Abstract
Different studies have already shown that the isolated inactivation of p21, p16, or p27 cyclin-dependent kinase inhibitors (CKIs) is associated with increased growth fraction, tumor progression, or decreased overall survival in cases of non-Hodgkin’s lymphoma. In this study we linked molecular study of the p53 and p16 genes with immunohistochemical analysis of p27 expression in a group of aggressive B-cell lymphomas [large B-cell lymphomas (LBCLs) and Burkitt’s lymphomas]. This was done to analyze the relationship between p53 and p16 silencing, p27 anomalous overexpression, and clinical follow-up, testing the hypothesis that the accumulation of CKI alterations could confer to the tumors a higher aggressivity. In a group of 62 patients, p53 inactivation as a result of mutation was observed in 11 cases (18%) and p16 silencing was seen in 27 cases (43.5%) as a result of methylation (20 of 62), 9p21 deletion (7 of 44), or p16 mutation (2 of 62). The simultaneous inactivation of p53 and p16 was detected exclusively in five LBCL cases. Anomalous expression of p27, which has been proven to be associated with the absence of p27/CDK2 complexes and the formation of p27/cyclin D3 complexes where p27 is inactivated, was detected in 19 of 61 cases (31%). Cases characterized by p27 anomalous expression display concurrent inactivation of p21 (provided by p53 mutations) and/or p16 CKIs in 11 of 14 LBCL cases (P = 0.040). When the relationship between the association of inactivated CKIs and overall survival was considered, a significant relationship was found between a lower overall survival probability and an increased number of inactivated CKIs in LBCL cases, with the worst prognosis for the cases displaying concurrent p53, p16, and p27 alterations. This proves that simultaneous inactivation of different tumor suppressor pathways does indeed take place, and that tumor aggressiveness takes advantage of this CKI-concerted silencing. In this same series of data, Burkitt’s lymphoma patients seem to behave in a different way than LBCLs, with p53 and p16 alteration being mutually exclusive and the association with p27 anomalous expression not being clinically significant. These facts seem to support that the additive effect of the inactivation of different CKIs could be dependent of the histological type.
Different genes working in multiple pathways tightly regulate cell cycle control. Two of these genes, p53 and p16, are the most frequently altered genes in human tumors. 1-4 The p16 protein is codified by INK4A/ARF located at the 9p21 locus, which also codifies for the p14/ARF (p14) protein. This locus is the nexus between the two main pathways that control cell cycle progression. The first pathway involves p16 cyclin-dependent kinase inhibitor (CKI), Rb tumor suppressor gene, CDK4 kinase, and cyclin D, and regulates passage through the G1 restriction point. p14 acts together with the p53 tumor suppressor gene, p21CKI, and MDM2, forming the pathway that regulates the G1 and G2 checkpoints or induces apoptosis in response to DNA damage or other cellular insults. MDM2 is also a nexus between both pathways, as p53 and p14 regulate it, and it inhibits p53 and regulates Rb, impeding its association with E2F transcription factors. Both pathways have been shown to be inactivated in a high percentage of human tumors, including lymphomas. 1-7 More frequently, alterations in the p53 and/or p16 genes are the cause of disruptions in both pathways, although anomalies in the other components have also been described. 8,9
In lymphomas, the p53 gene has been found to be mutated in 19 to 35% of cases, 10,11 whereas p16 has been found silenced in from 14 to 85% of cases, usually because of promoter hypermethylation or allelic loss, although rare mutations have been observed. 12-15
p27/KIP1 (p27) is another CKI, which also acts at the restriction point, blocking Rb phosphorylation by cyclin E/CDK2 inactivation. p27 overexpression in a group of aggressive B-cell lymphomas with a high proliferative index and adverse clinical outcome has been shown to occur in previous studies. 16-18 p27 overexpression has also been referred to in other types of tumors. 19,20 This anomalous expression of p27 was described in association with the absence of p27/CDK2 complexes and the formation of p27/cyclin D3 complexes where p27 is inactive, stabilized, and detectable by immunohistochemical techniques. 21,22
In vitro studies have demonstrated the existence of p27-cyclin D/CDK6 or p27-cyclin D/CDK4 complexes with Rb-kinase activity in cell lines of different types, 23-26 suggesting that p27 is not active when joined to cyclin D/CDK4-6 complexes. A redistribution of p27 from cyclin E/CDK2 to cyclin D/CDK4 complexes, which precludes its negative regulator action on CDK2 activity, has also been described in growing cells, where p27 is associated with cyclin D/CDK4. 21,22,27,28 p15 induction by transforming growth factor-β displaces p27 from these complexes to cyclin E/CDK2, co-operating in inducing cell cycle arrest. 23
The co-operation of the p16/Rb and p53/ARF pathways is suggested by evidence of the synergy found in in vitro analysis and the concomitant alteration in both pathways in many tumor types. 29-37 Initial studies in non-Hodgkin’s lymphomas have shown hints of concerted inactivation in both pathways, as has also been described in virally -induced cell transformation. 34 In mantle cell lymphomas it has recently been shown that simultaneous inactivation of the p16/Rb and p53/ARF pathways is associated with higher aggressivity, 38 but this has not yet been fully explored in more frequent types of lymphoma, such as large B-cell lymphoma (LBCL) and Burkitt’s lymphoma (BL). We therefore explore the status and correlation of the p53 and p16 pathways and the possible relationship with anomalous p27 expression, to find if the accumulation of alterations in CKIs in the same tumor could give more aggressiveness to the tumor. To this end, we performed a molecular analysis for the p53 and p16 genes, together with an immunohistochemical study of p27 and Ki67 proliferation index, in a group of 62 cases of LBCLs and BLs.
Materials and Methods
Tissue Samples and Clinical Study
Sixty-two samples of tumor specimens from aggressive B-cell lymphoma, including 46 cases of LBCL and 16 BL, were obtained from the routine files of the Virgen de la Salud Hospital, and diagnosed using the criteria of the revised European-American Classification (REAL) classification. 39
Cases were selected taking into account the existence of an adequate clinical follow-up and the availability of tissue for molecular studies. Some of the tumors in this series were included in previous studies of p53 mutations 10,11 and p16 alterations. 12
Molecular Studies
Allelic Loss Assays at the 9p21 Locus
Tumor and normal DNA were analyzed for loss of heterozygosity (LOH) or homozygous deletion (HD) by amplification of dinucleotide repeats containing sequence microsatellite markers, 40 under conditions previously described by Villuendas and colleagues. 12 These markers are at the 9p21 region, surrounding the p16 gene (IFN-α, D9S736, D9S1749, D9S1747, D9S974, D9S1748, D9S1752, D9S171). Briefly, 100 ng of DNA was amplified in a volume of 25 μl with 1× polymerase chain reaction (PCR) buffer, 200 μmol/L dNTPs, 10 pmol of each primer, 1 μCi of 32P-dCTP and 1 U of Taq polymerase. Amplified products were separated by electrophoresis in denaturing 7 mol/L urea-6% acrylamide gels, followed by autoradiography. LOH was considered to have occurred if an entire band was missing or was reduced by >40% of the normal band. If the band was reduced by <40% it was considered to be imbalanced. 12,41 The presence of HDs was assessed in all cases by comparative multiplex PCR assay, using primer sets from loci outside the 9p21 region (D9S934 and D7S1824, in chromosome 9q and 7q, respectively).
