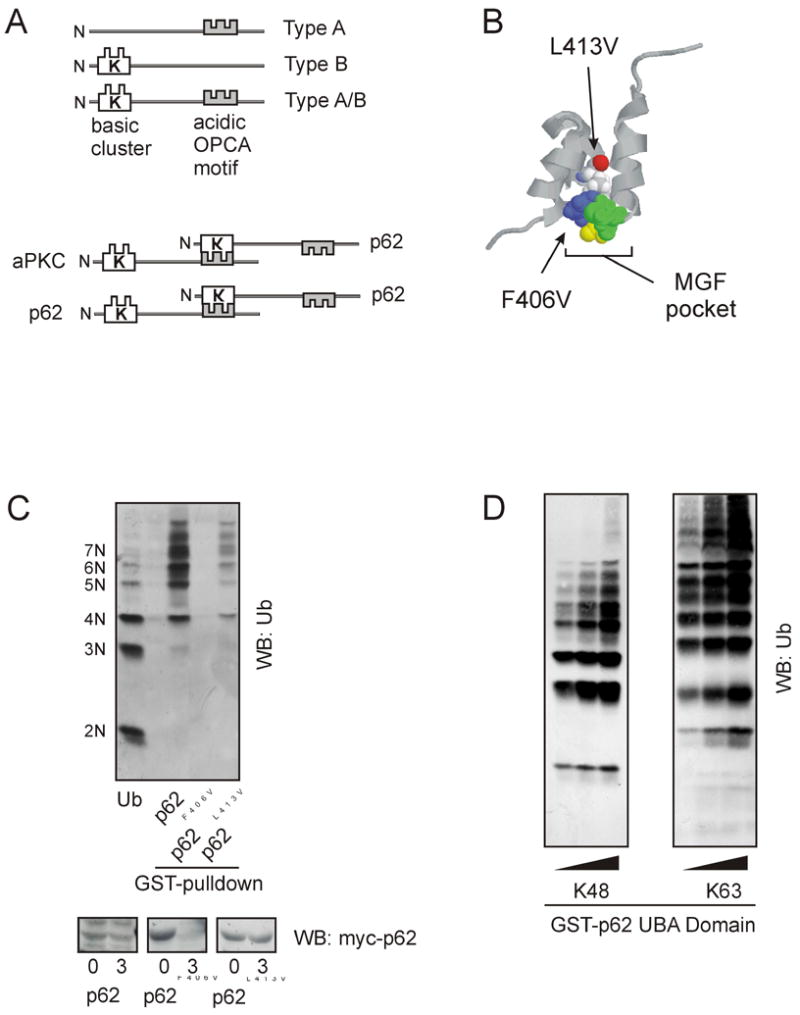
Functional properties of p62 UbL and UBA domains. a) PB1 domains can be one of three types: Type A, Type B, or Type A/B. p62 interacts with aPKC and can also dimerize with another p62 molecule via back-to-back binding of the basic cluster and the OPCA motif within the PB1 domain. b) The “MGF” binding pocket of p62’s UBA domain accommodates polyubiquitin binding. Location of p62 UBA mutations, F406V and L413V. c) UBA domain mutants were tested for their ability to bind polyubiquitin chains in a GST-UBA pulldown and Western blotting for ubiquitin (Ub), upper panel. p62 turnover was assessed by transiently transfecting HEK cells with myc-tagged p62 UBA domain mutants. Following overnight incubation, cells were treated with cyclohexamide for 24 hours. Lysates were prepared immediately after treatment 0, or after 3 hours the expression of p62 was monitored by Western blotting for myc. d) The UBA domain of p62 bind both K48 and K63 polyubiquitin chains [19]. GST-tagged p62 UBA domain was used in a pulldown assay along with increasing concentrations of either K48 or K63 polyubiquitin chains (kindly provided by Cecile Pickart).
