Abstract
The distal half of chromosome arm 18q is frequently lost in ovarian carcinoma. To define the putative tumor suppressor locus/loci more precisely we performed allelic analysis with 27 polymorphic microsatellite markers located at 18q12.3-q23 in 64 serous and 9 mucinous ovarian carcinomas. Fifty-nine percent of the serous carcinomas, but only one (11%) of mucinous carcinomas, showed allelic loss at one or more loci (P = 0.018). In serous carcinomas, deletions were found to be associated with tumor grade and poor survival. The highest frequency of losses was detected at the distal part, 18q22-q23. Two minimal common regions of loss (MCRL) were identified at this region: MCRL1 between D18S465 and D18S61 at 18q22 (3.9 cM) and MCRL2 between D18S462 and D18S70 at 18q23 (5.8 cM). At 18q21.1, proximal to the MCRLs, there are three candidate tumor suppressor genes: SMAD4 (DPC4), SMAD2, and DCC. Their protein expression was studied by immunohistochemistry in normal ovarian tissue and serous carcinomas. Lost or very weak expression of SMAD4, SMAD2 and DCC was found in 28, 28, and 30% of serous carcinomas, respectively. Comparison of allelic loss and protein expression status indicated that none of these genes alone could be the target for the frequent allelic loss at 18q21.1. Together, these genes may account for a substantial proportion of the events, but not all of them. Thus, we propose that the frequent allelic loss at 18q is because of the effect of multiple genes, and there is at least one as yet unidentified tumor suppressor gene at 18q residing distal to SMAD4, SMAD2, and DCC involved in serous ovarian carcinoma.
On the basis of comparative genomic hybridization analyses, the distal half of 18q is a site of frequent loss of genetic material in ovarian carcinoma. 1,2 Three candidate tumor suppressor genes have been identified at 18q21.1: SMAD4 (DPC4), SMAD2, and DCC, 3-5 but few mutations in these genes have been detected in ovarian carcinoma. 6-8 Ovarian carcinoma originates from the surface epithelium of the ovary. DCC has been shown to be expressed in the surface epithelium, 9 but it is not known if SMAD4 and SMAD2 are expressed in these cells. Furthermore, it is unknown if the expression of DCC, SMAD4, and SMAD2 is lost during malignant transformation. Previously, allelic loss has been studied with several markers at 18q21 in ovarian carcinoma, but fine allelotyping has not been performed at more distal regions of 18q (18q22-q23), which is the site of most frequent loss as suggested by comparative genomic hybridization results. 1,2
Ovarian carcinoma shows several different histological types, including serous, mucinous, endometrioid, and clear cell carcinomas. There is an increasing amount of biological and molecular evidence that different histological types of ovarian carcinoma should be regarded as distinct entities. 10-14 The most common form of ovarian carcinoma is the serous histological type, which accounts for ∼55% of all cases. In our previous comparative genomic hybridization studies, loss of distal 18q was characteristic of serous ovarian carcinoma, 2,15 suggesting the presence of a tumor suppressor gene(s) at distal 18q involved particularly in the serous histological type of ovarian carcinoma.
To define the putative tumor suppressor locus/loci more precisely we performed loss of heterozygosity (LOH) analysis with 27 polymorphic microsatellite markers located at 18q12.3-q23 in 64 serous ovarian carcinomas. The expression of SMAD4, SMAD2, and DCC was studied by immunohistochemistry in normal ovarian tissue and the tumor samples, and their expression was correlated with LOH results. To compare the pattern of allelic loss at this region in serous and mucinous ovarian carcinomas, we also performed allelic analysis with the same 27 markers in 9 microdissected mucinous ovarian carcinomas.
Materials and Methods
LOH
Tumor Samples, Microdissection, and DNA Extraction
Tumor and blood samples were taken from 73 patients undergoing primary surgery for ovarian carcinoma at the Department of Obstetrics and Gynecology, Helsinki University Central Hospital. The tumors were staged according to the classification scheme of the International Federation of Gynecologists and Obstetricians. All of the specimens were reviewed by the same investigator (RB) as regards histological subtype and grade. There were 64 tumor specimens of serous histology (13 stage I, 3 stage II, 42 stage III, and 6 stage IV; 15 grade 1, 18 grade 2, and 31 grade 3) and 9 specimens of mucinous histology (8 stage I and one stage II; 4 grade 1, three grade 2, and two grade 3). After removal, the tissues were snap-frozen. In mucinous carcinomas, as a rule the amount of nonneoplastic cells was high and a laser microbeam microdissection technique 16 was used to separate carcinoma cells before DNA extraction. In serous carcinoma, only tissue samples containing >40 to 50% of tumor cells were included in the study (range, 40 to 95%; median, 70%) and no microdissection was needed. Tumor DNA from serous carcinomas was extracted from fresh-frozen tumor tissue blocks after mechanical disruption and DNA from mucinous carcinomas from frozen sections. Normal DNA was extracted from blood lymphocytes of these patients. A standard proteinase K-phenol-chloroform method was used for DNA extraction.
Microsatellites
A set of 27 highly polymorphic microsatellite markers at 18q12.3-q23 were used. Primer sequences and reaction conditions for dinucleotide markers were obtained from the Genethon human linkage map (D18S58, D18S61, D18S64, D18S65, D18S68, D18S70, D18S462, D18S465, D18S468, D18S469, D18S474, D18S483, D18S1009, D18S1118, D18S1119, D18S1130, and D18S1131) and for tri- and tetranucleotide markers from Genome Database (D18S539, D18S815, D18S844, D18S845, D18S857, D18S858, D18S871, D18S969, D18S977, and D18S979). The genetic order of the markers was based on the Genethon map, the Genome Database, and GeneMap’99 (Figure 1) ▶ . The oligonucleotides were labeled fluorescently with one of three dyes (6-FAM, TET, HEX; Institute of Biotechnology, University of Helsinki, Finland). A fourth dye (TAMRA; Perkin-Elmer, Foster City, CA) was reserved for the size standard.
