Abstract
Lipoblastomas are rare soft tissue tumors that occur primarily in young children. They typically contain variably differentiated adipocytes, primitive mesenchymal cells, myxoid matrix, and fibrous trabeculae. Abnormalities in chromosome 8, leading to rearrangements of the PLAG1 gene, were demonstrated recently in four lipoblastomas. In the present report, we determine the frequency of PLAG1 alterations in 16 lipoblastomas from children aged 13 years or younger, and we also evaluate the stages of lipoblastoma differentiation at which PLAG1 genomic alterations are found. Eleven lipoblastomas (69%), including those with either classic or lipoma-like histology, had rearrangements of the 8q12 PLAG1 region. Another three lipoblastomas had polysomy for chromosome 8 in the absence of PLAG1 rearrangement. Only two cases (13%) lacked a chromosome 8 abnormality. Notably, the lipoblastomas with chromosome 8 polysomy had up to five copies of chromosome 8 as an isolated cytogenetic finding in an otherwise diploid cell. We also demonstrate that PLAG1 alterations are found in a spectrum of mesenchymal cell types in lipoblastomas, including lipoblasts, mature adipocytes, primitive mesenchymal cells, and fibroblast-like cells. This finding is consistent with neoplastic origin in a primitive mesenchymal precursor and with variable differentiation to a mature adipocyte end-point. Hence, our studies provide biological validation for the clinical observation that lipoblastomas can evolve into mature, lipoma-like, lesions. They also suggest that PLAG1 dosage alterations caused by polysomy 8 might represent an alternative oncogenic mechanism in lipoblastoma.
Lipoblastomas are fatty tumors primarily affecting children younger than 3 years of age. 1,2 Most lipoblastomas present in the superficial tissues of the arms and legs, but they may also arise in the head and neck regions, trunk, mediastinum, and retroperitoneum. Although lipoblastomas do not metastasize, they can recur locally after incomplete excision. Lipoblastomas feature a wide spectrum of adipocytic differentiation, in which mono- and multivacuolated lipoblasts are admixed with mature adipocytes. The adipocytic cells are often set in a variably myxoid matrix containing spindle or stellate mesenchymal cells, and the tumor is usually organized into lobules, separated by fibrous septae. The admixture of lipoblasts and mature adipocytes has given rise to the broadly held notion that lipoblastomas can mature to simple lipomas. 3 Recently, we reported chromosomal rearrangements involving band 8q12 in four lipoblastomas. Each of these alterations targeted the PLAG1 oncogene, which encodes a zinc finger transcription factor, expressed primarily in fetal tissues and only at very low levels postnatally. 4,5 Notably, the 8q12 rearrangements in lipoblastoma were associated with promoter swapping, in which the PLAG1 promoter element was replaced by those from the hyaluronic acid synthase 2 (HAS2) or collagen (COL1A2) genes. These promoter swapping events accomplish PLAG1 transcriptional up-regulation by substitution of an active promoter and are mechanistically similar to the PLAG1 rearrangements in pleomorphic adenomas of salivary gland. 4 In the present study, we sought to further characterize the role of PLAG1 alteration in lipoblastoma by determining the frequency of genomic PLAG1 rearrangements and by evaluating PLAG1 alterations in cells at different levels of differentiation.
Materials and Methods
Patients
Cytogenetic and molecular genetic data on cases 1 to 5 and 8 have been previously published. 5-7 Clinical and histopathological data for all cases are found in Table 1 ▶ .
Table 1.
Clinical, Histopathological, Cytogenetic, and in Situ Hybridization Data for 16 Lipoblastomas
| Case | Histology* | Age/sex | Location | Cytogenetics | In situ hybridization |
|---|---|---|---|---|---|
| 1 | Classic | 2.5 /M | Perineal | 8q12 rearrangement | PLAG1 split |
| 2 | Classic | 1.5 /F | Back | 8q12 rearrangement | PLAG1 split |
| 3 | Classic | 7 /M | Intramuscular | 8q12 rearrangement | PLAG1 split |
| 4 | Classic | 6.5 /M | Mediastinal | Ring chromosome 8 | PLAG1 split |
| 5 | Classic | 1 /M | Subscapular | 8q12 rearrangement | PLAG1 split |
| 6 | Classic | 3.5 /F | Neck | 8q12 rearrangement | PLAG1 split |
| 7 | Classic | 13 /M | Hip | 8q12 rearrangement | PLAG1 split |
| 8 | Classic | 1 /M | Axilla | 8q12 rearrangement | PLAG1 split |
| 9 | Classic | 1.5 /F | Hip | Normal | Disomy 8, tetrasomy 8 |
| 10 | Classic | 5 /M | Back | Normal | Disomy 8 |
| 11 | Classic | 2 /F | Leg | Normal | Disomy 8 |
| 12 | Myxoid | 1.5 /M | Neck | 8q12 rearrangement | PLAG1 split |
| 13 | Myxoid | 7 /M | Neck | Pentasomy 8 | Disomy 8, trisomy 8, tetrasomy 8, pentasomy 8, hexasomy 8 |
| 14 | Lipoma-like | 5.5 /F | Arm | 8q12 rearrangement | PLAG1 split |
| 15 | Lipoma-like | 8 mo /F | Neck | Normal | PLAG1 split |
| 16 | Hibernoma-like | 1 /M | Back | Not Done | Disomy 8, trisomy 8, tetrasomy 8 |
*Histological subclassification is defined in the text.