The structure of the PB1 domain reveals an ubiquitin-like fold topology similar to the structure of the UbL domain [8]. In yeast, Rad23 binds to the proteasome through interaction of its UbL domain with the UIM motif of the Rpn10 subunit of the proteasome [14]. Similarly, Dsk2 UbL domain interacts with the Rpn1 subunit of the 19S regulatory proteasomal subcomplex (14). p62 has been reported to interact with the proteasome by binding to the S5a subunit (homologous to Rpn10 in yeast) via its N-terminus [9]. Thus, p62 may function in the formation of multimeric protein complexes because of the interactions of its PB1 domain with other proteins and it may also play a role in protein turnover due to its association with the proteasome. The PB1 domain may possess a flexible functional role, either favoring formation of self-associating p62 molecules that promote formation of aggresomes, or by interaction with other molecules that define specific cellular functions.
2.2 Interaction with Polyubiquitin
The UBA domain was originally identified by a bioinformatics analysis and is found in many proteins in the ubiquitin/proteasome pathway [15]. Most proteins containing UBA domains bind ubiquitin and display a preference for polyubiquitin chains [16]. The UBA structure reveals a three-helix bundle (Fig. 1B) with a conserved hydrophobic patch, MGF, that is necessary for interaction with polyubiquitin [17]. The interaction of the UBA domain with polyubiquitin protects Rad23 from proteolytic degradation and extends the half-life of the protein [2]. Mutation of F406V within p62’s UBA core domain prevents interaction with polyubiquitin chains [9] and likewise shortens the half-life of p62 (Fig. 1C). In comparison, mutation of L413V which does not disrupt polyubiquitin binding ability of the UBA domain has little effect on p62’s half-life (Fig.1C). Therefore polyubiquitin binding may serve as a stabilization signal to extend the half-life of the protein and to prolong the functional role of p62. This may be a common and conserved effect that polyubiquitin binding has upon all proteins which possess a UBA domain.
While K48 polyubiquitin chains are the most common signal for proteasomal degradation, chains can form on any of the seven lysine residues in the ubiquitin molecule [18]. Rad23 and Dsk2 bind K48-linked chains in vivo and are intimately involved in the protein degradation pathway. However, a recent in vitro survey found little selectivity of either UBA domain for K48 compared to K63 chains [19]. This observation suggests that other structural determinants exist in the holo-protein that enables chain-selective interactions. In this regard, we have observed that p62’s UBA domain binds both K48 and K63 polyubiquitin chains in an in vitro pull down assay (Fig.1D). Nonetheless, p62 selectively regulates the extension of K63 polyubiquitin chains on to substrates in vivo by its interaction with the E3 ubiquitin ligase, TRAF6, an enzyme that synthesizes K63 chains in vivo [20]. Interaction of E3 ubiquitin ligases with their respective scaffolding proteins likely serves as a mechanism whereby chains are transferred to target substrates. Both p62 and TRAF6 have been co-localized to discrete punctate structures upon stimulation of p62 signaling pathways [21]. These sites may localize inactive complexes in the cell, whereas active signal complexes of TRAF6-p62 may reside in the cytoplasm [22].
2.3 Scaffolding
The neurotrophin receptor, TrkA, is a substrate of TRAF6 and upon binding of NGF, leads to formation of a multimeric signal complex consisting of TrkA, TRAF6 and p62 [23]. In fact, TRAF6 K63-polyubiquitination of TrkA is necessary for internalization and sorting of the receptor. These findings reveal that p62 serves as a scaffold to regulate K63 polyubiquitination via interaction with TRAF6. Interestingly, loss of p62 interaction/dimerization affects TRAF6 K63-autoubiquitination/activation as well [20]. Therefore, p62-TRAF6 interactions may induce a conformational change in p62’s UBA domain allowing the subsequent interaction with specific polyubiquitinated substrates. A number of factors likely contribute to the functional specificity of the UBA domain in vivo. This idea is further strengthened by recent evidence that when bound to substrate, polyubiquitin chains can change their conformation to accommodate multiple UBA domains [24].
Previously, p62 was found to be necessary for TRAF6-dependent activation of NF-κB [20]. The activity of TRAF6 is regulated by autoubiquitination. Cells devoid of p62 exhibit low basal TRAF6 polyubiquitination. However, when p62 is expressed, autoubiquitination of TRAF6 is enhanced [20]. This has also been confirmed using p62 −/− mice in which the absence of p62 reduced activation of TRAF6. Deletion constructs that remove the PB1 domain, UBA domain or TRAF6 binding domain reveal that each is necessary for activation of NF-κB or TRAF6 [20]. Dimerization of p62 through its N-terminus is also necessary for TRAF6 activation. Together, the three protein interaction modules, PB1, TRAF6, and UBA, play a coordinated role in the activation and regulation of TRAF6 (Fig. 2A).