Figure 1.
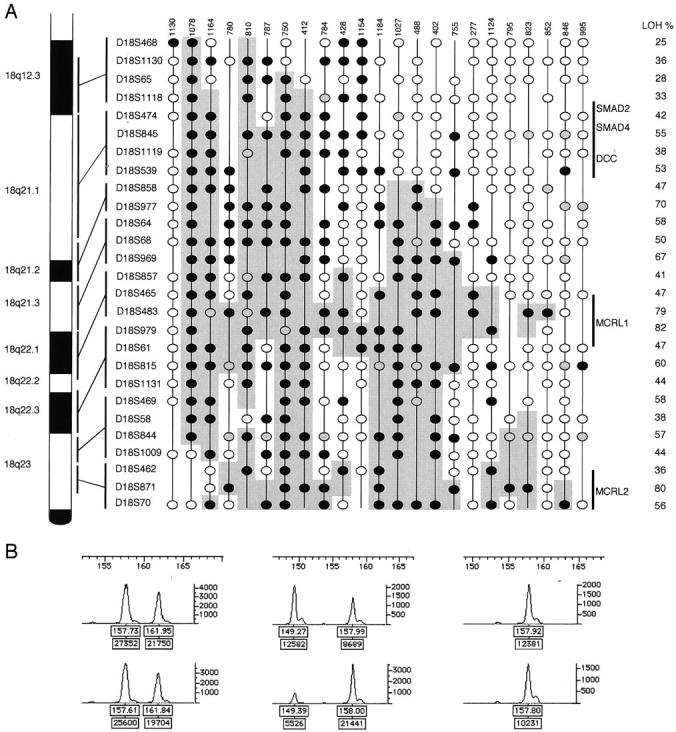
A: Deletion map of 23 serous ovarian carcinomas showing partial deletion (36%) of chromosome arm 18q. The 15 (23%) serous carcinomas with LOH at all informative markers and the 26 (41%) carcinomas with no LOH are not shown. The genetic order and the approximate loci of 27 microsatellite markers are shown on the right side of the chromosome 18q figure. Each vertical column represents one tumor sample; case number shown on the top. Frequency of allelic loss (LOH/informative) at each marker is presented on the right. Black circle, LOH; white circle, informative with no loss; gray circle, not interpretable; vertical line, not informative. Shaded area, potential deletions including the minimal common regions of loss (markers showing LOH and flanking noninformative markers). MCRL1 and MCRL2, minimal common regions of loss. B: Representative examples of LOH assessment, marker D18S979; left, informative with no loss, case 1130; middle, LOH, case 1078; right, not informative (homozygous), case 1164; size in bp is shown on the x axis at the top of each figure. The peak heights in fluorescence units are shown on the y axis on the right of each figure. In each figure the upper trace is amplification from normal tissue and the lower trace amplification from tumor tissue. Each peak has two labels (boxes): upper label, size in bp; lower label, peak area.
Polymerase Chain Reaction
The polymerase chain reaction reactions for genotyping were performed in a volume of 10 μl and included GeneAmp 1× polymerase chain reaction buffer (Perkin-Elmer), each dNTP at 50 μmol/L, 60 ng DNA (5 to 10 ng DNA from the microdissected samples), 0.5 U AmpliTaq Gold polymerase (Perkin-Elmer), and 5 pmol of each primer (one of them fluorescently labeled). The reaction mixtures were given 30 to 35 cycles of 5 seconds at 96°C, 59 seconds at 92°C, 1 minute 15 seconds at 55°C (60°C for D18S474, D18S815, D18S844, and D18S845), and 45 seconds at 72°C, preceded by a 10-minute hot start at 96°C for enzyme activation and followed by final extension at 72°C for 30 minutes.
Electrophoresis and Allele Scoring
The 27 products were pooled in three groups, each consisting of 9 μl. One μl of this mixture was added to 12.5 μl of formamide and 0.5 μl of TAMRA 500 size standard and denatured at 96°C for 3 minutes before loading the samples into an ABI Prism 310 Genetic Analyzer (Perkin-Elmer), which introduces the samples into a polymer-filled capillary for electrophoresis. Analysis of raw data and assessment of LOH were performed with GeneScan and Genotyper software (Perkin-Elmer). The peaks of the normal DNA sample were used to determine whether the sample was homozygous (one peak only) or heterozygous (two peaks). The sizes of the allele peaks were assigned according to the area under the highest peak. When two alleles were present in normal tissue and one was absent in the tumor, the result was determined to be LOH (Figure 1B) ▶ . In cases in which the assessment was not clear-cut, the ratio of alleles was calculated for each normal and tumor sample, and the tumor ratio was divided by the normal ratio, ie, T2:T1/N2:N1 (T1 and N1 are the area values for the shorter length alleles and T2 and N2 are the values for the longer length alleles, for tumor and normal tissue, respectively). If the ratio was <0.6 or >1.67, the result was determined to be LOH. 17 In ambiguous cases, the polymerase chain reaction, electrophoresis, and scoring were repeated.
