Abstract
Methylation of cytosines in CpG islands silences gene expression. CpG island methylator phenotype (CIMP) in colorectal cancers is characterized by abnormal methylation of multiple CpG islands including those in several tumor suppressor genes such as p16, hMLH1, and THBS1. CpG island methylation has not been well characterized in adenomas. We evaluated methylation status at p16, MINT2, and MINT31 loci, which are frequently methylated in colorectal carcinomas, in 108 colorectal adenomas from a prospective study of 50 patients without cancer. Methylation at one or more loci was present in 48% (52 of 108) of adenomas with 25% (19 of 76) CIMP-high (two or more methylated loci) and 32% (24 of 76) CIMP-low (one methylated locus). The p16 gene was methylated in 27% (19 of 71) of adenomas. Methylation status of different adenomas from the same patient was not correlated (odds ratio, 0.93; P = 0.77). Adenomas with tubulovillous or villous histology were frequently methylated: 73% (17 of 26) versus 41% (35 of 85) of tubular adenomas (odds ratio, 3.46; P = 0.02). High levels of microsatellite instability were more frequent in adenomas without methylation (13% versus 2%; odds ratio, 8.48; P = 0.05). Our results indicate that methylation plays an important role early in colorectal tumorigenesis. CpG island methylation is more common in adenomas with tubulovillous/villous histology, a characteristic associated with more frequent predisposition to invasive carcinoma. Methylation is distinct from microsatellite instability and develops in individual adenomas rather than resulting from a field defect in an individual patient.
Colorectal cancer is the second most common cause of cancer deaths in the United States. Most colorectal cancers develop from adenomatous polyps, and morphological and genetic progression in an adenoma-adenocarcinoma sequence and in hereditary colorectal cancer syndromes are well described. 1-5
The majority of colorectal cancers has truncating mutations or deletions of the adenomatous polyposis coli (APC) gene on chromosome 5q 6 or mutations of the β-catenin gene. Point mutations of the K-ras proto-oncogene, 7 loss of the deleted in colorectal cancer (DCC) gene and nearby SMAD2 and SMAD4 genes on chromosome 18q, 8 and mutations and/or deletions of the p53 gene on chromosome 17p are also common. 9
In a second pathway to colorectal neoplasia, microsatellite instability (MSI; also termed DNA replication errors and ubiquitous somatic mutations) is caused by alteration of a nucleotide mismatch repair gene, including hMSH2, hMLH1, PMS1, PMS2, or GTBP. 1-5 MSI is characterized by additions and deletions of nucleotides in numerous repeated nucleotide sequences (microsatellites). Germline mutation of a mismatch repair gene causes hereditary nonpolyposis colorectal cancer. Alterations of mononucleotide tracts present in genes such as transforming growth factor-β type II receptor and BAX genes are commonly found in MSI-positive carcinomas. 10,11
Another molecular defect commonly present in colorectal cancer is CpG island methylation. CpG islands are 0.5- to 2-kb regions rich in cytosine-guanine dinucleotides and are present in the 5′ region of approximately half of all human genes. 12 Methylation of cytosines within CpG islands is associated with loss of gene expression by repression of transcription and is observed in physiological conditions such as X chromosome inactivation 13 and aging, 14 but also in neoplasia. 15 Examples of this process in colorectal cancers include inactivation of the p16 cell-cycle regulator, 16 the estrogen receptor growth suppressor, 14 the THBS1 angiogenesis inhibitor, 17 the TIMP3 metastasis suppressor, 18 the O6-methylguanine DNA methyltransferase DNA repair gene, 19 and the hMLH1 nucleotide mismatch repair gene. 20
Recently, a distinct pathway of colorectal carcinogenesis was described, termed CpG island methylator phenotype (CIMP). 21 CIMP-positive colorectal cancers are characterized by a high degree of concordant CpG island methylation of genes in colorectal cancer but not in normal mucosa. CIMP phenotype is also observed in large colorectal adenomas removed with colorectal cancer, 22 but CIMP status in adenomas unassociated with cancer has not been reported.
In the present study, we examined methylation status in a prospective study of sporadic colorectal adenomas removed at colonoscopy from patients without cancer. The methylation status was compared with patient and adenoma characteristics including methylation of adenomas, and with other genetic alterations present in adenomas.
Materials and Methods
Characteristics of Patients and Specimens
This study includes 108 colorectal adenomas that were collected from 50 patients prospectively enrolled in the endoscopy unit of The Johns Hopkins Hospital. K-ras mutation, p53 overexpression, 18q loss, and microsatellite instability status of these adenomas have been described previously. 23 The demographics of the patient population and adenoma characteristics analyzed in the present study are summarized in Table 1 ▶ . All patients had given informed consent for the collection of tissue according to institutional guidelines.
Table 1.