Fig. 2.
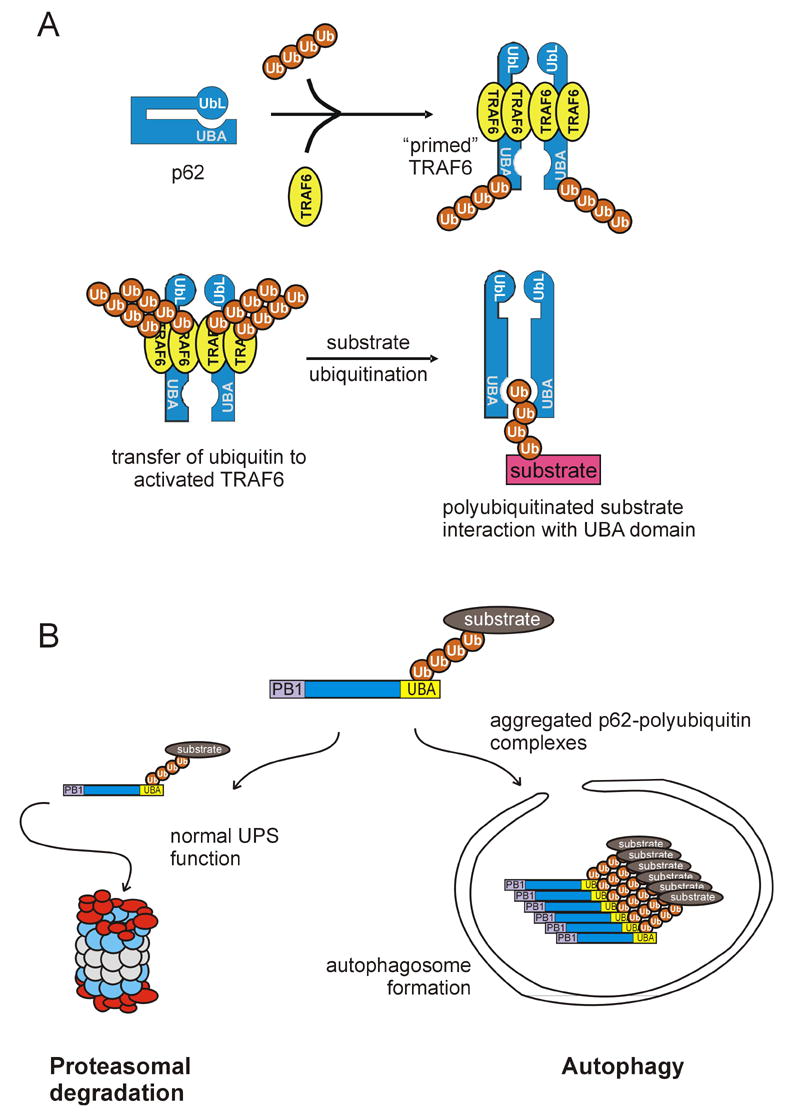
Models of p62 function. a) p62 can act as a scaffold for the K63 polyubiquitination of TRAF6 specific substrates. In a native form, the UbL domain of p62 may interact with its UBA domain, prior to interaction with ubiquitin. Next, the protein may “open” allowing access to multiple protein binding domains. Following p62 dimerization through the PB1 domain, dimerized TRAF6 binds to p62 at the TRAF6-binding site allowing the transfer of K63 polyubiquitin chains generated from TRAF6 to specific substrates. K63-tagged substrates can interact with the UBA domain of p62. b) p62 may reside at the intersection of two degradation pathways. Once the polyubiquitin chain of a substrate protein binds to the UBA domain of p62, the substrate can be trafficked to the proteasome for degradation. An example of this is the p62-dependent turnover of tau which requires p62 for proteasomal degradation [21]. p62 may also be involved the autophagic/lysosomal pathway by sequestering aggregated proteins prior to their inclusion in autophagosomes.
