Abstract
Immunohistochemistry is a method that can provide complementary diagnostic and prognostic information to morphological observations and soluble assays. Sensitivity, specificity, or requirements for arduous sample preparation or signal amplification procedures often limit the application of this approach to routine clinical specimens. Rolling circle amplification (RCA) generates a localized signal via an isothermal amplification of an oligonucleotide circle. The application of this approach to immunohistochemistry could extend the utility of these methods to include a more complete set of immunological and molecular probes. RCA-mediated signal amplification was successfully applied to the sensitive and specific detection of a variety of cell surface antigens (CD3, CD20, and epithelial membrane antigen) and intracellular molecules (vimentin and prostate-specific antigen) within a variety of routinely fixed specimens, as well as samples prepared for flow cytometry. RCA technology, which has an intrinsically wide dynamic range, is a robust and simple procedure that can provide a universal platform for the localization of a wide variety of molecules as a function of either antigenicity or nucleic acid sequence. The use of RCA in this way could enhance the use of markers of current interest as well as permit the integration of emerging information from genomics and proteomics intocell- and tissue-based analyses.
Analyses, such as immunohistochemistry and flow cytometry, provide information that is complementary to that obtained through the more traditional morphological descriptions provided by conventional cytology or histology. This general area of molecular tissue pathology has required adaptation of many of the antibody probes first used in homogenous cell-free preparations, and has required the development of techniques to refine the examination of cells and tissues. It is now generally accepted that a tissue-based assay can yield superior information to a soluble assay. For example, the determination of estrogen receptor status in breast biopsies is better accomplished using an immunohistochemical approach than the biochemical assay of receptors, which fails to evaluate the receptor status of neoplastic tissue in contrast to benign glands. 1 The ability to detect rare cells or foci of abnormal cells within a tissue is of increasing importance as biopsies become progressively smaller and subject to more refined analyses using the rapidly accumulating knowledge of the molecular basis of disease. The ability to investigate and apply new markers that are characteristic of specific genotypes and phenotypes in tissue could provide information regarding diagnosis, prognosis, and response to various treatment regimens. Emerging fields, such as pharmacogenomics, 2 depend on the ability to simultaneously monitor the expression profiles for several markers. Immunohistochemistry, as it is applied to fixed tissue, can provide critical material for the analyses that are necessary to validate these markers.
The application of antibodies to cells and tissues has presented specific difficulties beyond those encountered when these reagents are applied to purified proteins in solution. At least two general sorts of problems have been encountered. First, biopsies and cell samples may, through differences in fixation and preparation, vary in their preservation from specimen to specimen. This issue has been addressed using antigen retrieval techniques 3,4 and enzymatic digestion. An additional difficulty is the ability to detect analytes present at low levels. In common with soluble assays, this becomes a matter of increasing signal without raising the level of nonspecific background. The approach that has been most commonly explored is signal amplification, which is achieved by successive rounds of enzymatic reactions. Biotinyl tyramide 5,6 is commonly used to increase the signal of low abundance targets that are otherwise undetectable by conventional methods. Tyramide-based amplification has not been used for clinical applications, because of increased background during the multiple rounds of signal amplification. This problem is especially prevalent in clinical samples in which nonspecific binding of antibodies or nucleic acid probes cannot be controlled to the extent it can be with cultured cells. 7,8