Patient Demographics and Adenoma Characteristics
| Characteristics* | % (no.) of Patients (n = 50) | % (no.) of Polyps (n = 108) | ||
|---|---|---|---|---|
| Gender | ||||
| Female | 44 (22) | 37 (40) | ||
| Male | 56 (28) | 63 (68) | ||
| Number of polyps analyzed from each patient (range, 1–10; median, 2) | ||||
| 1 | 48 (24) | 22 (24) | ||
| 2–3 | 38 (19) | 40 (43) | ||
| >3 | 14 (7) | 38 (41) | ||
| Site† | ||||
| Right colon | 48 (24) | 43 (47) | ||
| Left colorectum | 80 (40) | 55 (59) | ||
| Not designated | 4 (2) | 2 (2) | ||
| Polyp size (mean, 0.74 cm± 0.79 STD)† | ||||
| 0.1–0.5 | 64 (32) | 60 (65) | ||
| >0.5–1 | 48 (24) | 26 (28) | ||
| >1–2 | 20 (10) | 9 (10) | ||
| >2 | 10 (5) | 5 (5) | ||
| Histopathology† | ||||
| Tubular | 86 (43) | 79 (85) | ||
| Tubulovillous | 32 (16) | 20 (22) | ||
| Villous | 2 (1) | 1 (1) | ||
*The age at polypectomy ranged from 28 to 91 years (mean, 63 ± 12 years).
†Total percentage for patients exceeds 100% because of multiple adenomas.
DNA Extraction
Genomic DNA was extracted after microdissection. 24 Each specimen was treated with 50 μl of buffer containing 0.5% Tween 20 (Boehringer Mannheim, Mannheim, Germany), 20 μg proteinase K (Boehringer Mannheim), 50 mmol/L Trizma base at pH 8.9, and 2 mmol/L ethylenediaminetetraacetic acid as previously described. 24 The samples were incubated at 56°C overnight. Proteinase K was inactivated by incubating the samples at 100°C for 10 minutes. The extracted DNA was stored at −80°C.
Bisulfite Treatment of DNA and Methylation-Specific Polymerase Chain Reaction (MSP) Followed by Restriction Enzyme Digest
The methylation status of p16, MINT2, and MINT31 was determined by bisulfite treatment of DNA followed by methylation-specific polymerase chain reaction (MSP-PCR), as described, with modification. 25 The selection of these loci was based on our previous study that showed these loci had high sensitivity and specificity for the detection of hypermethylation in cancer and offered excellent discrimination for CIMP status. 21 MINT2 corresponds to a CpG island that is in the 5′ region of a cDNA with an open reading frame that has no protein homology (J. P. Issa, unpublished data). MINT31 is 2 kb upstream of the CACNA1G, a T-type calcium channel gene. 26 In brief, 2 μg of microdissected genomic DNA was denatured with 2 mol/L NaOH at 37°C for 10 minutes, followed by incubation with 3 mol/L sodium bisulfite (pH 5.0) at 50°C for 16 hours in the dark. After treatment, DNA was purified using the DNA cleanup kit (Promega, Madison, WI) as recommended by the manufacturer, incubated with 3 mol/L NaOH at room temperature for 5 minutes, precipitated with 10 mol/L ammonium acetate and 100% ethanol, washed with 70% ethanol, and finally resuspended in 20 μl of distilled water. Methylation status of p16, MINT2, and MINT31 was determined using 5 μl of bisulfite-treated DNA for bisulfite-PCR followed by restriction enzyme digest of PCR product as described. 27 In brief, a 40 μl aliquot of the amplified products was digested with restriction enzymes that distinguish methylated from unmethylated sequences, electrophoresed on 5% acrylamide gels, and visualized by ethidium bromide staining. Primer sequences, conditions for PCR, and restriction enzymes used are tabulated in Table 2 ▶ . DNA from RKO colon cancer cell line (American Type Culture Collection, Manassas, VA) was used as a control for methylation. Methylation status was confirmed by MSP for p16 in 7 adenomas and for MINT31 in 10 adenomas as previously described. 25 In brief, 4 μl of DNA was used as template for PCR reactions using primers specific for methylated and unmethylated alleles. For quantitation of methylated alleles, gel images were digitized using a BioRad imager and quantitated using the accompanying software (BioRad, Hercules, CA). Both bisulfite-PCR and MSP provide semiquantitative results. The loci (p16, MINT2, and MINT31) used in this study are unmethylated (<1% methylation) in normal tissues. Therefore, any locus showing ≥1% methylation was considered positive.
Table 2.