Naturally occurring isoforms of p62 (ZIP2, ZIP3) exist [25]. ZIP2 lacks the TRAF6 binding site required for oligomerization and ZIP3 lacks a UBA domain needed for binding polyubiquitin [26]. It is plausible to infer that either of these isoforms could play a defined cellular role in regulating TRAF6 activation by preventing TRAF6 from being recruited to a p62 signaling scaffold or by disrupting the formation of K63 polyubiquitin chains necessary for TRAF6-dependent ubiquitination of targeted substrates. Diversity in regulation of TRAF6 activation may occur in a tissue-specific manner depending upon the expression of p62 isoforms. Adding further complexity to the situation, ZIP2 exerts a dominant-negative effect on p62/ZIP1 function [20].
2.4 Movement of Cargo
p62 also serves as a shuttling factor for delivery of polyubiquitinated substrates, like tau, to the proteasome [21]. We found that p62 binds to the S5a subunit of the proteasome [9] by interaction of its N-terminal PB1 domain with S5a, while its UBA domain interacts with polyubiquitinated tau [21]. This is the first reported example of p62 serving a shuttling function similar to other UbL/UBA family members [27]. Post-ubiquitination, the substrate complex is shuttled to the proteasome for degradation (Fig. 2B). The accumulation of hyperphosphorylated and aggregated tau is a hallmark of neurodegenerative disease. Interestingly, p62 has been localized with tau in tangles [12], which suggests a possible functional interaction in clearance of tau in vivo.
Recently, the N-terminus of p62 has also been shown to interact with LC3, a marker of the autophagocytic vesicle [reviewed in 28]. Autophagy is a bulk process whereby cytoplasm and organelles are delivered to the lysosome for degradation. It is possible that p62 may play a role in selective sequestration of polyubiquitinated proteins for autophagy. Thus, p62 may exist at the intersection of the endocytosis, proteasome, and autophagosome networks (Fig. 2B).
3.0 Questions that Remain
What defines the specificity of the interactions of p62? Originally identified as an aPKC interacting protein [4,5], p62 was shown to interact with aPKC through binding of their respective PB1 domains [7]. However, the p62 PB1 domain has also been shown to interact with other proteins such as MEK5, NBR1 and LC3. Also, p62 dimerizes through interactions of its PB1 domain with that of another p62 molecule [7]. A model could be envisioned where competition for PB1 domain binding between all these different players leads to diversity in signaling pathways and/or the intracellular localization of p62. However, any effects of p62 binding partners can not be discounted in regulation of interactions with the PB1 domain. Monoubiquitination of either Sts1or Sts2 regulates their ability to bind ubiquitinated cargo in trans [29]. Therefore, a similar mechanism may exist for p62 as a means to regulate interaction with its cargo. In this regard, several naturally occurring variants of the p62 UBA domain have been linked with Paget’s disease of the bone [30]. These mutants may cause pleiotropic effects with functional consequences upon polyubiquitin binding the UBA domain. For example, mutations in the UBA domain may alter binding of polyubiquitin [9], impair activation of TRAF6 [20], disrupt interaction with cargo, alter formation of aggresomes [9], or block the turnover of p62 (Fig. 1C). Moreover, the extension of polyubiquitin chain by TRAF6 onto a target substrate is dependent upon the ubiquitin binding capabilities of the UBA domain (Fig. 3). Deletion of p62’s UBA domain, or mutations within the UBA domain which impair interaction of p62 with polyubiquitin, impair TrkA substrate ubiquitination, whereas mutations that are without affect on polyubiquitin binding have no effect on TrkA polyubiquitination. These findings suggest that p62’s UBA domain carries polyubiquitin chains not only necessary for activation of TRAF6 [20], but that these chains are transferred to the substrate (Fig. 2A). In keeping with this observation we have observed that inhibition of p62/TRAF6 interaction prevented TrkA polyubiquitination as well [23].
Fig. 3.