Rolling circle amplification (RCA) is an isothermal nucleic-acid amplification method. 9-12 It differs from the polymerase chain reaction and other nucleic-acid amplification schemes in several respects. In addition to an exponential mode, which is capable of generating amplification in excess of 109-fold, a linear mode of RCA can generate 105-fold signal amplification during a brief enzymatic reaction that has been demonstrated in microarray assays. 11 Perhaps the most important feature of linear RCA is that the product of amplification remains tethered to the target molecule. In conjunction with the isothermal nature of the RCA reaction and the capability to localize multiple markers simultaneously, RCA seems well suited to cell- and tissue-based assays where it is critical to maintain morphological information. It has been demonstrated that RCA amplification permits the localization of signals, representing single molecules with specific genetic 9,13 or biochemical features. 10,11 This has been achieved in haloed nuclei 9 and on microarrays of proteins. 10,11
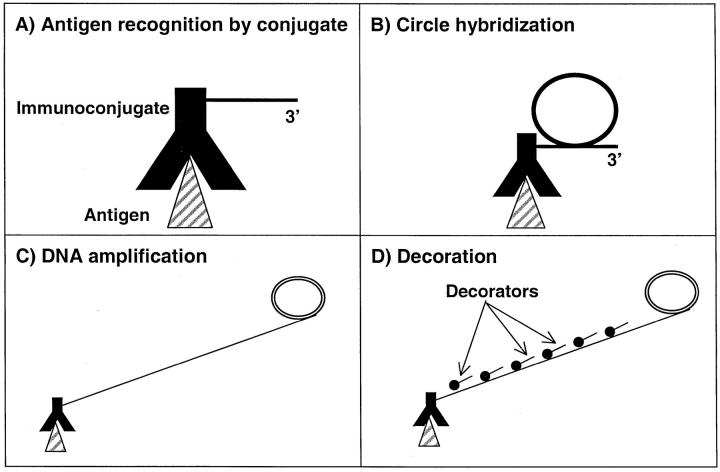
Localized signal amplification by means of RCA can recognize nucleic acid targets 9 through hybridization of nucleic acid probes. More recently it has been established 10,11 that RCA can be used to increase the sensitivity of immunoassays. In a manner analogous to immunopolymerase chain reaction 14,15 it has been possible to couple the specificity of antibodies to the signal amplification of RCA. However, in contrast to immunopolymerase chain reaction, signal amplification via immuno-RCA results in an easily detectable high-molecular weight nucleic-acid molecule at the site of antibody binding. Furthermore, because the product of an immuno-RCA reaction remains attached to the immune complex, it is compatible with localization of the product on a microarray or within a biological structure. As illustrated in Figure 1, a ▶ polyclonal antibody to species-specific immunoglobulins is a generic reagent with the capability to magnify the signal generated by standard immunohistochemistry. The present report describes experiments that demonstrate the ability of RCA to increase sensitivity as well as localize specific molecules in cells and tissues using antibodies. The goal of these experiments was to prove the feasibility and versatility of a generic platform using RCA with a variety of markers in cells and tissues.
Figure 1.
Schematic representation of immuno-RCA approach. A: Diagram of immunoconjugate bound to target antigen. B: Hybridization of circle to RCA primer. C: Enzymatic synthesis of high-molecular weight DNA by rolling circle replication. D: Detection of localized amplification at site of bound antibody with labeled decorator oligonucleotide.
Materials and Methods
Cells and Tissues
The λ light chain-producing human myeloma cell line U266 (ATCC TIB-196) and the acute T-cell leukemia-derived cell line Jurkat (ATCC TIB-152) were obtained from the American Type Culture Collection and propagated as suggested. Formalin-fixed paraffin-embedded tissues were obtained from DAKO (Carpinteria, CA) and Newcomer Supply Inc (Middleton, WI).
Oligonucleotide Sequences
Two sets of primers, circles and decorator sequences were used in these experiments: primer 1, 5′-AAAAAAAA-AAAAAAAATGATCACAGCTGAGGATAGGACATGC GA-3′; primer 4.2, 5′-AAAAAAAAAAAAAAACTTGTACATGTCTCAGTAGCTC GTCAGT-3′; circle 1, 5′-pCGCATGTCCTATCCTCAGCTGTGATCATCAGAACT CACCTGTTAGACGCCACCAGCTCCAACTGTGAAGATCGCTTAT-3′; circle 4.2, 5′-pACTGACGAGCTACTGAGACATGTACAATCGGACCTGTGAGGTACTACC CTAATCGGACCTGTGAGGTACTACCCTAACTT-3′; decorators for circle 1, 5′-XACTGTGAAGATCGCTTATX-3′ (X = biotin), 5′-FACTCTGAAGATCGCTTATF-3′ (F = fluorescein); decorators for circle 4.2, 5′-TCGGACCTGTGAGGTACTACCCTAA HRP-3′ (HRP = horseradish peroxidase) oligo obtained from Synthetic Genetics (San Diego, CA), 5′-FTCGGACCTGTGAGG TACTACCCTAAF-3′.