Primer Sequences, PCR Condition, and Restriction Enzymes Used for Bisulfite-PCR
| Locus | Primer set, forward/reverse | Annealing temperature (cycles) | Restriction enzyme |
|---|---|---|---|
| MINT2 | F:5′-YGTTATGATTTTTTTGTTTAGTTAAT-3′ | 60 (3), 58 (4), 56 (5), 54 (26) | TaqI |
| R:5′-TACACCAACTACCCAACTACCTC-3′ | |||
| MINT31 | F:5′-GAYGGYGTAGTAGTTATTTTGTT-3′ | 68 (3), 56 (4), 54 (5), 52 (26) | MaeII |
| R:5′-CATCACCACCCCTCACTTTAC-3′ | |||
| p16 | F:5′-GGTTTTGGYGAGGGTTGTTT-3′ | 60 (5), 57 (5), 54 (5), 51 (23) | TaqI |
| R:5′-ACCCTATCCCTCAAATCCTCTAAAA-3′ |
CIMP of Adenomas
CIMP status was determined for adenomas with two or more evaluated loci. Adenomas were classified as CIMP-negative if none of the evaluated loci were methylated; CIMP-low if one locus was methylated; and CIMP-high if two or more loci were methylated. Adenomas with one unmethylated evaluated locus were not classified. These criteria were based on our previous study in which CIMP-positive adenomas were methylated at the majority of MINT loci and p16, but CIMP-negative adenomas were rarely methylated at any MINT loci and never methylated at p16. 21 The sensitivity of any MINT locus to predict CIMP is 75% and specificity is 95%. The sensitivity of p16 to predict CIMP phenotype is 65% and specificity is 100%.
K-ras Mutations, p53 Overexpression, MSI, and Loss of Heterozygosity of Chromosome 18q in Adenomas
These alterations were reported previously. 23
Statistical Analysis
The primary statistical endpoint of this study was the determination of factors related to methylation status at the p16, MINT2, and MINT31 loci. All adenomas in the study with a successful determination of their methylation status for at least one locus were included in the analysis. Each adenoma was represented by a methylation index (number of loci methylated/number of loci evaluated). Patients with more than one adenoma were represented multiple times in this data set. To correctly model the within- and between-adenoma correlation as well as simultaneously partition out the effects of the various factors considered, marginal logistic regression models for correlated binary data were used to assess associations between methylation status and the various adenoma and patient characteristics. Estimates were obtained using the generalized estimating equation approach of Liang and Zeger. 28 An appropriate correlation structure was chosen to account for possible correlations between adenomas within patients, and also within adenomas between observations from different loci. Patient characteristics included age and sex. Adenoma characteristics included site, size, and histology, as well as presence or absence of K-ras mutations, p53 overexpression, MSI, and loss of heterozygosity of chromosome 18q in polyps. A factor was also included in the model to account for locus-specific methylation rates. Because there was no evidence in the data that any patient or adenoma characteristics had effects that differed according to locus, no locus-by-factor interactions were included in the final generalized estimating equation model. Relationships between adenomas within patients and within adenomas between loci were represented as odds ratios, in which an odds ratio of greater than one suggests positive correlation in methylation status within patients and within adenomas, respectively. Comparisons of adenoma size were done using Proc mixed in SAS (SAS Institute, Inc., Cary, NC).
Results
Figure 1 ▶ shows examples of methylation at p16, MINT2, and MINT31. Figure 2 ▶ summarizes the frequency in adenomas of methylation at p16, MINT2, and MINT31, and at any locus. Patient and adenoma characteristics are summarized with methylation status of adenomas in Tables 3 and 4 ▶ ▶ . Patient and adenoma characteristics are summarized with methylation status of adenomas in Table 5 ▶ .
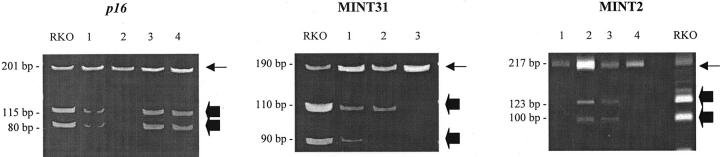
Figure 1.
Methylation analysis of CpG islands in adenomas. Methylation of p16, MINT2, and MINT31 was evaluated by bisulfite-PCR and restriction enzyme digestion by methylation-specific enzymes. Loci examined and adenoma numbers are indicated above each gel. Arrows indicate unmethylated alleles and block arrows indicate methylated alleles. RKO is a colon cancer cell line included as a positive control. Samples 1, 3, and 4 are methylated for p16; sample 1 for MINT31, and samples 2 and 3 for MINT2. Sample 2 shows nonspecific amplification for MINT31 as the other band present after restriction enzyme digest in methylated samples is not present.