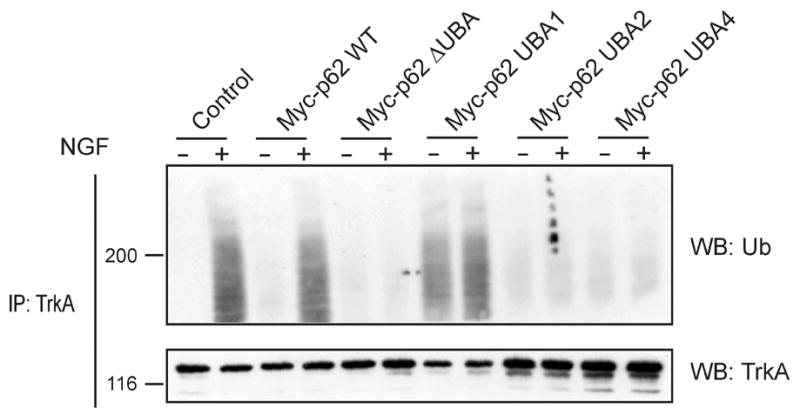
Interaction of polyubiquitin with p62 UBA is necessary for substrate ubiquitination. PC12 cells were transfected with HA-TrkA along with Myc-tagged wild-type (WT) p62, Myc-p62 ΔUBA (a construct lacking the UBA domain), Myc-p62 UBA mutant 1: L398V (no effect on polyubiquitin binding), Myc-p62 UBA mutant 2: F406V (inhibits polyubiquitin binding); or Myc-p62 UBA mutant 4: L417V (inhibits polyubiquitin binding). Complete characterization of the mutants and the effects on binding polyubiquitin has been previously described [9]. Thirty-six hours after transfection the cells were treated with NGF (50 ng/ml) for 15 min, lysed and immunoprecipitated with HA antibody [23], followed by Western blotting for either ubiquitin or TrkA as shown.
Recent studies have shown that Dsk2’s UbL domain interacts with its UBA domain [31]. While the UbL-UBA interaction is relatively weak compared to the binding of ubiquitin to the UBA domain, it is possible to propose a model for the regulatory mechanism of Dsk2’s adaptor/shuttling function. The protein is may be in a closed configuration until ubiquitin is introduced and the UbL domain is competed away from the UBA domain making it available for interaction with the proteasome, and allowing Dsk2 to carry its “cargo” for proteolytic degradation [31]. Indeed a similar model may enable p62 to adopt a closed conformation, where the ubiquitin fold of the PB1 domain competes for UBA binding, another level of specificity might be achieved. Displacement of the UbL domain from the UBA domain may then enable p62 to interact with its various binding partners with monoubiquitination terminating this interaction [29]. How might this conformational change be achieved? One obvious regulator is interaction with TRAF6. However, one should also consider the role of the atypical PKCs (iota/lambda and zeta) in this process. Studies by Puls et al. [4] revealed that p62 aggregates were increased in the absence of aPKC/p62 interaction, whereas aPKC regulated the cytosolic localization of p62. Therefore, aPKC emerges as a critical factor of aggresome formation and may regulate the functional localization of p62, as well as, its ability to interact with polyubiquitin. Further studies are needed to explore consequences of aPKC/TRAF6/p62 interactions.