Preparation of Conjugates
Immunoconjugates, in which RCA primers were covalently attached to antibodies, were prepared as previously described. 10 Either a polyclonal preparation of rabbit anti-mouse immunoglobulins (DAKO) or a mouse monoclonal antibody to fluorescein (Fitzgerald Industries, Concord, MA) were linked to primer 1 or 4.2. Desalted antibody (41 nmol) was treated with a 10-fold molar excess of sulfo-N-(4-maleimidobutyryloxy) sulfosuccinimide (GMBS) (Pierce Chemical, Rockford, IL) under nitrogen in the dark for 30 minutes at 37°C, followed by 30 minutes at room temperature. Unreacted sulfo-GMBS was removed by chromatography over a PD-10 column equilibrated with NaPhosphate, pH 7.5, 150 mmol/L NaCl. The antibody was concentrated in a Centricon YM-30 at 4°C. The number of maleimides per antibody was determined by using Ellman’s reagent (Pierce) to measure sulfhydryls after titration of β-mercaptoethanol by the activated antibody. Sulfo-GMBS, 28.1 nmol, of activated antibody and 142 nmol of 5′ thiol oligonucleotide were conjugated in a volume of 825 μl for 2 hours at room temperature, followed by overnight at 4°C. Antibody conjugated to oligonucleotide was purified by anion exchange chromatography on Q-Sepharose (Amersham Pharmacia Biotech, Piscataway, NJ) using a salt gradient. Fractions containing conjugate were pooled and subjected to size exclusion chromatography on Superdex-200 (Pharmacia, Piscataway, NJ) at 4°C to remove free oligonucleotide. A 10:1 ratio of oligonucleotide:antibody was typically used in conjugations, which resulted in a more reproducible production of conjugates with 3 to 5 oligonucleotides per antibody molecule. Size exclusion chromatography was used to remove unconjugated oligonucleotide, ensuring a final preparation free of unconjugated antibody or DNA. The overall efficiency of the conjugation procedure routinely gave 50% recovery of the starting antibody.
Immunohistochemistry
Before staining, tissue sections were baked for 1 hour at 65°C and deparaffinized through xylene and graded ethanol (100, 95, and 70%, two times at 2 minutes each). Sections were rinsed in phosphate-buffered saline (PBS), pH 7.4, and blocked for 15 minutes in 10% normal goat serum diluted in PBS. Primary antibodies to CD20 (clone L26), epithelial membrane antigen (EMA) (clone E29), and vimentin (clone V9) were obtained from DAKO. The anti-prostate-specific antigen (PSA) antibody (clone M212091) was obtained from Fitzgerald. All antibodies were routinely diluted in 50 mmol/L Tris, pH 7.5, 150 mmol/L NaCl, 0.1% Triton X-100, and 3% bovine serum albumin. Primary antibodies were incubated for 30 minutes at room temperature. Conventional immunohistochemistry was performed using a biotinylated rabbit anti-mouse secondary antibody (DAKO) followed by streptavidin-HRP (DAKO). For the detection of EMA in breast sections, a fluorescein isothiocyanate (FITC)-conjugated primary antibody was used followed by rabbit anti-FITC-alkaline phosphatase conjugate (Novocastra). Detection by immuno-RCA was performed in the same manner, except RCA conjugates were substituted for a biotinylated secondary antibody. After RCA, the product was decorated and detected by one of three methods. Tonsil sections were screened for CD20 by use of a biotinylated decorator followed by an additional 30-minute incubation with streptavidin-HRP conjugate. Breast tissue used for EMA analysis used the FITC-labeled decorator followed by an additional 30-minute incubation with rabbit anti-FITC-alkaline phosphatase conjugate. Vimentin and PSA analysis on tissues was performed via an oligonucleotide-HRP conjugate. Two 2× standard saline citrate/0.1% Triton X-100 washes followed each decoration step. Colorimetric detection used diaminobenzidine/hydrogen peroxide (DAKO) or Nova Red (Vector Laboratories, Burlingame, CA) for HRP detection, whereas AP detection used Vector Blue (Vector Laboratories). Slides were washed with deionized water, air-dried, and counterstained. Sections were mounted in VectaMount (Vector Laboratories).