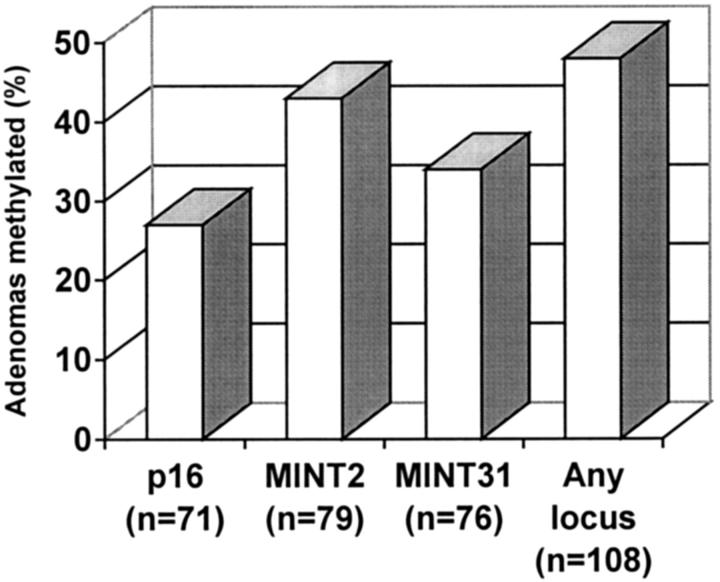
Figure 2.
Frequency of methylation of p16, MINT2, MINT31, and at any locus in adenomas.
Table 3.
Methylation Status of p16, MINT2, or MINT31 in 108 Adenomas Compared to Patient and Adenoma Characteristics
| Characteristic | Total (no.) | Unmethylated, all loci assayed (n = 56) % (no.) | Methylated, any locus (n = 52) % (no.) |
|---|---|---|---|
| Age (years)± SD | 63 ± 12 | 64 ± 11 | 62 ± 13 |
| Sex | |||
| Female | 40 | 50 (20) | 50 (20) |
| Male | 68 | 53 (36) | 47 (32) |
| Site | |||
| Right | 47 | 55 (26) | 45 (21) |
| Left | 59 | 49 (29) | 51 (30) |
| Not designated | 2 | 50 (1) | 50 (1) |
| Size in cm± SD | 0.74 ± 0.79 | 0.56 ± 0.42 | 0.92 ± 1.02 |
| Histopathology | |||
| Tubular | 85 | 59 (50) | 41 (35) |
| Tubulovillous/villous | 23 | 26 (6) | 73 (17) |
| Other genetic alterations | |||
| K-ras | 43 | 58 (25) | 41 (18) |
| p53 overexpression | 9 | 44 (4) | 56 (5) |
| MSI-high* | 7 | 85 (6) | 15 (1) |
| 18q loss* | 3 | 33 (1) | 67 (2) |
*Microsatellite instability and 18q loss was not assessed for nine adenomas without methylation and six adenomas with methylation.
Table 4.
Odds Ratio and 95% Confidence Intervals (CI) for Patient and Adenoma Characteristics Associated with Methylation, from Generalized Estimated Equation Marginal Regression Models
| Factor | Odds ratio | 95% CI | Chi-squared | P value |
|---|---|---|---|---|
| Patient characteristics | ||||
| Age | 0.97 | (0.94, 1.00) | 2.29 | 0.13 |
| Sex | ||||
| Female | 1.26 | (0.57, 2.77) | 0.31 | 0.58 |
| Male | 1.00 | |||
| Adenoma characteristics | ||||
| Site | ||||
| Left | 1.05 | (0.62, 1.80) | 0.03 | 0.86 |
| Right | 1.00 | |||
| Histology of adenoma* | ||||
| Villous/tubulovillous | 3.46 | (1.59, 7.56) | 5.48 | 0.02 |
| Tubular | 1.00 | |||
| K-ras mutation† | ||||
| Absent | 1.69 | (0.66, 4.30) | 1.16 | 0.28 |
| Present | 1.00 | |||
| Microsatellite instability | ||||
| Absent | 8.48 | (1.66, 43.25) | 3.84 | 0.05 |
| Present | 1.00 |
*Size is directly correlated with the villous component (r = 0.49) and is not included in the model.
†p53 overexpression is not related to methylation status (P = 0.98) and is not included in the model.
Table 5.