Methylation Studies
Methylation-specific PCR (MSP) assays were performed to analyze the methylation status of CpG islands of the first exon of p16, as described by Herman and colleagues. 42 Briefly, 1 μg of denatured genomic DNA was modified by reaction with sodium bisulfite under conditions that convert all unmethylated cytosines to uracils. Modified DNA was purified using the Wizard DNA clean-up system (Promega, Madison, WI). Modification was completed by NaOH 0.3 mol/L treatment for 5 minutes at room temperature, followed by ethanol precipitation.
Bisulfite-modified DNA was amplified using p16 unmethylated-specific primers (U), methylated-specific primers (M), and unmodified or wild-type primers (W). 42 Fifty ng of bisulfite-modified DNA was amplified using 1 U of AmpliTaq Gold (Applied Biosystems, Weiterstadt, Germany) under the following conditions: 30 seconds at 94°C, 30 seconds at 60°C (for U and W), or 65°C (for M) and 30 seconds at 72°C, for 35 cycles. Controls without DNA and positive controls for U and M reactions were performed for each set of PCRs. The PCR product was visualized under UV illumination in agarose gels stained with ethidium bromide. If a methylation-specific PCR product was detected, the whole procedure using sodium bisulfite and MSP was performed again to minimize the possible influence of contamination or incomplete bisulfite treatment.
DNA methylation was determined by the presence of a 150-pb fragment in those samples amplified with the M primers. DNA from nontumor samples (including peripheral blood lymphocytes from eight healthy donors and three reactive lymphoid nodes) was analyzed as a negative control. DNA from the Raji Burkitt cell line was used as a positive control of p16 methylation. 43
Mutation Study
Mutations in exons 5 to 8 of p53 10 and exon 2 of p16 44 genes were analyzed by direct sequencing with an Automated DNA sequencer ABI PRISM 310 genetic analyzer (Applied Biosystems) according to the manufacturer’s procedures. The primers and conditions for this sequencing have been previously described. 10,44
Immunostaining Techniques
All immunostaining techniques were performed in paraffin-embedded tissue sections, using a previous step of heat-induced antigen retrieval for all antibodies. Thus before incubation with the antibodies, the slides were heated in a pressure cooker for 3 minutes in a solution of 0.01 mol/L sodium citrate. After incubation with the primary antibodies, immunodetection was performed with biotinylated anti-mouse immunoglobulins, followed by peroxidase-labeled streptavidin (LSAB-DAKO, Glostrup, Denmark) with diaminobenzidine chromogen as substrate. All immunostaining was performed using the Techmate 500 (DAKO) automatic immunostaining device. Incubation omitting the specific antibodies was used as negative control of the technique.
p27 protein was detected with a monoclonal antibody, K25020 (1:1000 dilution; Transduction Laboratories, Lexington, KY), generated against the full-length mouse KIP1 protein. Expression in small lymphocytes was used as an internal control of immunostaining. p27 was scored in the following way: the mean value of the series, (17%), was chosen as a threshold for p27 reactivity. This is approximately equivalent to the level of expression of p27 within the germinal centers.
The proliferation index was evaluated using nuclear antigen Ki67 expression detected with MIB1 antibody from Novocastra (Newcastle on Tyne, UK). Cells showing nuclear staining were considered to be Ki67-positive. The Ki67 index was determined by scoring at least 300 tumor cells in well-preserved areas as a percentage of Ki67-positive cells. Ki67 reactivity was rounded to the nearest fifth percentile.
Statistical Analysis
Statistical study of the correlation between distributions was performed using either the chi-square test for categorical variables (Fisher’s exact test, Yates’ correction, or the Pearson test as appropriate) and the Kruskal-Wallis test for single ordered data. Survival curves were calculated by the Kaplan-Meier method and compared by log rank. Differences were considered significant if P < 0.05. We did not have appropriate clinical data for only 2 of 62 cases (two LBCL cases). The SPSS software package for Windows (SPSS, Inc., Chicago, IL) was used for all statistical analysis.
Results
Analysis of p53 Mutation
We searched for mutations in exons 5 to 8 of the p53 gene. Fourteen mutations were identified in 13 cases (Table 1) ▶ . Ten mutations were missense, and there was one nonsense mutation. Three silent mutations were identified in codon 213 (exon 6) in cases LB322, LB415, and BL324. In case BL324 two mutations were detected: one silent mutation in codon 213, and one missense mutation in codon 215 (Table 1) ▶ . In both codons, as well as in the mutations detected in cases LB194 and LB271, the peak corresponding to mutated nucleotide is higher than that corresponding to wild-type (wt), suggesting that one allele is mutated and the other deleted. The wt peak can be interpreted as being derived from the nontumoral cells present in the sample.
Table 1.
Cases with p53 Mutations Including Type of Mutation
| Case | Codon (exon) | Mutation | Change in amino acid |
|---|---|---|---|
| LB14 | 158 (5) | CGC→CAC | Arg→His |
| LB21 | 244 (7) | GGC→GAC | Gly→Asp |
| LB47 | 273 (8) | CGT→CAT | Arg→His |
| LB48 | 273 (8) | CGT→CAT | Arg→His |
| LB54 | 270 (8) | TTT→CTT | Phe→Leu |
| LB146 | 245 (7) | GGC→AGC | Gly→Ser |
| LB194 | 157 (5) | GTC→TTC | Val→Phe |
| LB271 | 135 (5) | TGC→TTC | Cys→Phe |
| LB322 | 213 (6) | CGA→CGG | Arg→Arg |
| LB415 | 213 (6) | CGA→CGG | Arg→Arg |
| BL13 | 196 (6) | CGA→TGA | Arg→STOP |
| BL172 | 248 (7) | CGG→CAG | Arg→Gln |
| BL324 | 215 (6) | AGT→CGT | Ser→Arg |
| 213 (6) | CGA→CGG | Arg→Arg |
Cases LB14, LB48, LB54, LB146, and BL172 showed both wt and mutant dinucleotides at similar intensity, suggesting that they also retained the normal allele. Another possibility is that one allele was deleted and the wt nucleotide detected was because of the nontumor cells present in the sample. In cases LB47 and BL13 the wt sequence was not detected at the mutation site, suggesting deletion of the wt allele. 10 In conclusion, p53 mutations were detected in 11 of 62 of the aggressive B-cell lymphomas analyzed (3 of 16 BL cases and 8 of 46 LBCL cases).