Flow Cytometry
Approximately 10 6 cells in a volume of 50 to 100 μl per sample were washed with PBS and incubated with a fluorescein-conjugated monoclonal antibody to CD3 (clone BMA 031; Caltag Laboratories, Burlingame, CA), diluted in DAKO diluent (DAKO). Cells were washed in PBS, followed by centrifugation and resuspension in Fixation medium (Caltag Laboratories). Cells were fixed for 15 minutes on ice, followed by a PBS wash. RCA amplification was performed in a similar manner to immuno-RCA on fixed tissues using the anti-fluorescein immunoconjugate and decorated with a fluorescein conjugated oligonucleotide decorator. Cells were analyzed using a Coulter EPICS/Profile II flow cytometer equipped with a He-Ne laser set to an excitation wavelength of 488 nm.
Rolling Circle Amplification
RCA amplification was performed as previously described. 9 Briefly, bound immunoconjugates were detected by incubation with a circle homologous to the RCA primer diluted to 200 nmol/L in 100 mmol/L of potassium glutamate. After incubation for 30 minutes at 37°C, slides were rinsed with 100 mmol/L of potassium glutamate. RCA reactions were performed for 45 minutes using φ29 DNA polymerase (Amersham Pharmacia Biotech) at 31°C. The products of these RCA reactions were then visualized by incubation at 37°C for 30 minutes with the appropriate oligonucleotide decorator diluted to 20 nmol/L in 2× standard saline citrate/0.1% Triton X-100.
Results
Protocol for RCA-Based Signal Amplification for Immunohistochemistry
Previous work 10,11 has established that coupling of an RCA primer to an antibody can be accomplished without significantly compromising the affinity or avidity of the conjugate. These immunoconjugates can be either primary antibodies specific for a molecule of interest, such as human immunoglobulin E, or a secondary antibody that reacts with a class of primary antibodies, eg, mouse monoclonal immunoglobulins. As illustrated in Figure 1 ▶ , when such a generic immunoconjugate is used it substitutes for conventional secondary detection antibodies. The subsequent steps, unique to RCA-mediated signal amplification, are annealing of circle (Figure 1B) ▶ , enzymatic amplification (Figure 1C) ▶ , and detection of the RCA product with a decorator probe (Figure 1D) ▶ . These steps added 60 to 90 minutes to the routine immunohistochemistry protocol that used a biotinylated secondary antibody followed by a streptavidin-enzyme conjugate. The general scheme presented here can be applied to monoclonal and polyclonal antibodies with a final readout that is either colorimetric or fluorescent.
Detection of Commonly Localized Antigen
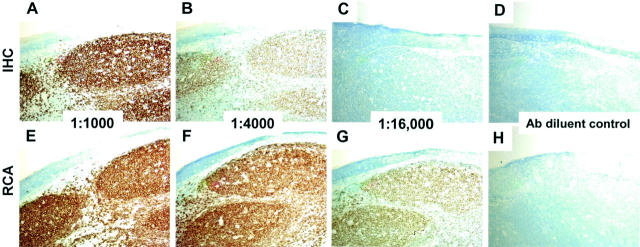
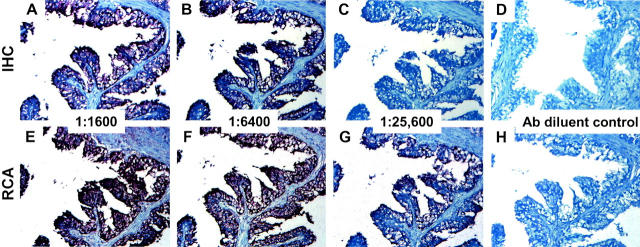
To evaluate the ability of RCA to provide localized signal amplification across a range of antibody concentrations, serial dilutions of monoclonal antibody that recognizes the B-cell antigen, CD20, were applied to tonsil sections. The binding of the antibody was determined using either a conventional detection scheme, a biotinylated secondary antibody followed by a streptavidin-horseradish peroxidase conjugate, or immuno-RCA in which an RCA primer, convalently linked to a polyclonal rabbit anti-mouse antibody, replaces the biotinylated secondary antibody. After RCA, the RCA reaction product was detected using a biotinylated oligonucleotide probe followed by the streptavidin-horseradish peroxidase conjugate. Comparison of the two sets of sections (Figure 2) ▶ shows that the slides developed by immuno-RCA have uniform signal, primarily within germinal centers, over a wider concentration range than the standard method. The increase in signal is at least fourfold with respect to antibody dilution. In addition to CD20, EMA was studied. EMA is also a membrane protein that is observed in the apical portion of duct-lining cells. In Figure 3 ▶ RCA provided a similar fourfold signal enhancement when compared to conventional immunohistochemistry.