CIMP Status in 76 Adenomas Compared to Patient and Adenoma Characteristics
| Characteristic | Total (no.) | CIMP-negative (n = 33) % (no.) | CIMP-low (n = 24) % (no.) | CIMP-high (n = 19) % (no.) |
|---|---|---|---|---|
| Age (years)± SD | 62 ± 10 | 64 ± 12 | 60 ± 14 | |
| <65 | 32 | 41 (13) | 34 (11) | 25 (8) |
| ≥65 | 44 | 45 (20) | 30 (13) | 25 (11) |
| Sex | ||||
| Female | 28 | 39 (11) | 36 (10) | 25 (7) |
| Male | 48 | 46 (22) | 29 (14) | 25 (12) |
| Site | ||||
| Right | 31 | 39 (12) | 32 (10) | 29 (9) |
| Left | 44 | 45 (20) | 32 (14) | 23 (10) |
| Undesignated | 1 | 100 (1) | 0 | 0 |
| Size in cm± SD | 0.45 ± 0.35 | 1.39 ± 1.31 | 0.45 ± 0.24 | |
| 0.1–0.5 | 49 | 53 (26) | 18 (9) | 29 (14) |
| >0.5–1.0 | 15 | 40 (6) | 27 (4) | 33 (5) |
| >1.0 | 12 | 8 (1) | 92 (11) | 0 |
| Histopathology | ||||
| Tubular* | 57 | 59 (30) | 41 (15) | 41 (12) |
| Tubulovillous/villous* | 19 | 26 (3) | 73 (9) | 73 (7) |
| Other genetic alterations | ||||
| K-ras | 25 | 52 (13) | 32 (8) | 16 (4) |
| p53 overexpression | 7 | 29 (2) | 43 (3) | 29 (2) |
| Microsatellite instability† | 3 | 67 (2) | 33 (1) | 0 |
| 18q loss† | 3 | 33 (1) | 0 | 67 (2) |
*Odds ratio, 3.57; 95% confidence interval 1.14, 11.12; chi-square, 4.51; P = 0.03.
†Microsatellite instability and 18q loss was not assessed for five CIMP-negative adenomas, two CIMP-low adenomas, and two CIMP-high adenomas.
Methylation at p16, MINT2, and MINT31
Methylation of MINT2 was present in 43% (34 of 79) of adenomas, of MINT31 in 34% (26 of 76) of adenomas, and p16 in 27 (19 of 71) of adenomas (P = 0.04; Figure 2 ▶ ). All three loci were methylated in 8 adenomas, two loci in 11 adenomas, one locus in 33 adenomas, and no locus in 56 adenomas. Thus, 48% (52 of 108) of adenomas had methylation at one or more loci, and 52 (56 of 108) of adenomas had no methylation at any of the evaluated locus. The methylation status for different loci of the same adenoma was positively correlated (odds ratio, 3.60; P = 0.0078), indicating that some adenomas had CIMP-high. In adenomas with two or more evaluated loci, 25% (19 of 76) of adenomas were CIMP-high, 32% (24 of 76) were CIMP-low, and 43% (33 of 76) were CIMP-negative.
Because 78% (84 of 108) of adenomas were from 26 patients with multiple adenomas, we were able to address within-patient correlation of methylation status. We found that different adenomas within the same patient were not correlated (odds ratio, 0.93; P = 0.77). This finding provides evidence against a field defect responsible for the development of methylation.
The age and sex of the patient or site of adenoma and methylation status of adenomas were not associated (Tables 3 and 4) ▶ ▶ . However, methylation was associated with villous histology and large size of adenomas. Methylation was present in 73% (17 of 23) of tubulovillous or villous adenomas versus 41% (35 of 85) of tubular adenomas (odds ratio, 3.46; P = 0.02). Methylation was present in 80% (12 of 15) of adenomas >1 cm in size, 40% (11 of 28) of adenomas between 0.5 cm to 1 cm in size, and 45% (29 of 65) adenomas <0.5 cm in size. The size of adenomas was not independently correlated with methylation status of adenoma as size was directly correlated with villous component (r = −0.49).
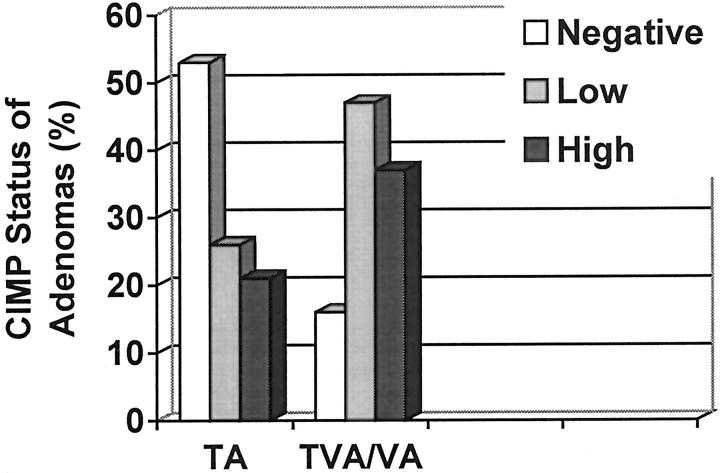
CIMP status of adenomas was also associated with the histological type of adenomas. The prevalence of CIMP-high, CIMP-low, and CIMP-negative was 37% (7 of 19), 47% (9 of 19), and 16% (3 of 19) in tubulovillous adenomas compared to 21% (12 of 57), 26% (15 of 57), and 53% (30 of 57) in tubular adenomas (odds ratio, 3.57; 95% confidence interval, 1.14, 11.12; chi-square, 4.51; P = 0.03) (Figure 3 ▶ and Table 5 ▶ ).
Figure 3.
Frequency of CIMP status in tubular adenomas (TA) and tubulovilluos/villous adenomas (TVA/VA).