SMAD4, SMAD2, and DCC Immunohistochemistry
Sixty of the 64 serous tumors analyzed for LOH and 20 normal ovarian samples were included in a tissue microarray, which was constructed as described previously. 18 In brief, core tissue biopsy specimens (diameter, 0.8 mm) were taken from representative areas of individual donor blocks and precisely arrayed into a new recipient paraffin block with a custom-built precision instrument (Beecher Instruments, Silver Spring, MD). Four core tissue biopsies were obtained from each carcinoma specimen. After block construction was completed, 5-μm sections were cut with a microtome. The presence of tumor tissue in the arrayed samples was verified in hematoxylin and eosin-stained sections.
The presence of SMAD4, SMAD2, and DCC protein in the samples was analyzed by immunohistochemistry using antibodies and protocols described earlier (R Salovaara, et al, Frequent loss of SMAD4/DPC4 protein in colorectal cancers, submitted). 19-21 The primary antibodies were: monoclonal anti-human SMAD4 (final concentration 2 μg/ml, sc-7966; Santa Cruz Biotechnology Inc., Santa Cruz, CA), goat polyclonal anti-SMAD2-peptide (6 μg/ml, sc-6200; Santa Cruz Biotechnology Inc.), and monoclonal anti-human DCC (5 μg/ml, clone G97-499; Pharmingen, San Diego, CA). The sections were pretreated in a microwave oven in buffered sodium citrate before SMAD4 and DCC immunohistochemistry. An avidin-biotin immunoperoxidase system was used to visualize the bound antibody. For SMAD4, the procedure was run in a Techmate automated machine (Peroxidase DAB detection kit; DAKO ChemMate, Glostrup, Denmark). For SMAD2 and DCC, the procedure was performed manually (Vectastain Elite ABC kit; Vector Laboratories, Burlingame, CA) and 3-amino-9-ethylcarbazole was used as the chromogen. The sections were counterstained with Mayer’s hematoxylin. Blocking of the antibody by peptide preincubation (SMAD2) or omitting the primary antibody were used as negative controls. Normal ovaries have previously been shown to express SMAD2 by Northern blotting 22 and DCC by immunohistochemistry and reverse transcriptase-polymerase chain reaction. 9 Thus, normal ovarian samples were used as positive controls for SMAD2 and DCC in the present study. For SMAD4, colon carcinoma cell lines shown to express SMAD4 by Western blotting and reverse transcriptase-polymerase chain reaction, were used as positive controls (R Salovaara, S Roth, A Loukola, V Launonen, P Sistonen, E Avizienyte, P Kristo, H Jarvinen, S Souchelnytsky, M Sarloma-Rikala, LA Aaltonen, submitted for publication). Absence of any reactivity or very weak staining that diverged from that observed in the surface epithelium of normal ovaries and the general pattern of positive staining of the tumor samples were interpreted as negative.
Statistical Analyses
Differences in LOH and lost expression were tested by using Fisher’s exact test (two-tailed P values) and allelic loss of informative markers by using the nonparametric Mann-Whitney U test. The product-limit method was used to construct survival curves and statistical significance was tested by log-rank analysis. Multivariate analysis was performed by using the Cox proportional hazards model.
Results
LOH at 18q12.3-q23 in Serous Ovarian Carcinoma
To refine the deletion map of the distal part of chromosome arm 18q we performed allelic analysis of 64 serous ovarian carcinomas using 27 microsatellite markers at 18q12.3-q23. The samples were informative on average at 18 loci (range, 13 to 25). Replication error of three microsatellite markers was seen in one tumor (case 1097) and three tumors (cases 810, 852, and 1106) showed replication error at only one marker each. Thirty-eight of 64 (59%) serous ovarian carcinomas studied showed allelic loss at one or more markers at distal 18q. Fifteen of the tumors (23%) showed loss of all informative markers, suggesting complete loss of distal 18q. Twenty-three samples showed partial deletion and they were used to construct a deletion map of this region (Figure 1) ▶ . In this deletion map 12 microsatellite markers showed allelic loss of >50% of informative alleles (D18S845, D18S539, D18S977, D18S64, D18S969, D18S483, D18S979, D18S815, D18S469, D18S844, D18S871, and D18S70) that were located at 18q21-23. There were three markers that showed LOH in >75% of informative alleles: D18S483 (18q22), D18S979 (18q22), and D18S871 (18q23). Two minimal common regions of loss could be defined around these markers with the highest percentage of LOH: MCRL1 between markers D18S465 and D18S61 (18q22) and MCRL2 between markers D18S462 and D18S70 (18q23).
Allelic Loss at Distal 18q in Mucinous Versus Serous Ovarian Carcinomas
In mucinous carcinomas LOH was found in one of the nine specimens (11%) and it showed LOH at 12 of 15 informative markers. The number of cases showing allelic loss in mucinous versus serous (59%) carcinomas was statistically significant (P = 0.018). The mean degree of LOH of informative alleles in mucinous carcinomas was 8.3% and in serous carcinomas it was 42% (P = 0.013). When only grade 1 and 2 (mucinous, 0%; serous, 36%) or stage I and II (mucinous, 8.3%; serous, 31%) tumors were taken into account, the difference in the degree of LOH of informative alleles still remained significant (P = 0.022 and 0.035, respectively).
Clinicopathological Characteristics and LOH at Distal 18q in Serous Ovarian Carcinomas
In serous carcinomas LOH at distal 18q was detected in 7.1% of grade 1 tumors, 72% of grade 2 tumors, and 77% of grade 3 tumors (grade 1 versus grades 2 and 3, P < 0.001). There was no correlation between LOH and stage of the serous tumors: LOH was detected in 56% of stage I and II tumors and in 60% of stage III and IV tumors.