Chromosome Analysis
Metaphase harvest and slide preparation from tumor cell cultures were as described previously, 8 with minor modifications. G banding was obtained by trypsin-Giemsa staining. For fluorescence in situ hybridization, chromosome preparations were pretreated in 2× standard saline citrate at 37°C for 1 hour, followed by 5 minutes of digestion with pepsin using 1:25 diluted Digest-All III (Zymed Laboratories, South San Francisco, CA). Biotinylated and digoxigenin-labeled α satellite probes for the centromeric regions of chromosomes 4, 8, 11, and 12 were from Oncor (Gaithersburg, MD). The yeast artificial chromosome probes 935e9 and 946b7, flanking PLAG1, were obtained from Center d’Etude Polymorphisme Humain, Paris, France. Yeast artificial chromosome DNA was isolated as previously described 8 and labeled by either digoxigenin or biotin by random octamer priming, using the BioPrime DNA labeling system (Life Technologies Inc., Rockville, MD). After labeling, DNA was purified by S-200HR spin column chromatography (Pharmacia, Uppsala, Sweden), co-precipitated with 5× Cot-1, and resuspended in hybridization buffer (50% formamide, 10% dextran sulfate, 2× standard saline citrate). Probes and chromosome preparations were co-denatured at 75°C for 2 minutes and hybridized overnight. Biotin- and digoxigenin-labeled probes were detected by strepatvidin-Alexa-498 (Molecular Probes, Eugene, OR) and fluorescein anti-digoxigenin (Roche Diagnostics, Indianapolis, IN), respectively. Chromosomes were counterstained using diamidinophenylindole (Vector Laboratories, Burlingame, CA).
Genetic Analyses of Tissue Sections
Chromogenic in situ hybridization (CISH) was performed on 4-μm formalin-fixed, paraffin-embedded, tissue sections on positively charged slides (SuperFrost Plus; Menzel-Gläser, Germany). The slides were baked overnight at 65°C and then deparaffinized in xylene for 3 × 15 minutes, followed by washing in 100% ethanol for 2 × 2 minutes. Air-dried slides were treated in 100 mmol/L Tris/50 mmol/L ethylenediaminetetraacetic acid (pH 7.0) in a microwave oven at 99°C for 15 minutes, washed briefly in phosphate-buffered saline (PBS), and digested with Digest-All III (Zymed Laboratories) for 1 to 7 minutes. The duration of the pepsin digestion was optimized for each case to obtain sufficient signal intensity without loss of tissue architecture. After washing in PBS, the slides were immersed in 10% formalin/PBS for 1 minute and dehydrated for 4 × 2 minutes in a 70 to 85% to 95 to 100% ethanol series. Probes for CISH were labeled with digoxigenin and fluorescein (New England Nuclear, Boston, MA), by random octamer priming, as described above. Tissue sections and probes were co-denatured at 94°C for 3 minutes and hybridized overnight at 37°C. Probe detection was with alkaline phosphatase anti-digoxigenin (Roche Molecular Biochemicals, Indianapolis, IN) and Fast Red substrate, and with horseradish peroxidase anti-fluorescein and H2O2-diaminobenzidine (Zymed Laboratories). Tissue sections were counterstained with hematoxylin and mounted with Glycerogel (DAKO, Carpinteria, CA). At least 50 cells (mean, 150) were analyzed from each tumor. Tumors with >10 cells showing one red-brown intact signal, one separate red, and one separate brown signal at yeast artificial chromosome probe hybridization were scored as positive for PLAG1 rearrangements. Tumors with >10 cells showing >2 signals for centromere 8 and ≤2 signals with a centromeric probe for another chromosome were scored as polysomic for chromosome 8.
Results
Histopathology
Review of the histology slides from the 16 lipoblastomas showed that all tumors were characterized by lobules of mature and immature fat cells including lipoblasts of the monovacuolated and multivacuolated variety. The fibrous septae varied in thickness and consisted of delicate and/or wiry collagen with interspersed spindle, fibroblast-like cells, some of which had plump nuclei and abundant cytoplasm and sometimes very small cytoplasmic vacuoles. There was substantial difference in the histological appearance of some tumors, prompting the classification described below. The classic lipoblastomas (n = 11; Table 1 ▶ ) usually had a minimal myxoid component–limited to rare microscopic foci in some tumors—consisting of intercellular mucin, spindle cells, and stellate primitive mesenchymal cells, which blended with the adipocytic component (Figure 1A) ▶ . The myxoid lipoblastomas (n = 2) contained abundant interstitial mucin that comprised >50% of the specimen (Figure 1B) ▶ . The lipoma-like lipoblastomas (n = 2) lacked a myxoid component and were composed predominantly of mature adipocytes with scattered monovacuolated and multivacuolated lipoblasts (Figure 1C) ▶ . The last tumor also lacked a myxoid component and was composed predominantly of multivacuolated lipoblasts, some of which had central nuclei and granular eosinophilic cytoplasm. This tumor was designated a “lipoblastoma with hibernoma-like features” (Figure 1D) ▶ . All tumors were categorized using the above-mentioned descriptors (Table 1) ▶ .