Figure 2.
Detection of CD20 by conventional immunohistochemistry or immuno-RCA. Serial sections of formalin-fixed paraffin-embedded tonsil were prepared for immunohistochemistry as described in the text and incubated with monoclonal antibody to CD20 diluted 1:1000 (A and E); 1:4000 (B and F); 1:16,000 (C and G); or antibody diluent alone (D and H). After development with a standard detection kit (A–D) or by immuno-RCA (E–H) using an RCA conjugate that recognizes mouse immunoglobulins, sections were incubated with diaminobenzidine/hydrogen peroxide (brown stain) and counterstained with methylene blue. All panels, showing the same region of the tonsil specimen, were photographed at an original magnification of ×10. Uniform staining was observed in the germinal centers, whereas the interfollicular regions of the tonsil showed little staining. IHC, immunohistochemistry.
Figure 3.
Detection of EMA. Serial sections of formalin-fixed paraffin-embedded breast tissue were prepared for immunohistochemistry as described in the text and incubated with monoclonal antibody to EMA diluted 1:1600 (A and E); 1:6400 (B and F); 1:25,600 (C and G); or antibody diluent alone (D and H). After development with either standard immunohistochemistry techniques (A–D) or immuno-RCA (E–H), sections were screened for alkaline phosphatase activity (blue stain) and counterstained with Nuclear Fast Red. All panels, representing the same region of the breast tissue, were photographed at an original magnification of ×20. Epithelial cells of the breast ducts and lobules were uniformly positive for EMA. IHC, immunohistochemistry.
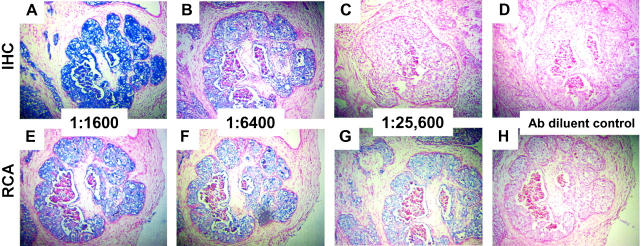
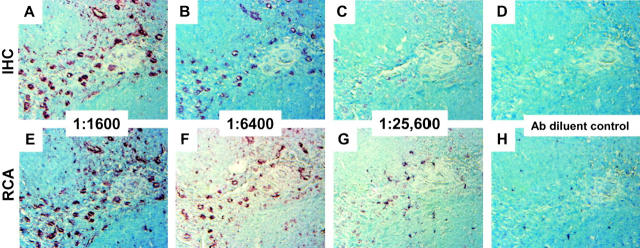
To evaluate the ability of immuno-RCA to localize an intracellular antigen, sections of tonsil and prostate tissue were analyzed with antibodies to vimentin or PSA, respectively. With respect to antibody dilution, RCA provides at least a fourfold increase in signal without compromising the discrete intracytoplasmic localization of vimentin or PSA (Figures 4 and 5) ▶ ▶ . In each set of experiments, tonsil, breast, and prostate biopsies demonstrated the reproducibility and sensitivity of immuno-RCA across the section at low concentrations of primary antibody.
Figure 4.