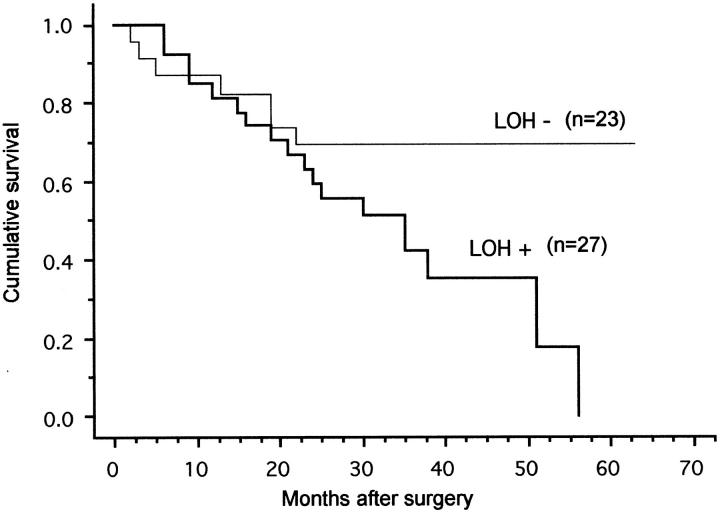
Follow-up data for >24 months was available for 52 of the 64 serous ovarian carcinomas analyzed for allelic loss at 18q. Two patients were excluded from the analysis: the cause of death of one patient was uncertain and the other patient died of a postoperative complication. The remaining 50 patients that were included in the analysis were either alive or had died of the ovarian carcinoma (Figure 2) ▶ . There were 27 carcinomas that showed LOH at 18q and 18 (67%) of these patients had died of the carcinoma, whereas of the 23 patients showing no LOH at 18q, 7 (30%) had died of their disease. The mean follow-up times of live patients were 37 and 38 months, respectively. The difference in survival between these two groups was statistically significant (P = 0.044). Tumor grade was also associated with survival (P = 0.0009), but the association between tumor stage and survival did not reach statistical significance (P = 0.058). In multivariate analysis, including grade, stage, and LOH status, only grade was an independent prognostic factor.
Figure 2.
Survival of patients with serous ovarian carcinoma according to allelic loss at 18q12.3-q23.
Immunohistochemistry of SMAD4, SMAD2, and DCC in Serous Ovarian Carcinomas
In normal ovarian tissue positive immunoreactivity of SMAD4 (moderate to strong), SMAD2 (weak), and DCC (focally weak to moderate) was observed in surface epithelial cells and a proportion of stromal cells. Forty-two of 60 serous carcinomas (70%) showed positive SMAD4 immunostaining, 17 (28%) were negative, and one not interpretable. SMAD2 expression was positive in 42 (70%), negative in 17 (28%), and not interpretable in one tumor. DCC expression was positive in 41 (68%), negative in 18 (30%), and not interpretable in one tumor (Table 1) ▶ . Comparison of allelic loss results and loss of expression of SMAD4, SMAD2, or DCC is shown in Table 2 ▶ . There was a tendency toward a higher amount of lost expression of SMAD4, SMAD2, and DCC in tumors with LOH at 18q21.1 compared with the tumors with no LOH at 18q21.1 (42 to 46% and 17 to 20%, respectively). However, only 41 to 46% of tumors with LOH at 18q21.1 had lost SMAD4, SMAD2, or DCC expression, suggesting that none of these genes alone could be the sole target of the frequent allelic loss at 18q21.1. When analyzing the additive effect of all three factors, a total of 83% of the tumors with LOH at 18q21.1 had lost SMAD4, SMAD2, and/or DCC expression, whereas 40% of the tumors with no LOH at 18q21.1 had lost expression of one or more of these proteins. Examples of SMAD4, SMAD2, and DCC immunohistochemistry are shown in Figure 3 ▶ .
Table 1.
Allelic Loss at 18q21.1 (D18S4747–D18S539) and Expression of SMAD4, SMAD2, and DCC in 60 Serous Ovarian Carcinomas
| Case no. | 18q21.1* | SMAD4† | SMD2† | DCC† |
|---|---|---|---|---|
| 256 | − | + | + | + |
| 405 | − | + | + | + |
| 784 | − | + | + | + |
| 1154 | − | + | + | + |
| 428 | − | + | + | − |
| 1121 | − | + | + | − |
| 859 | − | + | − | − |
| 780 | − | + | − | − |
| 1217 | − | + | − | − |
| 761 | − | + | − | + |
| 981 | − | + | − | + |
| 787 | − | + | − | + |
| 810 | − | + | − | + |
| 1182 | − | + | − | + |
| 983 | − | − | + | + |
| 412 | − | − | + | + |
| 223 | − | − | + | − |
| 723 | − | − | + | − |
| 1117 | − | − | + | − |
| 1183 | − | − | + | − |
| 1164 | − | − | + | − |
| 755 | − | − | − | + |
| 1078 | − | − | − | + |
| 750 | − | − | − | − |
| 1151 | − | 0 | 0 | 0 |
| 277 | + | + | + | + |
| 402 | + | + | + | + |
| 1027 | + | + | + | + |
| 823 | + | + | + | + |
| 1130 | + | + | + | + |
| 852 | + | + | + | + |
| 250 | + | + | + | + |
| 261 | + | + | + | + |
| 793 | + | + | + | + |
| 796 | + | + | + | + |
| 818 | + | + | + | + |
| 821 | + | + | + | + |
| 849 | + | + | + | + |
| 984 | + | + | + | + |
| 960 | + | + | + | + |
| 989 | + | + | + | + |
| 994 | + | + | + | + |
| 1053 | + | + | + | + |
| 1129 | + | + | + | + |
| 1180 | + | + | + | + |
| 1192 | + | + | + | + |
| 1106 | + | + | + | − |
| 779 | + | + | + | − |
| 846 | + | + | − | + |
| 201 | + | + | − | + |
| 828 | + | + | − | + |
| 1097 | + | + | − | + |
| 1184 | + | + | − | − |
| 1185 | + | − | + | + |
| 1195 | + | − | + | + |
| 795 | + | − | + | − |
| 769 | + | − | + | − |
| 995 | + | − | + | − |
| 1136 | + | − | + | − |
| 1124 | + | − | − | + |
*−, LOH at 18q21.1; +, no LOH at 18q21.1.