Analysis of p16 Alterations
9p21 Deletion
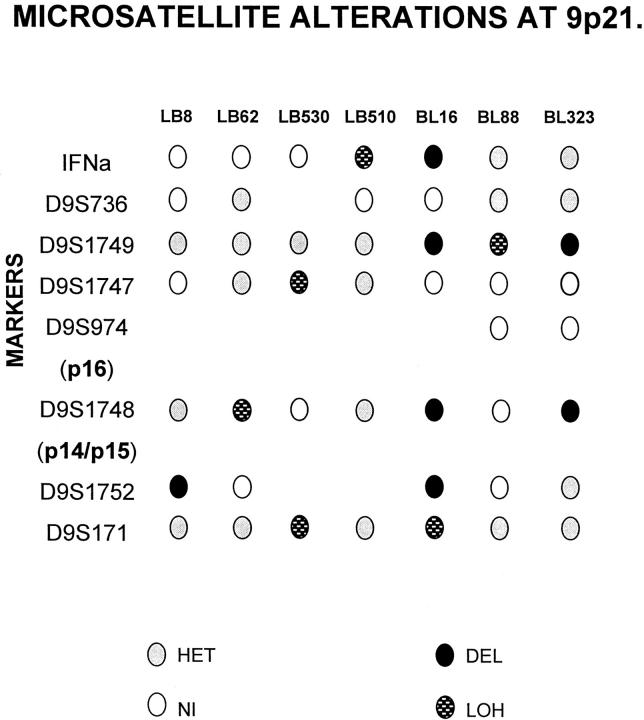
Forty-four cases, for which DNA from nontumor tissue was available, were analyzed using markers for the microsatellites surrounding the CDKN2A locus at 9p21. Allelic losses were detected in 7 of 44 cases (16%). The results of allelic loss analysis are shown in Figure 1 ▶ , and representative cases are shown in Figure 2 ▶ .
Figure 1.
Summary of data corresponding to allelic loss at the 9p21 region in aggressive B-cell lymphomas. The relative locations of the microsatellite markers analyzed and the p16 and p14 (exon 1β) genes are included. Only cases showing LOH or HD are illustrated. HET: Retention of heterozygosity; NI, noninformative.
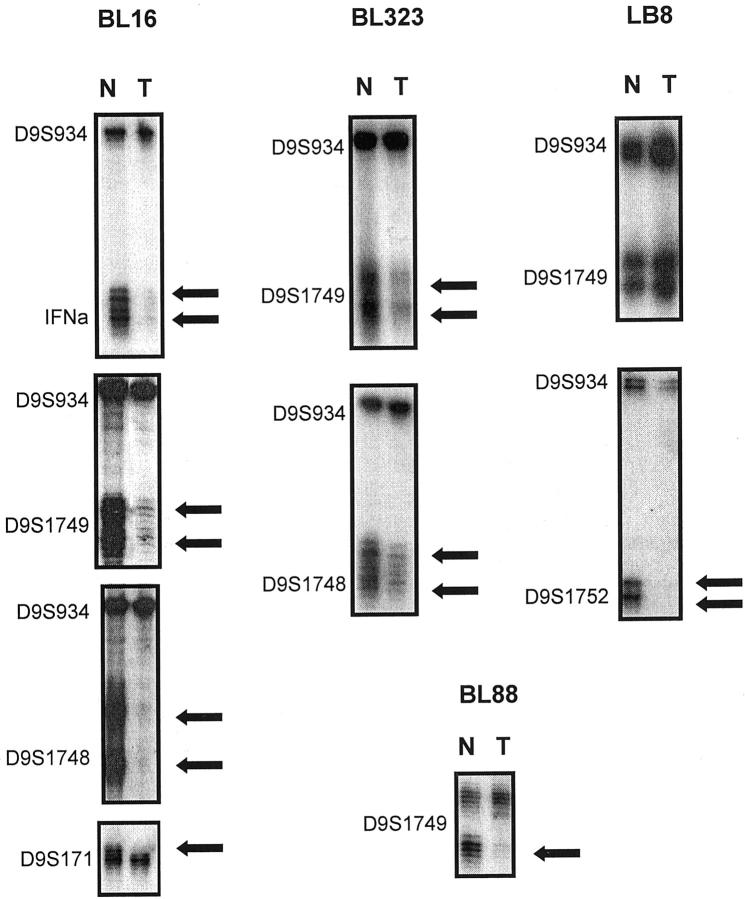
Figure 2.
Representative examples of allelic loss at the 9p21 region. The presence of allelic loss in tumor (T) in comparison with matched normal (N) DNA is shown by arrows. Case BL16 showed HD in microsatellites IFN-α, D9S1749, and D9S1748 detected by multiplex PCR with the microsatellite marker D9S934; and LOH at the D9S171 marker. Case BL323 also showed HD at microsatellites D9S1749 and D9S1748. Case LB8 showed HD at the D9S1752 marker, but there was retention of both alleles in the other microsatellites studied, such as the marker shown here, ie, D9S1749. Case BL88 showed LOH at the D9S1749 locus.
Five cases (LB62, LB510, LB530, and BL88) showed LOH in one or more microsatellite markers. Case BL16 showed a clear HD affecting D9S1752, D9S1748, D9S1749, and IFN-α loci and LOH at the D9S171 marker (Figure 2) ▶ . In case BL323 a decrease in the intensity of both alleles of tumor DNA at D9S1749 and D9S1748 markers was detected by multiplex PCR of these markers with the control locus D9S934, so we considered this case to show HD. Case LB8 showed HD of marker D9S1752, identified by multiplex PCR (Figure 2) ▶ .
Two cases (LB81 and LB66) showed instability at the D9S1749. Case LB1 showed imbalance at markers D9S1748 and IFN-α, and another case (LB54) showed imbalance at marker D9S1748. These four cases were considered to have no deleted 9p21 locus.
p16 Methylation
The MSP technique was used to analyze the hypermethylation status of the CpG island in the promoter region of the p16 gene. Hypermethylation was found in 20 of 62 cases studied (32.2%) (Figure 3) ▶ including 3 of 16 (18.7%) BLs and 17 of 46 (36.9%) LBCLs.
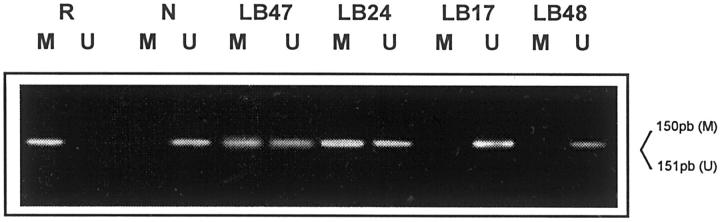
Figure 3.