Methylation of adenomas was inversely associated with MSI but was not associated with K-ras mutation, p53 overexpression, or 18q loss (Tables 3 and 4) ▶ ▶ . MSI-high was present in 2% (1 of 46) of adenomas with methylation, and 13% (6 of 47) of adenomas with no methylation (odds ratio, 8.48; P = 0.05). MSI-high adenomas without methylation were from three hereditary nonpolyposis colorectal cancer patients who had one adenoma each and from three patients without hereditary nonpolyposis colorectal cancer patient who had one, one, and two adenomas, respectively. K-ras mutations were present in 37% (18 of 52) of adenomas with methylation and 47% (25 of 56) of adenomas with no methylation (odds ratio, 1.69; P = 0.28). Adenomas with K-ras mutation and methylation were larger in size compared to adenomas with K-ras mutation but no methylation (1.1 ± 0.16 versus 0.58 ± 0.13, F = 6.92, P = 0.02).
Discussion
Aberrant methylation of CpG islands in the promoter region of tumor suppressor genes is associated with transcriptional inactivation of the genes and is thought to play an important role in carcinogenesis. 15,29 The recently discovered CIMP is a novel pathway characterized by methylation of multiple CpG islands in colorectal carcinomas and adenomas. 21 The molecular genetics of colorectal neoplasms, including adenomas, have been studied extensively, but few studies have addressed clinical and pathological associations in prospectively defined patient populations. We, therefore, studied methylation in adenomas from patients without cancer in a cohort undergoing colonoscopy.
Methylation of one or more CpG island(s) was present in 52% of adenomas, and methylation of two or more CpG islands (CIMP-high) was present in 25% of adenomas. In contrast, in our previous study the prevalence of CIMP-high in adenomas was 49%. 22 The adenomas in our previous study were from patients with synchronous cancers and were larger. In addition, methylation was studied using an extended set of markers. In both studies, however, CIMP-high was more common in larger adenomas. In the present study, we also show that methylation is more common in adenomas with tubulovillous or villous histology. Thus, methylation is more common in larger adenomas and adenomas with tubulovillous or villous histology, two adenoma characteristics associated with more frequent predisposition to invasive carcinoma.
p16 methylation was present in 27% of adenomas in the present study. We have previously reported that K-ras mutation was the most frequent genetic alteration in colorectal adenomas occurring in 35% of adenomas, but other genetic alterations are infrequent. 23 Thus, p16 methylation is one of the most common early genetic alterations in the adenoma-carcinoma sequence.
CpG island methylation of adenomas in our present study was associated with lack of K-ras mutation or MSI. Colorectal cancers with methylator phenotype have a distinct genetic profile with frequent K-ras mutation, microsatellite instability because of methylation of hMLH1, but less frequent p53 mutation. 21,22,30,31 K-ras mutation was also frequent in large (>1.5 cm) colorectal adenomas with CpG island methylation in our previous study. 22 In our present study, the colorectal adenomas with CpG island methylation and K-ras mutation are larger as compared to colorectal adenomas with K-ras mutation but no CpG island methylation. O6-Methylguanine DNA methyltransferase, a DNA repair protein that removes alkyl groups and adducts at the O 6 position of guanine, is frequently hypermethylated in colorectal carcinomas and adenomas. 19,32 Hypermethylation of O6-methylguanine DNA methyltransferase is associated with G to A transitions in the K-ras gene. Hypermethylation of O6-methylguanine DNA methyltransferase is also present in smaller adenomas but G to A transitions in K-ras gene are not present in smaller adenomas. 32 The prevalence of smaller adenomas (94%) in our present study may explain these paradoxical results.
MSI-high was present in six adenomas with no methylation and only one adenoma with methylation from three patients with and three without hereditary nonpolyposis colorectal cancer in our study. MSI is more frequent in sporadic colorectal carcinomas than colorectal adenomas, 17,23,31,33-35 and MSI in sporadic colorectal carcinomas is principally because of methylation of hMLH1. 17,31,33 Although, based on a limited number of samples, our data suggest that most of MSI in colorectal adenomas is because of mutation or allelic loss of hMLH1 or hMSH2 in contrast to sporadic colorectal carcinomas, and methylation of hMLH1 is a late event in the adenoma-carcinoma sequence.