†+, positive immunoreactivity; −, negative immunoreactivity; 0, not interpretable.
Table 2.
Loss of Expression of SMAD4, SMAD2, or DCC in Tumors with LOH (n = 24) and without LOH at 18q21.1 (D18S474–D18S539) (n = 35)
| LOH | No LOH | P value | |
|---|---|---|---|
| SMAD4 | 10/24 = 42% | 7/35 = 20% | 0.086 |
| SMAD2 | 11/24 = 46% | 6/35 = 17% | 0.022 |
| DCC | 11/24 = 46% | 7/35 = 20% | 0.046 |
| SMAD4, SMAD2, and/or DCC | 20/24 = 83% | 14/35 = 40% | 0.0012 |
Figure 3.
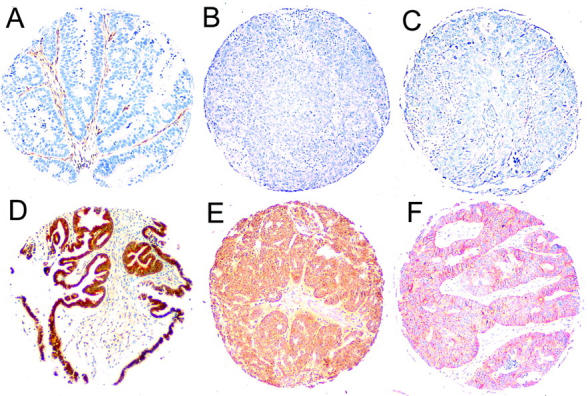
Immunohistochemical staining of SMAD4 (A and D), SMAD2 (B and E), and DCC (C and F) in serous ovarian carcinoma specimens on tissue microarray (original magnification, ×20). Top: Examples with negative or weak immunoreactivity (A–C). Bottom: Examples with positive immunoreactivity of the carcinoma cells (D–F).
Discussion
In the present study 59% of the serous ovarian carcinomas showed allelic loss at one or more markers located at 18q12.3-q23. This is among the highest frequencies observed at any chromosomal arm in ovarian carcinoma. 23,24 At 18q LOH has been previously detected in 41 to 60% of ovarian carcinomas. 7,24,25 In these studies all histological types of ovarian carcinoma have been combined. When only the serous tumors in these studies are considered, the frequency of LOH was found to be 45% (15 of 33; six markers) by Takakura and colleagues 7 and 61% (22 of 36; six markers) by Chenevix-Trench and colleagues. 25 In the study by Takakura and colleagues 7 all the markers were located at 18q21, whereas in the study by Chenevix-Trench and colleagues 25 two markers were located distal to 18q21. In the latter study the highest frequency of allelic loss was detected distal to 18q21, which is in accordance with the results of the present study.
In contrast to the frequent allelic loss (59%) found in serous carcinomas, only one of nine (11%) mucinous carcinomas showed LOH at 18q. Despite the limited number of mucinous carcinomas included in the study, the present results showed a significant difference in the frequency of allelic loss at 18q in these two types of ovarian carcinoma. Previously, serous and mucinous ovarian carcinomas have been found to differ, for example in respect to LOH at 17q and 8p, mutations of p53 and K-ras, genomic alterations as observed in comparative genomic hybridization, and cytogenetic changes. 10-14,26 Our finding of a distinct pattern of LOH at 18q adds to the evidence of a different molecular pathogenesis of these common types of ovarian carcinoma.
To our knowledge this is the first detailed mapping of distal 18q in ovarian carcinoma. Fifteen of 38 (40%) serous tumors showing LOH at 18q presented with loss of all informative markers, suggesting loss of the whole of distal 18q. A deletion map of 18q12.3-q23 was constructed based on the 23 tumors showing partial or interstitial losses (Figure 1) ▶ . The pattern of allelic loss at distal 18q was complex, but based on the LOH frequencies and the deletion map of tumors showing partial losses, two minimal common regions of loss could be defined: MCRL1 between markers D18S465 and D18S61 at 18q22 (3.9 cM) and MCRL2 between markers D18S462 and D18S70 at 18q23 (5.8 cM). Both of the MCRLs are located distal to SMAD4, SMAD2, and DCC loci (18q21.1). There are no well-known tumor suppressor genes at these regions, but the cadherin 7 gene (CDH7), at 18q22.1, could be one candidate. 27 CDH7 has not been studied in cancers, but a mutation in the E-cadherin gene (CDH1) has been detected in ovarian carcinoma. 28 In a previous allelotype study of ovarian carcinoma, using six markers at 18q, the smallest region of overlap was also found distal to 18q21. In that study only two markers were located at 18q22-q23 and more detailed mapping of the distal region was not possible. 25 Our results suggest that there are as yet unknown tumor suppressor genes at 18q22-q23 involved in serous ovarian carcinoma and we have defined two loci for further studies.