Methylation-specific PCR analysis of p16 promoter. The Raji cell line (R) was used as positive control, and DNA from volunteer healthy donor (N) was used as negative control. LB24 and LB47 are examples of methylated cases, showing both the M and U bands. LB17 and LB48 are two negative cases. Only the U band could be detected in these cases.
All of the controls used (reactive lymphoid tissue and normal peripheral blood lymphocytes) showed amplification with unmethylated-specific primers (U) but not with methylated-specific primers (M). DNA from the Raji cell line was used as a positive control for the amplification reaction with p16-M primers, and no amplification was seen in the PCR reaction with p16-U primers. PCR reactions with wild-type primers (W) were used as a control of the efficacy of chemical modification.
p16 Mutation
To determine whether mutations of the p16 gene were present, we analyzed exon 2 of this gene by direct sequencing in 62 cases. Five mutations in exon 2 were identified in four different cases. Three of them correspond to the change that substitutes ACGThr for GCGAla, which has previously been described as a polymorphism (Table 2) ▶ .
Table 2.
Cases with p16 Mutations Showing Type of Mutation
| Case | Codon | Mutation | Change in amino acid |
|---|---|---|---|
| LB20 | 129 | TAC→TAG | Tyr→STOP |
| 148 | ACG→GCG | Thr→Ala | |
| LB47 | 73 | GCC→GGC | Ala→Gly |
| LB66 | 148 | ACG→GCG | Thr→Ala |
| LB169 | 148 | ACG→GCG | Thr→Ala |
p27 Expression
We investigated p27 expression by immunohistochemistry in 61 of 62 cases. Forty-two of 61 lymphoma cases showed absent or low reactivity, below the chosen threshold. All other cases (19 of 61, 31%; 14 LBCLs and 5 BLs) were considered to be positive for p27 (Figure 4 ▶ and Table 3 ▶ ).
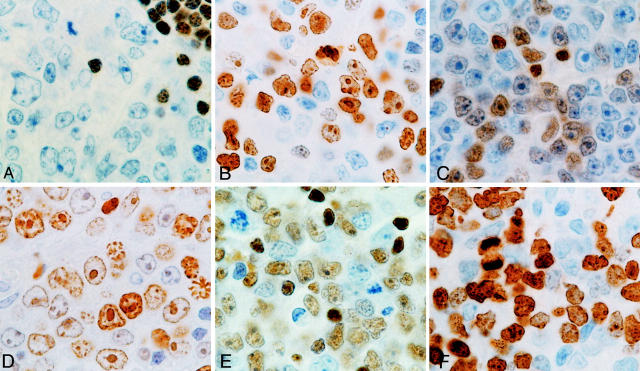
Figure 4.
Different patterns of p27 and Ki67 protein expression in aggressive B-cell lymphomas. A: p27 expression of case LB17, considered to be negative. P27 expression could only be seen in small lymphocytes. B: Ki67 expression in the same case (LB17). C: p27-positive (+) case (LB47) with the corresponding Ki67 immunostaining (D). E: A highly p27-positive (++) case (LB169), also displaying high Ki67 protein expression (F).
Table 3.
Summary of p53 and p16 Molecular Alterations, p27 and Ki67 Expression and Clinical Outcome Data in Aggressive B-Cell Lymphomas (46 LBCL and 16 BL)
| Case | p53 status | p16 status | p27 IHQ | Ki67 IHQ | Clinical outcome | |
|---|---|---|---|---|---|---|
| OS | Status | |||||
| LBCL | ||||||
| LB1 | + | 50 | 59 | Death | ||
| LB4 | MET | − | 60 | 125 | Death | |
| LB8 | HD | − | 50 | 77 | Life | |
| LB14 | MUT | MET | − | 80 | 2 | Death |
| LB17 | − | 70 | 9 | Death | ||
| LB20 | MUT | ++ | 95 | 2 | Death | |
| LB21 | MUT | MET | + | 95 | 3 | Death |
| LB24 | MET | − | 95 | 69 | Life | |
| LB47 | MUT | MET/MUT | + | 90 | N.A. | N.A. |
| LB48 | MUT | − | 90 | 23 | Death | |
| LB51 | − | 90 | 94 | Life | ||
| LB52 | − | 85 | 9 | Death | ||
| LB53 | − | 60 | 19 | Life | ||
| LB54 | MUT | MET | − | 75 | 50 | Life |
| LB55 | − | 50 | 39 | Death | ||
| LB61 | − | 65 | 36 | Life | ||
| LB62 | LOH | + | 75 | 78 | Life | |
| LB65 | − | 45 | 4 | Death | ||
| LB66 | + | 80 | 3 | Death | ||
| LB68 | MET | + | 60 | 81 | Death | |
| LB69 | − | 90 | 3 | Death | ||
| LB81 | − | 75 | 41 | Life | ||
| LB139 | MET | + | 95 | N.A. | N.A. | |
| LB146 | MUT | − | 90 | 12 | Death | |
| LB168 | − | 85 | 180 | Life | ||
| LB169 | MET | ++ | 80 | 22 | Death | |
| LB173 | MET | + | 95 | 28 | Death | |
| LB174 | MET | − | 50 | 9 | Life | |
| LB183 | MET | − | 100 | 75 | Life | |
| LB184 | − | 80 | 7 | Life | ||
| LB188 | − | 50 | 10 | Life | ||
| LB193 | − | 60 | 3 | Death | ||
| LB194 | MUT? | MET | + | 70 | 2 | Death |
| LB196 | + | 60 | 48 | Life | ||
| LB198 | − | 60 | 115 | Death | ||
| LB211 | MET | + | 95 | 28 | Life | |
| LB271 | MUT | + | 100 | 122 | Life | |
| LB311 | − | 60 | 42 | Life | ||
| LB322 | − | 50 | 22 | Life | ||
| LB340 | − | 60 | 8 | Death | ||
| LB342 | − | 70 | 26 | Death | ||
| LB415 | MET | − | 90 | 7 | Life | |
| LB510 | MET/LOH | − | 60 | 3 | Death | |
| LB530 | LOH | − | 70 | 3 | Death | |
| LB540 | MET | − | 25 | 52 | Life | |
| LB568 | − | 75 | 2 | Death | ||
| BL | ||||||
| BL1 | ++ | 100 | 3 | Death | ||
| BL7 | MET | ++ | 95 | 4 | Death | |
| BL10 | − | 90 | 6 | Death | ||
| BL13 | MUT | ++ | 90 | 96 | Life | |
| BL15 | MET | − | 95 | 55 | Life | |
| BL16 | HD | N.A. | N.A. | N.A. | N.A. | |
| BL18 | ++ | 100 | 28 | Death | ||
| BL82 | ++ | 100 | 10 | Life | ||
| BL88 | LOH | − | 90 | 111 | Life | |
| BL172 | MUT | − | 90 | 2 | Death | |
| BL181 | − | 100 | 25 | Death | ||
| BL414 | − | 100 | 42 | Life | ||
| BL323 | HD | − | 90 | 45 | Life | |
| BL324 | MUT | − | 85 | 110 | Life | |
| BL343 | MET | − | 95 | 8 | Death | |
| BL3035 | − | 90 | 71 | Life | ||
N.A., data not available; MUT, mutated; MET, methylated; HD, homozygous deletion; LOH, loss of heterozygosity.