There was no correlation between the methylation status of adenomas within a given patient. These data suggest that in the majority of patients with colorectal adenomas, methylation is an epigenetic alteration in an adenoma and is not because of a field defect. We have previously shown lack of intrapatient correlation for K-ras in colorectal adenomas, but weak intrapatient association for MSI and p53 overexpression. 23
In conclusion, our study demonstrates the early role of methylation in colorectal tumorigenesis. Methylation is not correlated in multiple adenomas from an individual patient. Factors related to villous histology and absence of K-ras mutations or MSI are involved in the methylation of colorectal adenomas. The mechanism of methylation of multiple CpG islands is not known. It is postulated that the defect could be either aberrant de novo methylation because of mutation in a DNA-methyltransferase or loss of protection against de novo methylation through the loss of a trans-activating factor. 36-38 This latter defect is exemplified by the adenine phosphoribosyltransferase gene in mice, in which an unidentified protein that binds to the Sp1 sites seems to prevent methylation of the CpG island. It has been proposed that other such proteins may prevent methylation of multiple CpG islands, and expression and/or action of such proteins may be regulated in a tissue-specific manner. Loss of such a protein in cancer may result in hypermethylation of multiple CpG islands.
Footnotes
Address reprint requests to Asif Rashid M.D., Ph.D., Department of Pathology, Box 85, University of Texas M.D. Anderson Cancer Center, 1515 Holcombe Blvd., Houston, TX 77030-4095. E-mail: arashid@mdanderson.org.
References
- 1.Kinzler KW, Vogelstein B: Lessons from hereditary colorectal cancer. Cell 1996, 87:159-170 [DOI] [PubMed] [Google Scholar]
- 2.Reale MA, Fearon ER: Gene defects in colorectal tumorigenesis. Young GP Rozen P Levin B eds. Prevention and Early Detection of Colorectal Cancer. 1996, :pp 63-86 WB Saunders Company Ltd., London [Google Scholar]
- 3.Fearon ER, Dang CV: Cancer genetics: tumor suppressor meets oncogene. Curr Biol 1999, 9:R62-R65 [DOI] [PubMed] [Google Scholar]
- 4.Perucho M: Microsatellite instability: the mutator that mutates the other mutator. Nat Med 1996, 2:630-631 [DOI] [PubMed] [Google Scholar]
- 5.Chung DC: The genetic basis of colorectal cancer: insights into critical pathways of tumorigenesis. Gastroenterology 2000, 119:854-865 [DOI] [PubMed] [Google Scholar]
- 6.Powell SM, Zilz N, Beazer-Barclay Y, Bryan TM, Hamilton SR, Thibodeau SN, Vogelstein B, Kinzler KW: APC mutations occur early during colorectal tumorigenesis. Nature 1992, 359:235-237 [DOI] [PubMed] [Google Scholar]
- 7.Morris RG, Curtis LJ, Romanowski P, Hardcastle JD, Jenkins DA, Robinson M, Wyllie AH, Bird CC: Ki-ras mutations in adenomas: a characteristic of cancer-bearing colorectal mucosa. J Pathol 1996, 180:357-363 [DOI] [PubMed] [Google Scholar]
- 8.Thiagalingam S, Lengauer C, Leach FS, Schutte M, Hahn SA, Overhauser J, Willson JK, Markowitz S, Hamilton SR, Kern SE, Kinzler KW, Vogelstein B: Evaluation of candidate tumour suppressor genes on chromosome 18 in colorectal cancers. Nat Genet 1996, 13:343-346 [DOI] [PubMed] [Google Scholar]
- 9.Nigro JM, Baker SJ, Preisinger AC, Jessup JM, Hostetter R, Cleary K, Bigner SH, Davidson N, Baylin S, Devilee P: Mutations in the p53 gene occur in diverse human tumour types. Nature 1989, 342:705-708 [DOI] [PubMed] [Google Scholar]
- 10.Markowitz S: TGF-beta receptors and DNA repair genes, coupled targets in a pathway of human colon carcinogenesis. Biochim Biophys Acta 2000, 1470:M13-M20 [DOI] [PubMed] [Google Scholar]
- 11.Parsons R, Myeroff LL, Liu B, Willson JK, Markowitz SD, Kinzler KW, Vogelstein B: Microsatellite instability and mutations of the transforming growth factor beta type II receptor gene in colorectal cancer. Cancer Res 1995, 55:5548-5550 [PubMed] [Google Scholar]
- 12.Bird AP: CpG-rich islands and the function of DNA methylation. Nature 1986, 321:209-213 [DOI] [PubMed] [Google Scholar]
- 13.Latham KE: X chromosome imprinting and inactivation in the early mammalian embryo. Trends Genet 1996, 12:134-138 [DOI] [PubMed] [Google Scholar]
- 14.Issa JP, Ottaviano YL, Celano P, Hamilton SR, Davidson NE, Sidransky D, Baylin SB: Methylation of the oestrogen receptor CpG island links ageing and neoplasia in human colon. Nat Genet 1994, 7:536-540 [DOI] [PubMed] [Google Scholar]
- 15.Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JPJ: Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res 1998, 72:141-196 [PubMed] [Google Scholar]
- 16.Herman JG, Merlo A, Mao L, Lapidus RG, Issa JPJ, Davidson NE, Sidransky D, Baylin SB: Inactivation of the CDKN2/p16/MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res 1995, 55:4525-4530 [PubMed] [Google Scholar]