We found that LOH at distal 18q was associated with high tumor grade. Furthermore, it was also found to be associated with poor survival: 67% of patients with tumors showing LOH at distal 18q had died of the ovarian carcinoma, in contrast to 30% of patients who had tumors showing no LOH at 18q. Previously, LOH at 18q has been associated with tumor progression and poor survival in other types of cancer, including colorectal and gastric carcinomas. 29,30 In an allelotype study of ovarian tumors, which included use of one marker at 18q, there was frequent allelic loss at D18S50 (18q23) in high-grade and -stage ovarian carcinomas, but not in well-differentiated carcinomas, borderline or benign tumors. 31 Interestingly, in a recent genome-wide analysis of copy number changes in ovarian carcinoma, an association between loss of 18q and reduced survival duration was found. 32 However, multivariate analysis of our data showed only tumor grade to be an independent prognostic factor, not tumor stage or LOH status. Because tumor grade was a strong predictor of survival, the association between LOH and grade may partly explain the correlation of LOH with poor survival. In addition, LOH at distal 18q may be associated with other characteristics of the tumors (eg, fractional allelic loss or genomic instability) that may directly affect tumor behavior and patient survival. Thus, loss at distal 18q seems to be associated with a more aggressive phenotype of serous ovarian carcinoma, but to evaluate the role of LOH at 18q as an independent prognostic factor, studies with larger numbers of cases are needed.
To date, three candidate tumor suppressor genes have been identified on chromosome arm 18q (18q21.1): SMAD4 (DPC4), SMAD2, and DCC. 3-5 SMAD2 and SMAD4 are known to reside in a pathway of transforming growth factor-β signaling. SMAD4 inactivation is frequent in pancreatic carcinomas, but relatively uncommon in other cancer types. 6 In ovarian carcinoma, mutations of SMAD4 have been observed in 3 of 78 (3.8%) primary tumors studied 6-8 and deletion in intron 3 of SMAD2 has been detected in 12 of 32 ovarian carcinomas. 8 DCC has been identified as a gene frequently deleted in colorectal carcinomas, 5 and it encodes a transmembrane protein that functions as a receptor for the axonal chemoattractant netrin-1. 33 No DCC mutations have been reported in ovarian carcinoma. We found allelic loss at SMAD4, SMAD2, and DCC loci (18q21.1) in ∼40% of serous ovarian carcinomas, which is similar to previous findings. 7 The present study, to our knowledge, is the first in which the expression of SMAD4 and SMAD2 in ovarian carcinoma has been evaluated by immunohistochemistry. We found lost or very weak expression of SMAD4, SMAD2, and DCC in 28, 28, and 30% of tumor samples, respectively. Our results are in agreement with earlier studies in which decreased expression of DCC in a subset of ovarian carcinomas has been reported. 9,34 In contrast to our results, in one study in which Western blotting was used, no abnormal expression of SMAD2 in ovarian carcinomas was found. 8 We found expression of SMAD2 in the stromal cells of normal ovaries and carcinomas. Thus, possible normal cell contamination in samples used for Western blotting would result in a positive signal, which might explain the discrepancy between the present and previous results.
Mutant SMAD2 and SMAD4 proteins are degraded more rapidly than their wild-type counterparts, 35 and SMAD4 immunohistochemistry has been found to be a sensitive and specific marker for gene alterations detected in SMAD4. 19 If one allele of these genes is inactivated by mutation and the other allele by allelic loss, one would expect to see decreased or lost protein expression. We found a tendency toward a higher amount of lost expression of SMAD4, SMAD2, and DCC in tumors showing LOH compared with tumors showing no LOH at 18q21.1. However, ∼20% of tumors with no LOH showed negative immunostaining of SMAD4, SMAD2, or DCC. In these tumors small deletions may reside between the markers used in the study, or down-regulated expression may be because of other mechanisms such as biallelic mutations, altered transcriptional regulation, or epigenetic events. Furthermore, <50% of the tumors showing LOH at 18q21.1 had lost SMAD4, SMAD2, or DCC expression, which suggests that none of these genes alone is the main target of frequent allelic loss at 18q. Together these genes could explain up to 80% of the cases showing LOH at 18q21.1. On the basis of these results there are still at least 20% of cases in which LOH at 18q21.1 cannot be explained by the effect of SMAD4, SMAD2, and DCC, suggesting the existence of other tumor suppressor gene(s). Consistent with this, the deletions in several tumors were large and also included more distal parts of 18q. Furthermore, the highest frequencies and minimal common regions of allelic loss were found at 18q22-q23.
In conclusion, we found frequent allelic loss at 18q in serous, but not in mucinous ovarian carcinomas. In serous carcinomas the highest frequency of losses was detected at the distal part, 18q22-q23, and two minimal common regions of loss could be defined at this region. Deletions were found to be associated with high-grade tumors and poor survival. The expression of three putative tumor suppressor genes, SMAD4, SMAD2, and DCC (18q21.1), was found to be lost in a subset of serous carcinomas, but none of these genes alone could be the target of the frequent allelic losses at distal 18q. The three genes could account for a substantial proportion of the cases showing LOH at 18q21.1, but not all of them. Thus we propose that the frequent allelic loss at 18q is because of the effect of multiple genes and there is at least one as yet unidentified tumor suppressor gene at 18q residing distal to SMAD4, SMAD2, and DCC involved in serous ovarian carcinoma.
Acknowledgments
We thank Ms. Gynel Arifdshan for excellent technical assistance.
Footnotes
Address reprint requests to Dr. Ralf Butzow, Department of Pathology, University of Helsinki, P.O. Box 21, FIN-00014 Helsinki, Finland. E-mail: ralf.butzow@hus.fi.