Interrelations of p16, p53, and p27 Alterations
A summary of alterations of p16 and p53 together with p27 immunohistochemistry is shown in Table 3 ▶ . A total of 33 cases (24 of 46 LBCLs and 9 of 16 BLs) had inactivated p16 and/or p53 genes (53.2% of cases). Five cases (5 LBCLs) showed concurrent p53 mutation and p16 inactivation (8.1%), all of which had p16 inactivated by methylation, and one by mutation together with methylation. Remarkably, although it is not statistically significant, none of the seven cases showing 9p21 locus deletion (where p14, a member of the p53/ARF pathway is located) had p53 mutation. Another striking finding is the absence in the group of BLs of double p53/p16 inactivation, only observed in the LBCL group.
The majority of the cases (11 of 14 LBCLs and 2 of 5 BLs) with a high level of expression of p27 have simultaneous p53 or p16 alterations. When the relationship between p27 overexpression and alterations in p53 and/or p16 were investigated in LBCLs, a significant association was found (chi-square, 4.202; P = 0.040; Yates’ correction test), which was absent in the BL group. Nevertheless, not all of the p53/p16 alterations were associated with p27 overexpression. Thus, there were 13 LBCLs and 6 BLs with p53 and/or p16 alteration, not showing p27 overexpression.
The Effect of the Simultaneous Inactivation of Different CKIs on Overall Survival
Survival analysis was performed to investigate the impact on the survival probability of the alterations in the p53, p16, and p27 gene/protein here analyzed. Additionally to p21 and p16 silencing, p27 overexpression was included here as an unfavorable marker, because it has been previously shown that, in aggressive lymphomas, it is the result of the formation of complexes with cyclin D3, where p27 is sequestered. 21 In the group of BLs none of the alterations was significantly related with the survival probability, thus this group was not considered for this objective.
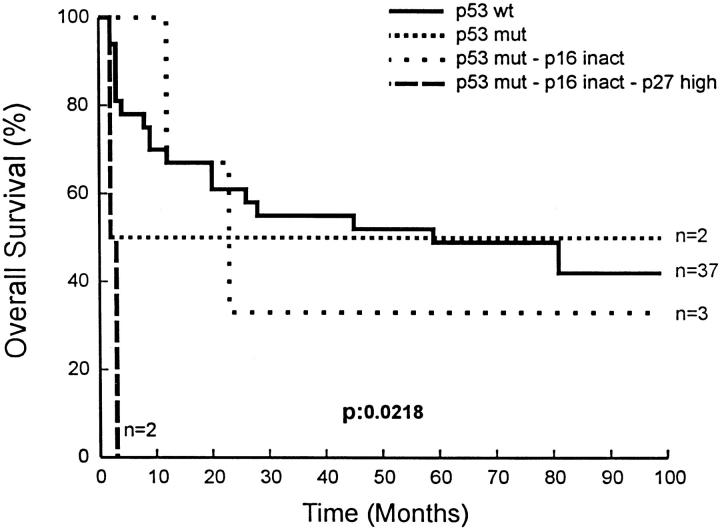
In the LBCL group, four different subgroups were compared: group A with wt p53; group B, composed of those cases with p53 mutations; group C with simultaneous p53 and p16 inactivation; and group D that included cases with accumulation of p53, p16, and p27 alterations. Patients with alteration in the three gene/proteins had a significantly worse prognosis than those with p53 mutations or without alterations (P = 0.0218) (Figure 5) ▶ .
Figure 5.
Overall survival of LBCL patients according to accumulation of alterations in p53, p16, and p27. Inact, inactivated; Mut, mutated. —, p53 wt; · · · · · ·, p53 mut-p16 inact; ······, p53 mut; – – –, p53 mut-p16 inact-p27 high.
Discussion
This study examined the status of p53 and p16 tumor suppressor genes in a series of aggressive B-cell lymphomas, together with p27 protein expression, in an attempt to elucidate if the accumulation of alterations cooperate in the pathogenesis of aggressive lymphomas.
p53 mutations and p16 inactivation have been described as the most frequent alterations in human cancer, having been found in approximately half of tumors studied. 4,5 In this study, p53 mutations were detected in 17.7% (11 of 62) of the aggressive B-cell lymphomas analyzed [3 of 16 BL cases (18.8%) and 8 of 46 (17.4%) LBCL cases], this being approximately consistent with previous reports that have shown a percentage of p53 mutations in non-Hodgkin’s lymphoma ranging from 19 to 35% of cases, most of them in aggressive tumors. 10,11 Alterations of p16 were observed in 43.5% of cases [6 of 16 BLs (37.5%) and 21 of 46 LBCLs (45.6%)]. In 20 of 62 (32.2%) we found promoter hypermethylation; 9p21 homozygous or hemizygous deletion were found in 7 of 44 cases (16%) and point mutations in 2 of 62 cases (3.2%). These results are consistent with previous reports on p16 alterations. 12-15,45-49 p27 anomalous overexpression was detected in 19 of 61 cases (30.6%), 14 of 46 LBCLs (30.4%), and 5 of 15 BLs (31.3%), a percentage comparable to those described previously. 16,21,50
A total of 33 cases (53.2%) showed inactivation of p16 and/or p53 (Table 3) ▶ . Five of these (8.1%, all LBCLs) showed concurrent p53 mutation and p16 inactivation, and all of these cases had p16 inactivated by methylation (one case also shows a mutation in p16 together with promoter hypermethylation), thereby confirming the existence of a subset of cases with simultaneous inactivation of the p53 and Rb pathways, as was recently shown by Pinyol and colleagues. 38 Strikingly, none of the analyzed BLs showed concurrent p53 and p16 alterations, and the frequency of loss of 9p21 was higher in this group, this showing that the inactivation of these growth suppressor pathways (ARF/p53 and p16/Rb) is dependent of the histological type and the underlying specific molecular alterations, such as c-myc overexpression and others.