- 17.Ahuja N, Mohan AL, Li Q, Stolker JM, Herman JG, Hamilton SR, Baylin SB, Issa JP: Association between CpG island methylation and microsatellite instability in colorectal cancer. Cancer Res 1997, 57:3370-3374 [PubMed] [Google Scholar]
- 18.Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB: Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet 1999, 21:103-107 [DOI] [PubMed] [Google Scholar]
- 19.Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG: Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res 1999, 59:793-797 [PubMed] [Google Scholar]
- 20.Kane MF, Loda M, Gaida GM, Lipman J, Mishra R, Goldman H, Jessup JM, Kolodner R: Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res 1997, 57:808-811 [PubMed] [Google Scholar]
- 21.Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JPJ: CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci USA 1999, 96:8681-8686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Toyota M, Ohe-Toyota M, Ahuja N, Issa JPJ: Distinct genetic profiles in colorectal tumors with or without the CpG island methylator phenotype. Proc Natl Acad Sci USA 2000, 97:710-715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rashid A, Zahurak M, Goodman SN, Hamilton SR: Genetic epidemiology of mutated K-ras oncogene, altered suppressor genes, and microsatellite instability in colorectal adenomas. Gut 1999, 44:826-833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moskaluk CA, Kern SE: Microdissection and polymerase chain reaction amplification of genomic DNA from histological tissue sections. Am J Pathol 1997, 150:1547-1552 [PMC free article] [PubMed] [Google Scholar]
- 25.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB: Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA 1996, 93:9821-9826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toyota M, Ho C, Ohe-Toyota M, Baylin SB, Issa J-PI: Inactivation of CAGNA1G, a T-type calcium channel gene, by aberrant methylation of its 5′ CpG island in human tumors. Cancer Res 1999, 59:4535-4541 [PubMed] [Google Scholar]
- 27.Xiong Z, Laird PW: COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res 1997, 25:2532-2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang K-Y, Zeger SL: Longitudinal data analysis using generalized linear models. Biometrika 1986, 73:13-22 [Google Scholar]
- 29.Jones PA: DNA methylation errors and cancer. Cancer Res 1996, 56:2463-2467 [PubMed] [Google Scholar]
- 30.Guan RJ, Fu Y, Holt PR, Pardee AB: Association of K-ras mutations with p16 methylation in human colon cancer. Gastroenterology 1999, 116:1063-1071 [DOI] [PubMed] [Google Scholar]
- 31.Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa J-PJ, Markowitz S, Willson JKV, Hamilton SR, Kinzler KW, Kane MF, Kolodner RD, Vogelstein B, Kunkel TA, Baylin SB: Incidence and functional consequences of hMLH1 promoter hypermethylation in colon carcinoma. Proc Natl Acad Sci USA 1998, 95:6870-6875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Esteller M, Toyota M, Sanchez-Cespedes M, Capella G, Peinado MA, Watkins DN, Issa JPJ, Sidransky D, Baylin SB, Herman JG: Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is associated with G to A mutation in K-ras in colorectal tumorigenesis. Cancer Res 2000, 60:2368-2371 [PubMed] [Google Scholar]
- 33.Thibodeau SN, French AJ, Cunningham JM, Tester D, Burgart LJ, Roche PC, McDonnell SK, Schaid DJ, Vockley CW, Michels VV, Farr GH, Jr, O’Connell MJ: Microsatellite instability in colorectal cancers: different mutator phenotypes and the principal involvement of hMLH1. Cancer Res 1998, 58:1713-1718 [PubMed] [Google Scholar]
- 34.Young J, Leggett B, Gustafson C, Ward M, Searle J, Thomas L, Buttenshaw R, Chenevix-Trench G: Genomic instability occurs in colorectal carcinomas but not in adenomas. Hum Mutat 1993, 2:351-354 [DOI] [PubMed] [Google Scholar]
- 35.Samowitz WS, Slattery ML: Microsatellite instability in colorectal adenomas. Gastroenterology 1997, 112:1515-1519 [DOI] [PubMed] [Google Scholar]
- 36.Macleod D, Charlton J, Mullins J, Bird AP: Sp1 sites in the mouse aprt gene promoter are required to prevent methylation of the CpG island. Genes Dev 1994, 8:2282-2292 [DOI] [PubMed] [Google Scholar]
- 37.Chen ZJ, Pikaard CS: Epigenetic silencing of RNA polymerase I transcription: a role for DNA methylation and histone modification in nucleolar dominance. Genes Dev 1997, 11:2124-2136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mummaneni P, Yates P, Simpson J, Rose J, Turker MS: The primary function of a redundant Sp1 binding site in the mouse aprt gene promoter is to block epigenetic gene inactivation. Nucleic Acids Res 1998, 26:5163-5169 [DOI] [PMC free article] [PubMed] [Google Scholar]