Supported by grants from Helsinki University Central Hospital, the Finnish Medical Foundation, the Emil Aaltonen Foundation, Helsinki University Science Fund, and the Cancer Society of Finland.
References
- 1.Arnold N, Hägele L, Walz L, Schempp W, Pfisterer J, Bauknecht T, Kiechle M: Overrepresentation of 3q and 8q material and loss of 18q material are recurrent findings in advanced human ovarian cancer. Gene Chromosom Cancer 1996, 16:46-54 [DOI] [PubMed] [Google Scholar]
- 2.Tapper J, Sarantaus L, Vahteristo P, Nevanlinna H, Hemmer S, Seppälä M, Knuutila S, Butzow R: Genetic changes in inherited and sporadic ovarian carcinomas by comparative genomic hybridization: extensive similarity except for a difference at chromosome 2q24–q32. Cancer Res 1998, 58:2715-2719 [PubMed] [Google Scholar]
- 3.Hahn SA, Schutte M, Hoque ATMS, Moskaluk CA, da Costa LT, Rosenblum E, Weinstein CL, Fischer A, Yeo CJ, Hruban RH, Kern SE: DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science 1996, 271:350-353 [DOI] [PubMed] [Google Scholar]
- 4.Eppert K, Scherer SW, Ozcelik H, Pirone R, Hoodless P, Kim H, Tsui L-C, Bapat B, Gallinger S, Andrulis IL, Thomsen GH, Wrana JL, Attisano L: MADR2 maps to 18q21 and encodes a TGFβ-regulated MAD-related protein that is functionally mutated in colorectal carcinoma. Cell 1996, 86:543-552 [DOI] [PubMed] [Google Scholar]
- 5.Fearon ER, Cho KR, Nigro JM, Kern SE, Simons JW, Ruppert MJ, Hamilton SR, Preisinger AC, Thomas G, Kinzler KW, Vogelstein B: Identification of a chromosome 18q gene that is altered in colorectal cancers. Science 1990, 247:49-56 [DOI] [PubMed] [Google Scholar]
- 6.Schutte M, Hruban RH, Hedrick L, Cho KR, Nadasdy GM, Weinstein CL, Bova GS, Isaacs WB, Cairns P, Nawroz H, Sidransky D, Casero RA, Jr, Meltzer PS, Hahn SA, Kern SE: DPC4 gene in various tumor types. Cancer Res 1996, 56:2527-2530 [PubMed] [Google Scholar]
- 7.Takakura S, Okamoto A, Saito M, Yasuhara T, Shinozaki H, Isonishi S, Yoshimura T, Ohtake Y, Ochiai K, Tanaka T: Allelic imbalance in chromosome band 18q21 and SMAD4 mutations in ovarian cancers. Gene Chromosom Cancer 1999, 24:264-271 [DOI] [PubMed] [Google Scholar]
- 8.Wang D, Kanuma T, Mizunuma H, Takama F, Ibuki Y, Wake N, Mogi A, Shitara Y, Takenoshita S: Analysis of specific gene mutations in the transforming growth factor-β signal transduction pathway in human ovarian cancer. Cancer Res 2000, 60:4507-4512 [PubMed] [Google Scholar]
- 9.Saegusa M, Machida D, Okayasu I: Loss of DCC gene expression during ovarian tumorigenesis: relation to tumour differentiation and progression. Br J Cancer 2000, 82:571-578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pierretti M, Cavalieri C, Conway PS, Gallion HH, Powell DE, Turker MS: Genetic alterations distinguish different types of ovarian tumors. Int J Cancer 1995, 64:434-440 [DOI] [PubMed] [Google Scholar]
- 11.Enomoto T, Weghorst CM, Inoue M, Tanizawa O, Rice JM: K-ras activation occurs frequently in mucinous adenocarcinomas and rarely in other common epithelial tumors of the human ovary. Am J Pathol 1991, 139:777-785 [PMC free article] [PubMed] [Google Scholar]
- 12.Milner BJ, Allan LA, Eccles DM, Kitchener HC, Leonard RCF, Kelly KF, Parkin DE, Haites NE: p53 mutation is a common genetic event in ovarian carcinoma. Cancer Res 1993, 53:2128-2132 [PubMed] [Google Scholar]
- 13.Tapper J, Butzow R, Wahlström T, Seppälä M, Knuutila S: Evidence for divergence of DNA copy number changes in serous, mucinous and endometrioid ovarian carcinomas. Br J Cancer 1997, 75:1782-1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diebold J, Siegert S, Baretton GB, Suchy B, Meier W, Haas CJ, Lohrs U: Interphase cytogenetic analysis of mucinous ovarian neoplasms. Lab Invest 1997, 76:661-670 [PubMed] [Google Scholar]
- 15.Pere H, Tapper J, Seppälä M, Knuutila S, Butzow R: Genomic alterations in fallopian tube carcinoma: comparison to serous uterine and ovarian carcinomas reveals similarity suggesting likeness in molecular pathogenesis. Cancer Res 1998, 58:4274-4276 [PubMed] [Google Scholar]
- 16.Schütze K, Lahr G: Identification of expressed genes by laser-mediated manipulation of single cells. Nature Biotechnol 1998, 16:737-742 [DOI] [PubMed] [Google Scholar]
- 17.Canzian F, Salovaara R, Hemminki A, Kristo P, Chadwick RB, Aaltonen LA, de la Chapelle A: Semiautomated assessment of loss of heterozygosity and replication error in tumors. Cancer Res 1996, 56:3331-3337 [PubMed] [Google Scholar]