The INK4a/ARF locus in the 9p21 chromosomal region codifies for p14 and p16 proteins; this locus is the nexus between the two main pathways that control cell cycle progression, the Rb/p16 and p53/ARF pathways. 51 None of the nine cases with LOH at 9p21 included in this series showed p53 mutation, which confirms previous observations that these two alterations may be mutually exclusive. 52
Anomalous p27 protein overexpression, detected in 19 of 61 cases of aggressive B-cell lymphomas analyzed, tends to occur in cases with p16 and/or p53 alteration in aggressive lymphomas (11 of 14 LBCLs). This association is statistically significant (P = 0.040). The cause of the anomalous overexpression of p27 in lymphomas with a high growth fraction has previously been demonstrated to be the existence of cyclin D3/p27 complexes that lead to increased stabilization of the protein. 21,22 In experimental assays it has also been shown that p15 induces p27 displacement from the cyclin D/CDK4 complex to cyclin E/CDK2 in response to transforming growth factor-β. 23 The presence of p16 has also been shown to modulate the reassortment of p27 from cyclin D/CDK4 to cyclin E/CDK2 complexes. 26 The results of this series seem to support the hypothesis that in LBCLs p27 accumulation is secondary to loss of the p16 and p21 CKIs, and therefore extends the findings of in vitro observations to a specific subset of human tumors. The absence of p16 or p21 precludes the redistribution of p27 from complexes containing CDK4/cyclin D, where p27 is stabilized and probably inactive, to others containing CDK2/cyclin E, where p27 is active in arresting cell proliferation. However, there are 17 cases with p53 or p16 inactivation that do not show simultaneous p27 overexpression, thus indicating the existence of additional factors (c-myc or other possible targets) to explain p27 accumulation. 53,54
Several in vitro studies have provided evidence of the synergy between p53 and p16, and concurrent alterations of p16 and p53 have been shown to confer a higher aggressiveness on different tumor types. 35,38 This series analyzing LBCLs shows that the concurrent inactivation of these CKIs [p21 (provided by p53 mutations), p16, and p27], operating in different but co-operative pathways has an accumulative effect on overall survival of LBCL patients (P = 0.0218). Whether this phenomenon is valid only in LBCLs or could be extrapolated to other tumors is a debatable point. Thus, in this same series of data, BL patients seem to behave in a different way than LBCLs, p53 and p16 alteration being mutually exclusive and the association with p27 anomalous expression is not clinically significant. Nevertheless, more comprehensive studies in BLs and other tumoral types could bring new data concerning the existence and characteristics of the simultaneous inactivation of different CKIs, and clarify the role that c-myc or cyclin D could play in this complex interaction.
Table 4.
Correlation between p27 Protein Expression and Status of p16 and p53 Genes in LBCL
| p27+ | p27− | |
|---|---|---|
| WT | ||
| p53 WT–p16 WT | 3 | 19 |
| ALT | ||
| p53 MUT–p16 WT | 1 | 2 |
| p16 ALT–p53 WT | 7 | 9 |
| p53 MUT–p16 ALT | 3 | 2 |
| Total ALT | 11 | 13 |
P = 0.040; n = 46.
WT, wild type; MUT, mutated; ALT, altered (p16 altered by mutation, deletion, or promoter hypermethylation).
Acknowledgments
We thank Lidia Sánchez-Verde (DAKO) for her technical assistance in immunohistochemical techniques; Cristina Romero, Isabel Fernández and Mercedes Navarrete for their excellent help in laboratory work; and Antonio José Sáez-Castillo of Jaen University for his help in setting up statistical analysis.
Footnotes
Address reprint requests to Margarita Sánchez-Beato, Programa de Patología Molecular, Centro Nacional de Investigaciones Oncológicas Carlos III, Ctra Majadahonda-Pozuelo, Km 2, 28220 Majadahonda, Madrid, Spain. E-mail: msbeato@cnio.es.
This study was supported by a grant from the Fondo de Investigaciones Sanitarias (FIS 98/993), Ministerio de Sanidad y Consumo, and, from the Comisión Interministerial de Ciencia y Tecnología (1FD97-0431), Spain.
M. S.-B. and A. I. S. contributed equally to this work.
References
- 1.Nobori T, Miura K, Wu DJ, Lois A, Takabayashi K, Carson DA: Deletions of the cyclin-dependent kinase-4 inhibitor gene in multiple human cancers. Nature 1994, 368:753-756 [DOI] [PubMed] [Google Scholar]
- 2.Liggett WH, Jr, Sidransky D: Role of the p16 tumor suppressor gene in cancer. J Clin Oncol 1998, 16:1197-1206 [DOI] [PubMed] [Google Scholar]
- 3.Hirama T, Koeffer HP: Role of the cyclin-dependent kinase inhibitors in the development of cancer. Blood 1995, 86:841-854 [PubMed] [Google Scholar]
- 4.Hollstein M, Shomer B, Greenblatt M, Soussi T, Hovig E, Montesano R, Harris CC: Somatic point mutations in the p53 gene of human tumors and cell lines. Nucleic Acids Res 1996, 24:141-146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pollock PM, Pearson JV, Hayward NK: Compilation of somatic mutations of the CDKN2 human cancers: non-random distribution of base substitutions. Genes Chromosom Cancer 1996, 15:77-88 [DOI] [PubMed] [Google Scholar]
- 6.Merlo A, Herman JG, Mao L, Lee DJ, Grabielson E, Burger PC, Baylin SB, Sidransky D: 5′CpG island methylation is associated with transcriptional silencing of the tumor suppressor p16/CDKN2/MTS1 in human cancers. Nat Med 1995, 1:686-692 [DOI] [PubMed] [Google Scholar]
- 7.Ruas M, Peters G: The p16INK4A/CDKN2A tumor suppressor and its relatives. Biochim Biophys Acta 1998, 1378:115-177 [DOI] [PubMed] [Google Scholar]
- 8.Gronbaek K, Nedergaard T, Andersen MK, Thor Straten P, Guldberg P, Moller P, Zeuthen J, Hansen EN, Hou-Jensen K, Ralfkiaer E: Concurrent disruption of cell cycle associated genes in mantle cell lymphoma: a genotypic and phenotypic study of cyclin D1, p16, p15, p53 and pRb. Leukemia 1998, 12:1266-1271 [DOI] [PubMed] [Google Scholar]