Acknowledgments
Our work on TrkA and p62 is supported by grants from the NINDS to M.W.W. We thank members of the Wooten lab for fruitful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Verma R, Oania R, Graumann J, Deshaies RJ. Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell. 2004;118:99–110. doi: 10.1016/j.cell.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 2.Heessen S, Masucci MG, Dantuma NP. The UBA2 domain functions as an intrinsic stabilization signal that protects Rad23 from proteasomal degradation. Mol Cell. 2005;18:225–235. doi: 10.1016/j.molcel.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 3.Kang Y, Vossler RA, Diaz-Martinez LA, Winter NS, Clarke DJ, Walters KJ. UBL/UBA ubiquitin receptor proteins bind a common tetraubiquitin chain. J Mol Biol. 2006;356:1027–1035. doi: 10.1016/j.jmb.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 4.Puls A, Schmidt S, Grawe F, Stabel S. Interaction of protein kinase C zeta with ZIP, a novel protein kinase C-binding protein. Proc Natl Acad Sci U S A. 1997;94:6191–6196. doi: 10.1073/pnas.94.12.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanchez P, De Carcer G, Sandoval IV, Moscat J, Diaz-Meco MT. Localization of atypical protein kinase C isoforms into lysosome-targeted endosomes through interaction with p62. Mol Cell Biol. 1998;18:3069–3080. doi: 10.1128/mcb.18.5.3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geetha T, Wooten MW. Structure and functional properties of the ubiquitin binding protein p62. FEBS Lett. 2002;512:19–24. doi: 10.1016/s0014-5793(02)02286-x. [DOI] [PubMed] [Google Scholar]
- 7.Lamark T, Perander M, Outzen H, Kristiansen K, Overvatn A, Michaelsen E, Bjorkoy G, Johansen T. Interaction codes within the family of mammalian Phox and Bem1p domain-containing proteins. J Biol Chem. 2003;278:34568–34581. doi: 10.1074/jbc.M303221200. [DOI] [PubMed] [Google Scholar]
- 8.Hirano Y, Yoshinaga S, Ogura K, Yokochi M, Noda Y, Sumimoto H, Inagaki F. Solution structure of atypical protein kinase C PB1 domain and its mode of interaction with ZIP/p62 and MEK5. J Biol Chem. 2004;279:31883–31890. doi: 10.1074/jbc.M403092200. [DOI] [PubMed] [Google Scholar]
- 9.Seibenhener ML, Babu JR, Geetha T, Wong HC, Krishna NR, Wooten MW. Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol Cell Biol. 2004;24:8055–8068. doi: 10.1128/MCB.24.18.8055-8068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson MI, Gill DJ, Perisic O, Quinn MT, Williams RL. PB1 domain-mediated heterodimerization in NADPH oxidase and signaling complexes of atypical protein kinase C with Par6 and p62. Mol Cell. 2003;12:39–50. doi: 10.1016/s1097-2765(03)00246-6. [DOI] [PubMed] [Google Scholar]
- 11.Shin J. p62 and the sequestosome, a novel mechanism for protein metabolism. Arch Pharm Res. 1998;21:629–633. doi: 10.1007/BF02976748. [DOI] [PubMed] [Google Scholar]
- 12.Kuusisto E, Salminen A, Alafuzoff I. Early accumulation of p62 in neurofibrillary tangles in Alzheimer's disease: possible role in tangle formation. Neuropathol Appl Neurobiol. 2002;28:228–237. doi: 10.1046/j.1365-2990.2002.00394.x. [DOI] [PubMed] [Google Scholar]
- 13.Zatloukal K, Stumptner C, Fuchsbichler A, Heid H, Schnoelzer M, Kenner L, Kleinert R, Prinz M, Aguzzi A, Denk H. p62 is a common component of cytoplasmic inclusions in protein aggregation diseases. Amer J Pathol. 2002;160:255–263. doi: 10.1016/S0002-9440(10)64369-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saeki Y, Sone T, Toh-e A, Yokosawa H. Identification of ubiquitin-like protein-binding subunits of the 26S proteasome. Biochem Biophys Res Commun. 2002;296:813–819. doi: 10.1016/s0006-291x(02)02002-8. [DOI] [PubMed] [Google Scholar]
- 15.Hofmann K, Bucher P. The UBA domain: a sequence motif present in multiple enzyme classes of the ubiquitination pathway. Trends Biochem Sci. 1996;21:172–173. [PubMed] [Google Scholar]