Detection of vimentin. Serial sections of formalin-fixed paraffin-embedded tonsil were prepared for immunohistochemistry as described in the text and incubated with monoclonal antibody to vimentin diluted 1:1600 (A and E); 1:6400 (B and F); 1:25,600 (C and G); or antibody diluent alone (D and H). After development with standard immunohistochemistry techniques (A–D) or immuno-RCA (E–H), sections were incubated with Vector Nova Red/hydrogen peroxide (brown stain) and counterstained with methylene blue. All panels, showing the same region of the tonsil specimen, were photographed at an original magnification of ×20. In contrast to CD20 (Figure 2) ▶ , lymphoid and endothelial cells in the interfollicular areas stain positively for vimentin. IHC, immunohistochemistry.
Figure 5.
Detection of PSA. Sections of formalin-fixed paraffin-embedded prostate tissue were incubated with a monoclonal antibody to PSA diluted at 1:1600 (A and E); 1:6400 (B and F); 1:25,600 (C and G); or antibody diluent alone (D and H). After development with standard immunohistochemistry techniques (A–D) or by immuno-RCA (E–H), sections were incubated with Vector Nova Red/hydrogen peroxide (dark stain) and counterstained with methylene blue. All panels, showing the same region of the prostate specimen, were photographed at an original magnification of ×20 and demonstrate that epithelial cells of the prostate glands and lobules were uniformly positive. IHC, immunohistochemistry.
Application of ImmunoRCA to Flow Cytometry
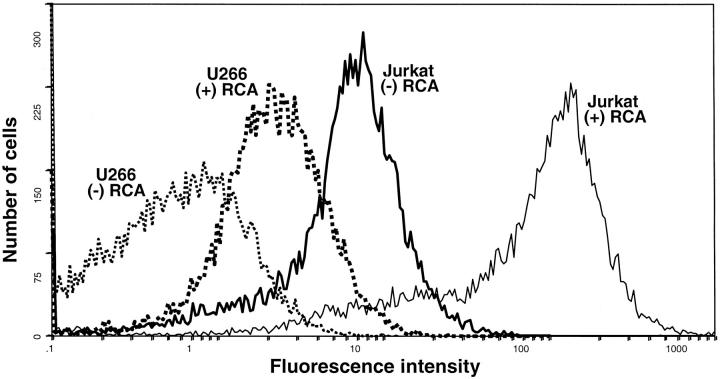
The use of antibodies to characterize cellular samples via flow cytometry shares with immunohistochemistry the need for minimizing background. Currently, flow cytometry is predominantly used to monitor relatively abundant cell surface molecules. The application of a suitable signal amplification approach to flow cytometry could permit the quantitative analysis of dispersed cell samples. To address the feasibility of using RCA technology, the presence of CD3 antigen was examined on the surface of lymphoid cells. The data presented in Figure 6 ▶ demonstrates the use of immuno-RCA to detect CD3 on Jurkat, a CD3-positive T-cell leukemia-derived cell line, in distinction to U266, a B-cell line derived from a myeloma patient. In this experiment these two cell lines were incubated with a FITC-labeled monoclonal antibody to CD3 under conditions routinely used for flow cytometry. A portion of each cell line was then subjected to immuno-RCA using a conjugate that reacts with FITC, thereby amplifying the signal from the bound FITC-labeled primary antibody. Flow analysis of the samples treated only with the primary antibody demonstrated a 10-fold greater fluorescence intensity for the CD3-positive Jurkat cells, than the CD3-negative U266 cells. After immuno-RCA, the fluorescent intensity of the Jurkat cells was increased by ∼40-fold. Most significantly, the relative signal for CD3-positive cells was at least 60-fold greater than that for the negative-control cell line. Thus, the discrimination between these two populations is improved by immuno-RCA. Similar results have been obtained by applying the same approach to the flow analysis of intracellular IgE in myeloma cells, discrimination between κ and λ light chain proteins in immunoglobulin-producing B cells as well as CD4 in a T-lymphocyte cell line (data not shown).
Figure 6.