- 9.Lukas J, Aagaard L, Strauss M, Bartek J: Oncogenic aberrations of p16INK4/cdkn2 and cyclin D1 cooperate to deregulate G1 control. Cancer Res 1995, 55:4818-4823 [PubMed] [Google Scholar]
- 10.Villuendas R, Pezzella F, Gatter K, Algara P, Sanchez-Beato M, Martinez P, Martinez JC, Muñoz E, García P, Sánchez L, Kocialkowsky S, Campo E, Orradre JL, Piris MA: p21 WAF1/CIP1 and MDM2 expression in non-Hodgkin’s lymphoma and their relationship to p53 status: a p53+, MDM2−, p21− immunophenotype associated with missense p53 mutations. J Pathol 1997, 181:51-61 [DOI] [PubMed] [Google Scholar]
- 11.Mateo M, Mollejo M, Villuendas R, Algara P, Sanchez-Beato M, Martinez-Delgado B, Martinez P, Piris MA: Absence of microsatellite instability and low frequency of p53 gene mutation in splenic marginal zone lymphoma. Mol Pathol 1998, 51:262-267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Villuendas R, Sánchez-Beato M, Martínez JC, Sáez AI, Martínez-Delgado B, García JF, Mateo MS, Sánchez-Verde L, Benítez J, Martínez P, Piris MA: Loss of p16/INK4A protein expression in non-Hodgkin’s lymphomas is a frequent finding associated with tumor progression. Am J Pathol 1998, 153:887-897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martínez-Delgado B, Fernandez-Piqueras J, García MJ, Arranz E, Gallego J, Rivas C, Robledo M, Benitez J: Hypermethylation of a 5′CpG island of p16 is a frequent event in non-Hodgkin’s lymphoma. Leukemia 1997, 11:425-428 [DOI] [PubMed] [Google Scholar]
- 14.Herman JG, Civin CI, Issa J-P J, Collector MI, Sharkis SJ, Baylin SB: Distinct patterns of inactivation of p15INK4b and p16INK4a characterize the major types of hematological malignancies. Cancer Res 1997, 57:837-841 [PubMed] [Google Scholar]
- 15.Pinyol M, Cobo F, Bea S, Jares P, Nayach I, Fernández PL, Montserrat E, Cardesa A, Campo E: p16INK4a gene inactivation by deletions, mutations and hypermethylation is associated with transformed and aggressive variants of non-Hodgkin’s lymphomas. Blood 1998, 91:2977-2984 [PubMed] [Google Scholar]
- 16.Sáez AI, Sánchez E, Sánchez-Beato M, Cruz MA, Chacon I, Muñoz E, Camacho FI, Martínez-Montero JC, Mollejo M, García JF, Piris M: P27KIP1 is abnormally expressed in diffuse large B-cell lymphomas, and is associated with an adverse clinical outcome. Br J Cancer 1999, 80:1427-1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vrhorac R, Delmer A, Tang R, Marie JP, Zittoun R, Ajchenbaum-Cymbalista F: Prognostic significance of the cell cycle inhibitor p27KIP1 in chronic B-cell lymphocytic leukemia. Blood 1998, 91:4694-4700 [PubMed] [Google Scholar]
- 18.Erlanson M, Portin C, Linderholm B, Lindh J, Roos G, Landberg G: Expression of cyclin E and the cyclin-dependent kinase inhibitor p27 in malignant lymphomas—prognostic implications. Blood 1998, 92:770-777 [PubMed] [Google Scholar]
- 19.Yatabe Y, Masuda A, Koshikawa T, Nakamura S, Kuroishi T, Osada H, Takahashi T, Mitsudomi T, Takahashi T: p27KIP1 in human lung cancers: differential changes in small cell and non-small cell carcinomas. Cancer Res 1998, 58:1042-1047 [PubMed] [Google Scholar]
- 20.Anayama T, Furihata M, Ishikawa T, Ohtsuki Y, Ogoshi S: Positive correlation between p27KIP1 expression and progression of human esophageal squamous cell carcinoma. Int J Cancer 1998, 79:439-443 [DOI] [PubMed] [Google Scholar]
- 21.Sánchez-Beato M, Camacho FI, Martínez-Montero JC, Sáez AI, Villuendas R, Sánchez-Verde L, García JF, Piris MA: Anomalous high p27/KIP1 expression in a subset of aggressive B-cell lymphomas is associated with cyclin D3 over-expression. p27/KIP1-cyclin D3 colocalization in tumor cells. Blood 1999, 12:765-772 [PubMed] [Google Scholar]
- 22.Barnouin K, Fredersdorf S, Eddaoudi A, Mittnacht S, Pan LX, Du MQ, Lu X: Antiproliferative function of p27kip1 is frequently inhibited in highly malignant Burkitt’s lymphoma cells. Oncogene 1999, 18:6388-6397 [DOI] [PubMed] [Google Scholar]
- 23.Reynisdóttir I, Polyak K, Iavarone A, Massagué J: Kip/Cip and Ink4 Cdk inhibitors cooperate to induce cell cycle arrest in response to TGF-β. Genes Dev 1995, 9:1831-1837 [DOI] [PubMed] [Google Scholar]
- 24.Soos TJ, Kiyokawa H, Yan JS, Rubin MS, Giordano A, DeBlasio A, Bottega S, Wong B, Mendelsohn J, Koff A: Formation of p27-CDK complexes during the human mitotic cell cycle. Cell Growth Differ 1996, 7:135-139 [PubMed] [Google Scholar]
- 25.Cheng M, Sexl V, Sherr CJ, Roussel MF: Assembly of cyclin D-dependent kinase and titration of p27KIP1 regulated by mitogen-activated protein kinase (MEK!). Proc Natl Acad Sci USA 1998, 95:1091-1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McConnell BB, Gregory FJ, Stott FJ, Hara E, Peters G: Induced expression of p16 INK4a inhibits CDK4- and CDK2-associated kinase activity by reassortment of cyclin-CDK-inhibitor complexes. Mol Cell Biol 1999, 19:1981-1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitra J, Dai CY, Somasundaram K, El-Deiry W, Satyamoorthy K, Herlyn M, Enders GH: Induction of p21WAF1/CIP1 and inhibition of Cdk2 mediated by tumor suppressor p16INK4a. Mol Cell Biol 1999, 19:3916-3928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bagui TK, Jackson RJ, Agrawal D, Pledger WJ: Analysis of cyclin D3-cdk4 complexes in fibroblasts expressing and lacking p27(kip1) and p21. Mol Cell Biol 2000, 20:8748-8757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanding V, Brand K, Herwing S, Lukas J, Bartek J, Strauss M: Adenovirally transferred p16INK4a/CDKN2 and p53 genes cooperate to induce apoptotic tumor cell death. Nat Med 1997, 3:313-319 [DOI] [PubMed] [Google Scholar]
- 30.Shapiro GI, Edwards CD, Ewen ME, Rollins BJ: p16INK4a, participates in a G1 arrest checkpoint in response to DNA damage. Mol Cell Biol 1998, 18:378-387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW: Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 1997, 88:593-602 [DOI] [PubMed] [Google Scholar]
- 32.Greenberg R, Chin L, Femino A, Lee KH, Gottlieb GJ, Singer RH, Greider CW, DePinho RA: Short dysfunctional telomeres impair tumorigenesis in the INK4aΔ2/3 cancer-prone mouse. Cell 1999, 97:515-525 [DOI] [PubMed] [Google Scholar]