- 18.Kononen J, Bubendorf L, Kallioniemi A, Bärlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G, Kallioniemi OP: Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med 1996, 4:844-847 [DOI] [PubMed] [Google Scholar]
- 19.Wilentz RE, Su GH, Dai JL, Sparks AB, Argani P, Sohn TA, Yeo CJ, Kern SE, Hruban RH: Immunohistochemical labeling for Dpc4 mirrors genetic status in pancreatic adenocarcinomas. Am J Pathol 2000, 156:37-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shibata D, Reale MA, Lavin P, Silverman M, Fearon ER, Steele G, Jessup JM, Loda M, Summerhayes IC: The DCC protein and prognosis in colorectal cancer. N Engl J Med 1996, 23:1727-1732 [DOI] [PubMed] [Google Scholar]
- 21.Wang R-A, Zhao G-Q: Transforming growth factor β signal transducer Smad2 is expressed in mouse meiotic germ cells, Sertoli cells, and Leydig cells during spermatogenesis. Biol Reprod 1999, 61:999-1004 [DOI] [PubMed] [Google Scholar]
- 22.Nakao A, Röijer E, Imamura T, Souchelnytskyi S, Stenman G, Heldin C-H, ten Dijke P: Identification of Smad2, a human Mad-related protein in the transforming growth factor β signaling pathway. J Biol Chem 1997, 272:2896-2900 [DOI] [PubMed] [Google Scholar]
- 23.Sato T, Saito H, Morita R, Koi S, Lee JH, Nakamura Y: Allelotype of human ovarian cancer. Cancer Res 1991, 51:5118-5122 [PubMed] [Google Scholar]
- 24.Cliby W, Ritland S, Hartmann L, Dodson M, Halling KC, Keeney G, Podratz KC, Jenkins RB: Human epithelial ovarian cancer allelotype. Cancer Res 1993, 53:2393-2398 [PubMed] [Google Scholar]
- 25.Chenevix-Trench G, Leary J, Kerr J, Michel J, Kefford R, Hurst T, Parsons PG, Friedlander M, Khoo SK: Frequent loss of heterozygosity on chromosome 18 in ovarian adenocarcinoma which does not always include the DCC locus. Oncogene 1992, 7:1059-1065 [PubMed] [Google Scholar]
- 26.Lassus H, Laitinen MP, Anttonen M, Heikinheimo M, Aaltonen LA, Ritvos O, Butzow R: Comparison of serous and mucinous ovarian carcinomas: distinct pattern of allelic loss at distal 8p and expression of transcription factor GATA-4. Lab Invest 2001, 81:517-526 [DOI] [PubMed] [Google Scholar]
- 27.Kremmidiotis G, Baker E, Crawford J, Eyre HJ, Nahmias J, Callen DF: Localization of human cadherin genes to chromosome regions exhibiting cancer-related loss of heterozygosity. Genomics 1998, 49:467-471 [DOI] [PubMed] [Google Scholar]
- 28.Risinger JI, Berchuck A, Kohler MF, Boyd J: Mutations of the E-cadherin gene in human gynecologic cancers. Nat Genet 1994, 7:98-102 [DOI] [PubMed] [Google Scholar]
- 29.Jen J, Kim H, Piantadosi S, Liu Z-F, Levitt RC, Sistonen P, Kinzler KW, Vogelstein B, Hamilton SR: Allelic loss of chromosome 18q and prognosis in colorectal cancer. N Engl J Med 1994, 331:214-221 [DOI] [PubMed] [Google Scholar]
- 30.Inoue T, Uchino S, Shiraishi N, Adachi Y, Kitano S: Loss of heterozygosity on chromosome 18q in cohesive-type gastric cancer is associated with tumor progression and poor prognosis. Clin Cancer Res 1998, 4:973-977 [PubMed] [Google Scholar]
- 31.Zborovskaya I, Gasparian A, Karseladze A, Elcheva I, Trofimova E, Driouch K, Trassard M, Tatosyan A, Lidereau R: Somatic genetic alterations (LOH) in benign, borderline and invasive ovarian tumors: intratumoral molecular heterogeneity. Int J Cancer 1999, 82:822-826 [DOI] [PubMed] [Google Scholar]
- 32.Suzuki S, Moore DH, II, Ginzinger DG, Godfrey TE, Barclay J, Powell B, Pinkel D, Zaloudek C, Lu K, Mills G, Berchuck A, Gray JW: An approach to analysis of large-scale correlations between genome changes and clinical endpoints in ovarian cancer. Cancer Res 2000, 60:5382-5385 [PubMed] [Google Scholar]
- 33.Fazeli A, Dickinson SL, Hermiston ML, Tighe RV, Steen RG, Small CG, Stoeckli ET, Keino-Masu K, Masu M, Rayburn H, Simons J, Bronson RT, Gordon JI, Tessier-Lavigne M, Weinberg RA: Phenotype of mice lacking functional Deleted in colorectal cancer (Dcc) gene. Nature 1997, 386:796-804 [DOI] [PubMed] [Google Scholar]
- 34.Enomoto T, Fujita M, Cheng C, Nakashima R, Ozaki M, Inoue M, Nomura T: Loss of expression and loss of heterozygosity in the DCC gene in neoplasms of the human female reproductive tract. Br J Cancer 1995, 71:462-467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu J, Attisano L: Mutations in the tumor suppressors Smad2 and Smad4 inactivate transforming growth factor β signaling by targeting Smads to the ubiquitin-proteasome pathway. Proc Natl Acad Sci USA 2000, 97:4820-4825 [DOI] [PMC free article] [PubMed] [Google Scholar]