RCA-amplified detection of CD3 antigen by flow cytometry. Jurkat and U266 cells were treated with FITC-labeled CD3 antibody as described in Methods and Materials. Cell suspensions were then divided into two aliquots. One portion was analyzed directly by flow cytometry. The second was subjected to amplification by immuno-RCA using a conjugate that recognizes FITC. The RCA products generated on the cells were detected using a FITC-labeled decorator probe, and the cells were subjected to flow cytometry. All cell suspensions were analyzed in the same run, with identical settings. The flow cytometer was set so the fluorescence intensity of the U266 (CD3-negative) cells, not subjected to RCA amplification, was 1.
Discussion
The features of RCA demonstrated in this work are applicable to analyzing the molecular characteristics of individual cells. Increased sensitivity without compromising cellular and tissue morphology has been demonstrated for immunohistochemical and flow cytometry applications. ImmunoRCA provides a universal approach to detect a variety of targets that differ in relative levels and localization.
In each of the cell-based assays presented in this report, the increase in sensitivity derived from use of RCA technology came with no loss of specificity. This is consistent with the application of RCA technology to soluble assays 9 and microarrays. 10,11 In general, the advantage of RCA is a significant increase in signal that is achieved through a single round of enzymatic amplification of a nucleic acid substrate. This is in contrast to biotinyl tyramide, which requires a multilayered amplification process. ImmunoRCA derives its specificity from an antibody-antigen interaction, and its signal amplification from nucleic acid hybridization and synthesis. As a result, it circumvents the problem of using multiple steps of antibody-mediated recognition and amplification that will suffer from similar nonspecific interactions.
The detection of rare cells with clinically relevant genotypes or phenotypes can be accomplished through in situ hybridization or immunohistochemistry. The challenge in both cases is to have increased sensitivity of detection, so the desired nucleic-acid or immunological probe can be used under sufficiently stringent conditions to prevent nonspecific reactions that confound analysis of clinical samples. The advantage of RCA, perhaps illustrated best by the flow cytometry data presented here, is that a single round of RCA can increase the signal from a specific target recognition event, leading to improvements in relative discrimination as well as absolute amounts of measurable signal. In contrast polymerase chain reaction or other nucleic-acid-based amplification approaches involve multiple rounds of amplification that can magnify background as well as specific signal. As a result of the RCA product being tethered to the conjugate, the signal is localized. This localized signal provides RCA with a significant advantage over biotinyl tyramide and polymerase chain reaction whose products would not remain localized on the cell surface in a flow cytometry assay.
The data presented are consistent with the feasibility of using RCA as a universal platform for detection of a variety of targets in cells and tissues. Because the kinetics of the RCA reactions used in the current approach are linear, it should be possible to adjust sensitivity of detection by varying incubation time or the concentration of immunoconjugates. Moreover, because the amplification is relatively simple, there are no additional steps yielding antibody-like background. Therefore, it should be possible to obtain reasonable sensitivity in a short time with greater ease than with conventional techniques or signal amplification techniques that involve serial incubations with detection reagents. Thus, the length of time or concentration of primary antibodies involved in the immunohistochemistry or flow protocols used in our experiments could be varied to permit rapid detection of targets expressed at moderate levels, and increased to detect rare antigens. The reagents used for immuno-RCA are relatively universal among the various approaches described here. The same circles, conjugates, and DNA polymerase can be used for cells and tissues fixed in different ways, and probed with a range of antibodies. The use of additional pairs of circles and conjugates can permit the simultaneous detection of multiple targets. 10 The capability to multiplex in this way should serve to extend the applicability of immuno-RCA and other variations on RCA technology.
The detection of single nucleotide polymorphisms in at least one locus has been accomplished using RCA in conjunction with padlock probes 9 on nuclear halo cytological preparations. This illustrates the potential of RCA to provide allele discrimination within individual cells, and amplify the product of this specific hybridization event. The immuno-RCA protocols described here, as applied to recognition of a specific small molecule (FITC), have been extended to hapten-labeled nucleic acid probes for nuclear DNA sequences or cytoplasmic mRNA molecules (unpublished data). RCA holds the promise of providing a universal platform for integrating current techniques in molecular tissue pathology with the vast amounts of information emerging from genomics and proteomics.