- 33.Williams BO, Remington L, Albert DM, Mukai S, Bronson RT, Jacks T: Cooperative tumorigenic effects of germline mutations in Rb and p53. Nat Genet 1994, 7:480-484 [DOI] [PubMed] [Google Scholar]
- 34.Levine AJ, Momand J: Tumor suppressor genes: the p53 and gene products. Biochim Biophys Acta 1990, 1032:119-136 [DOI] [PubMed] [Google Scholar]
- 35.Gorgoulis V, Zacharatos P, Kotsinas A, Liloglou T, Nyroudi A, Veslemes M, Rassidakis A, Halazonetis T, Meld J, Kittas C: Alterations of the locus p16-Rb pathway and the chromosome locus 9p21–22 in non-small-cell lung carcinomas. Relationship with p53 and MDM2 protein expression. Am J Pathol 1998, 153:1749-1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Markl ID, Jones PA: Presence and location of TP53 mutation determines pattern of CDKN2A/ARF pathway inactivation in bladder cancer. Cancer Res 1998, 58:5348-5353 [PubMed] [Google Scholar]
- 37.Kinoshita I, Dosaka-Akita H, Mishina T, Akie K, Nishi M, Hiroumi H, Hommura F, Kawakami Y: Altered p16INK4a and retinoblastoma protein status in non-small cell lung cancer: potential synergistic effect with altered p53 protein on proliferative activity. Cancer Res 1996, 56:5557-5562 [PubMed] [Google Scholar]
- 38.Pinyol M, Hernandez L, Martinez A, Cobo F, Hernandez S, Bea S, Lopez-Guillermo A, Nayach I, Palacin A, Nadal A, Fernandez PL, Montserrat E, Cardesa A, Campo E: INK4a/ARF locus alterations in human non-Hodgkin’s lymphomas mainly occur in tumors with wild-type p53 gene. Am J Pathol 2000, 156:1987-1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harris NL, Jaffe ES, Stein H, Banks PM, Chan JKC, Cleary ML, Delsol G, De Wolf-Peters C, Falini B, Gatter KC, Grogan TM, Isaacson PG, Knowles DM, Mason DY, Muller-Hermenlink H-K, Pileri SA, Piris MA, Ralfkiaer E, Warnke RA: A revised European-American classification of lymphoid neoplasms: a proposal from the international lymphoma study group. Blood 1994, 84:1361-1369 [PubMed] [Google Scholar]
- 40.Cairns P, Polascik TJ, Eby Y, Tokino K, Califano J, Merlo AA, Mao L, Herath J, Jenkins R, Westra W, Rutter JL, Buckler A, Gabrielson E, Tockman M, Cho KR, Hedrick L, Bova GS, Isaacs W, Koch W, Schwab D, Sidransky D: Frequency of homozygous deletion at p16/CDKN2 in primary human tumors. Nat Genet 1995, 11:210-212 [DOI] [PubMed] [Google Scholar]
- 41.Mateo M, Mollejo M, Villuendas R, Algara P, Sanchez-Beato M, Martinez P, Piris MA: 7q31–32 allelic loss is a frequent finding in splenic marginal zone lymphoma. Am J Pathol 1999, 154:1583-1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB: Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA 1996, 93:9821-9826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garcia JF, Villuendas R, Algara P, Saez AI, Sanchez-Verde L, Martinez-Montero JC, Martinez P, Piris MA: Loss of p16 protein expression associated with methylation of the p16INK4A gene is a frequent finding in Hodgkin’s disease. Lab Invest 1999, 79:1453-1459 [PubMed] [Google Scholar]
- 44.Kamb A, Gruis NA, Weaver-Feldhaus J, Liu Q, Harshman K, Tavtigian SV, Stockert E, Day RS, Johnson BE, Skolnick MH: A cell cycle regulator potentially involved in genesis of many tumor types. Science 1994, 264:436-440 [DOI] [PubMed] [Google Scholar]
- 45.Baur AS, Shaw P, Burri N, Delacrétaz F, Bosman FT, Chaubert P: Frequent methylation silencing of p15Ink4b (MTS2) and p16 Ink4a (MTS1) in B-cell and T-cell lymphomas. Blood 1999, 94:1773-1781 [PubMed] [Google Scholar]
- 46.Drexler HG: Review of alterations of the cyclin dependent kinase inhibitor INK4 family genes p15, p16, p18 and p19 in human leukemia-lymphoma cells. Leukemia 1998, 12:845-849 [DOI] [PubMed] [Google Scholar]
- 47.Elenitoba-Johnson KS, Gascoyne RD, Lim MS, Chhanabai M, Jaffe ES, Raffeld M: Homozygous deletions at chromosome 9p21 involving p16 and p15 are associated with histologic progression in follicle center lymphoma. Blood 1998, 91:4677-4685 [PubMed] [Google Scholar]
- 48.Ogawa S, Hangaishi A, Miyawaki S, Hirosawa S, Miura Y, Takeyama K, Kamada N, Ohtake S, Uike N, Shimazaki CH, Toyama K, Hirano M, Mizoguchi H, Kobayashi Y, Furusawa S, Saito M, Emi N, Yazaki Y, Ueda R, Hirai H: Loss of the cyclin-dependent kinase 4-inhibitor (p16;MTS1) gene is frequent in and highly specific to lymphoid tumors in primary human hematopoietic malignancies. Blood 1995, 86:1548-1556 [PubMed] [Google Scholar]
- 49.Stranks G, Height SE, Mitchell P, Jadayel D, Yuille MA, De Lord C, Clutterbuck RD, Treleaven JG, Powles RL, Nacheva E, Oscier DG, Karpas A, Lenoir GM, Smith SD, Millar JL, Catovsky D, Dyer MJS: Deletions and rearrangement of CDKN2 in lymphoid malignancy. Blood 1995, 85:893-901 [PubMed] [Google Scholar]
- 50.Sánchez-Beato M, Sáez AI, Martínez-Montero JC, Mateo MS, Sánchez-Verde L, Villuendas R, Troncone G, Piris MA: Cyclin dependent kinase inhibitor p27kip1 in lymphoid tissue: p27kip1 expression is inversely proportional to the proliferative index. Am J Pathol 1997, 151:151-160 [PMC free article] [PubMed] [Google Scholar]
- 51.Sharpless EN, DePinho RA: The INK4/ARF locus and its two gene products. Curr Opin Genet Dev 1999, 9:22-30 [DOI] [PubMed] [Google Scholar]
- 52.Schmitt CA, McCurrach ME, Stanchina E, Wallace-Brodeur RR, Lowe SW: INK4a/ARF mutations accelerate lymphomagenesis and promote chemoresistance by disabling p53. Genes Dev 1999, 13:2670-2677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bouchard C, Thieke K, Maier A, Saffrich R, Hanley-Hyde J, Ansorge W, Reed S, Sicinski P, Baryek J, Eilers M: Direct induction of cyclin D2 by Myc contributes to cell cycle progression and sequestration of p27. EMBO J 1999, 18:5321-5333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perez-Roger I, Kim S-H, Griffiths B, Sewing A, Land H: Cyclins D1 and D2 mediate Myc-induced proliferation via sequestration of p27/KIP1 and p21/Cip1. EMBO J 1999, 18:5310-5320 [DOI] [PMC free article] [PubMed] [Google Scholar]