- 16.Wilkinson CR, Seeger M, Hartmann-Peterson R, Stone M, Wallace M, Semple C, Gordon C. Proteins containing the UBA domain are able to bind to multi-ubiquitin chains. Nature Cell Biol. 2001;3:939–943. doi: 10.1038/ncb1001-939. [DOI] [PubMed] [Google Scholar]
- 17.Dieckmann T, Withers-Ward ES, Jarosinski MA, Liu CF, Chen ISY, Feigon J. Structure of a human DNA repair protein UBA domain that interacts with HIV-1 Vpr. Nat Struct Biol. 1998;12:1042–1047. doi: 10.1038/4220. [DOI] [PubMed] [Google Scholar]
- 18.Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nat Biotech. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 19.Raasi S, Varadan R, Fushman D, Pickart C. Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nature Struc Mol Biol. 2005;12:708–714. doi: 10.1038/nsmb962. [DOI] [PubMed] [Google Scholar]
- 20.Wooten MW, Geetha T, Seibenhener ML, Babu JR, Diaz-Meco MT, Moscat J. The p62 scaffold regulates nerve growth factor-induced NF-kappaB activation by influencing TRAF6 polyubiquitination. J Biol Chem. 2005;280:35625–35629. doi: 10.1074/jbc.C500237200. [DOI] [PubMed] [Google Scholar]
- 21.Babu JR, Geetha T, Wooten WM. Sequestosome 1/p62 shuttles polyubiquitinated tau for proteasomal degradation. J Neurochem. 2005;94:192–203. doi: 10.1111/j.1471-4159.2005.03181.x. [DOI] [PubMed] [Google Scholar]
- 22.Wang KZ, Wara-Aswapati N, Boch JA, Yoshida Y, Hu CD, Galson DL, Auron PE. TRAF6 activation of PI 3-kinase-dependent cytoskeletal changes is cooperative with Ras and is mediated by an interaction with cytoplasmic Src. J Cell Sci. 2006;119:1579–1591. doi: 10.1242/jcs.02889. [DOI] [PubMed] [Google Scholar]
- 23.Geetha T, Jiang J, Wooten MW. Lysine 63 polyubiquitination of the nerve growth factor receptor TrkA directs internalization and signaling. Mol Cell. 2005;20:301–312. doi: 10.1016/j.molcel.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 24.Varadan R, Walker O, Pickart C, Fushman D. Structural properties of polyubiquitin chains in solution. J Mol Biol. 2003;324:637–647. doi: 10.1016/s0022-2836(02)01198-1. [DOI] [PubMed] [Google Scholar]
- 25.Gong J, Xu J, Bezanilla M, van Huizen R, Derin R, Li M. Differential stimulation of PKC phosphorylation of potassium channels by ZIP1 and ZIP2. Science. 1999;285:1565–1569. doi: 10.1126/science.285.5433.1565. [DOI] [PubMed] [Google Scholar]
- 26.Croci C, Brandstatter JH, Enz R. ZIP3, a new splice variant of the PKC-zeta-interacting protein family, binds to GABAC receptors, PKC-zeta, and Kv beta 2. J Biol Chem. 2003;278:6128–6135. doi: 10.1074/jbc.M205162200. [DOI] [PubMed] [Google Scholar]
- 27.Chen L, Madura K. Rad23 promotes the targeting of proteolytic substrates to the proteasome. Mol Cell Biol. 2002;22:4902–4913. doi: 10.1128/MCB.22.13.4902-4913.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Komatsu M, Kominami E, Tanaka E. Autophagy and neurodegeneration. Autophagy. 2006;2:315–317. doi: 10.4161/auto.2974. [DOI] [PubMed] [Google Scholar]
- 29.Hoeller D, Crosetto N, Blagoev B, Raiborg C, Tikkanen R, Wagner S, Kowanetz K, Breitling R, Mann M, Stenmark H, Dikic I. Regulation of ubiquitin-binding proteins by monoubiquitination. Nature Cell Biol. 2006;8:163–169. doi: 10.1038/ncb1354. [DOI] [PubMed] [Google Scholar]
- 30.Layfield R, Ciani B, Ralston SH, Hocking LJ, Sheppard PW, Searle MS, Cavey JR. Structural and functional studies of mutations affecting the UBA domain of SQSTM1 (p62) which cause Paget's disease of bone. Biochem Soc Transact. 2004;32:728–730. doi: 10.1042/BST0320728. [DOI] [PubMed] [Google Scholar]
- 31.Lowe ED, Hasan N, Trempe JF, Fonso L, Noble ME, Endicott JA, Johnson LN, Brown NR. Structures of the Dsk2 UBL and UBA domains and their complex. D Biol Crystallogr. 2006;62:177–188. doi: 10.1107/S0907444905037777. [DOI] [PubMed] [Google Scholar]