Footnotes
Address reprint requests to Vanessa Wheeler, Molecular Staging, Inc., 300 George St., 7th Floor, New Haven CT 06511. E-mail: vanessaw@molecularstaging.com.
References
- 1.Esteban JM, Ahn C, Battifora H, Felder B: Predictive value of estrogen receptors evaluated by quantitative immunohistochemical analysis in breast cancer. Am J Clin Pathol 1994, 102:S9-S12 [PubMed] [Google Scholar]
- 2.Evans WE, Relling MV: Pharmacogenomics: translating functional genomics into rational therapeutics. Science 1999, 286:487-491 [DOI] [PubMed] [Google Scholar]
- 3.Shi SR, Key ME, Kalra KL: Antigen retrieval in formalin-fixed, paraffin-embedded tissues: an enhancement method for immunohistochemical staining based on microwave oven heating of tissue sections. J Histochem Cytochem 1991, 39:741-748 [DOI] [PubMed] [Google Scholar]
- 4.Leong AS, Milios J: Accelerated immunohistochemical staining by microwaves. J Pathol 1990, 161:327-334 [DOI] [PubMed] [Google Scholar]
- 5.de Haas RR, Verwoerd NP, van der Corput MP, van Gijlswijk RP, Siitari H, Tanke HJ: The use of peroxidase-mediated deposition of biotin-tyramide in combination with time-resolved fluorescence imaging of europium chelate label in immunohistochemistry and in situ hybridization. J Histochem Cytochem 1996, 44:1091-1099 [DOI] [PubMed] [Google Scholar]
- 6.Malisius R, Merz H, Heinz B, Gafumbegete E, Koch BU, Feller AC: Constant detection of CD2, CD3, CD4, and CD5 in fixed and paraffin-embedded tissue using the peroxidase-mediated deposition of biotin-tyramide. J Histochem Cytochem 1997, 45:1665-1672 [DOI] [PubMed] [Google Scholar]
- 7.Mengel M, Werner M, von Wasielewski R: Concentration dependent and adverse effects in immunohistochemistry using the tyramine amplification technique. Histochem J 1999, 31:195-200 [DOI] [PubMed] [Google Scholar]
- 8.Kaufmann O, Kother S, Dietel M: Use of antibodies against estrogen and progesterone receptors to identify metastatic breast and ovarian carcinomas by conventional immunohistochemical and tyramide signal amplification methods. Mod Pathol 1998, 11:357-363 [PubMed] [Google Scholar]
- 9.Lizardi PM, Huang X, Zhu Z, Bray-Ward P, Thomas DC, Ward DC: Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nat Genet 1998, 19:225-232 [DOI] [PubMed] [Google Scholar]
- 10.Schweitzer B, Wiltshire S, Lambert J, O’Malley S, Kukanskis K, Zhu Z, Kingsmore S, Lizardi P, Ward D: Immunoassays with rolling circle DNA amplification: a versatile platform for ultrasensitive antigen detection. Proc Natl Acad Sci USA 2000, 97:10113-10119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wiltshire S, O’Malley S, Lambert J, Kukanskis K, Edgar D, Kingsmore S, Schweitzer B: Detection of multiple allergen-specific IgE on microarrays by immunoassay with rolling circle amplification. Clin Chem 2000, 46:1990-1993 [PubMed] [Google Scholar]
- 12.Schweitzer B, Kingsmore S: Combining nucleic acid amplification and detection. Curr Opin Biotech 2001, 12:21-27 [DOI] [PubMed] [Google Scholar]
- 13.Zhong X, Lizardi PM, Huang X, Bray-Ward PL, Ward DC: Visualization of oligonucleotide probes and point mutations in interphase nuclei and DNA fibers using rolling circle DNA amplification. Proc Natl Acad Sci USA 2001, 98:3940-3945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sano T, Smith CL, Cantor CR: Immuno-PCR: very sensitive antigen detection by means of specific antibody-DNA conjugates. Science 1992, 258:120-122 [DOI] [PubMed] [Google Scholar]
- 15.Ruzicka V, Marz W, Russ A, Gross W: Immuno-PCR with a commercially available avidin system. Science 1993, 260:698-699 [DOI] [PubMed] [Google Scholar]