Figure 1.
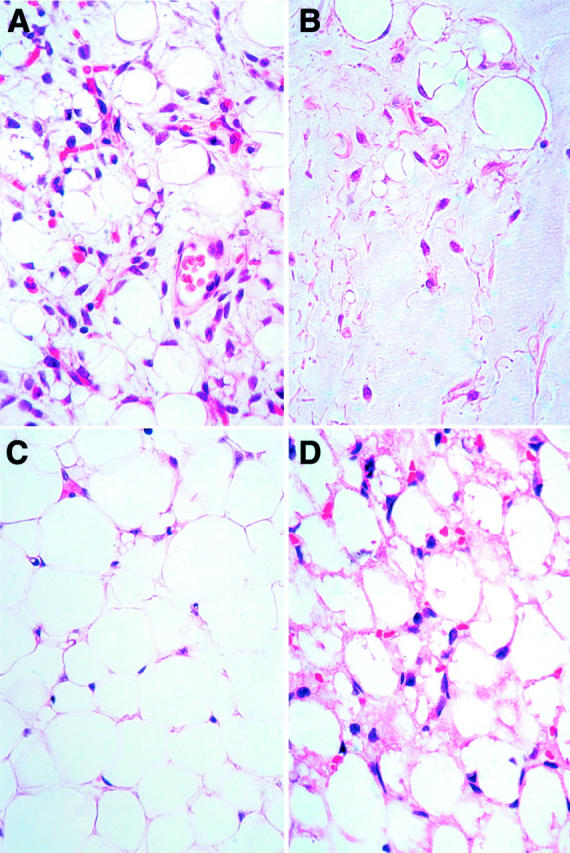
Photomicrographs of representative lipoblastoma histologies. A: Classic lipoblastoma has stellate and spindle mesenchymal cells, monovacuolated and multivacuolated lipoblasts, adipocytes, and intercellular mucin. B: Myxoid lipoblastoma is characterized by abundant intercellular mucin. C: Lipoma-like lipoblastoma has mostly mature adipocytes with only a rare lipoblast. D: Hibernoma-like lipoblastoma has polyvacuolated adipocytes with granular cytoplasm with some cells having central nuclei.
G Band and Fluorescence in Situ Hybridization/CISH Analyses
G-banding analyses revealed rearrangement of the 8q12 chromosome region in 9 of 16 lipoblastomas, and a ring chromosome 8 with unassignable cytogenetic breakpoints in the chromosome 8 long arm in another tumor (Table 2) ▶ . PLAG1 abnormalities were evaluated in all 16 tumors by metaphase cell fluorescence in situ hybridization (12 cases) and/or paraffin-section interphase cell CISH (10 cases). CISH was used for the paraffin sections because lipid-related autofluorescence interfered with fluorescence in situ hybridization detection methods, particularly when attempting to evaluate the adipocytic populations in the sections (data not shown). After optimization of the enzymatic digestion procedure, the paraffin sections from each of the 10 lipoblastomas could be analyzed with a simultaneous appreciation of tissue architecture, cellular morphology, and chromosomal abnormalities. The PLAG1 in situ hybridization analyses confirmed rearrangement of the PLAG1 region (Figure 2, a and b) ▶ in all lipoblastomas with cytogenetic 8q rearrangement and in one lipoblastoma that had been diploid cytogenetically. In addition, the in situ hybridization analyses revealed three to six copies of chromosome 8 in three lipoblastomas, including one (case 13) that had pentasomy 8 by G banding (Figure 3) ▶ . Neither chromosome banding nor molecular cytogenetic analyses showed evidence of a general increase in DNA ploidy in the polysomy 8 lipoblastomas. Not only was the karyotype indicative of an isolated chromosome 8 polysomy in case 13, but there were no polysomies for four comparison chromosomes (1, 4, 11, and 12, by α-satellite probes) in any of the polysomy 8 tumors (Figure 2c) ▶ . None of the cells in the polysomy 8 tumors had molecular cytogenetic evidence of PLAG1 rearrangement.
Table 2.
G-Banded Karyotypes for 16 Lipoblastomas
| Case | Karyotype |
|---|---|
| 1 | 46,XY,ins(6;8)(p25;q12q24.1)*† |
| 2 | 46,XX,der(1)(7pter->7p22::8q24.1->8q22::1p13->1qter),der(7)(1pter->1p13::8q22>8q13::7p22->7qter),der(8)(pter->q13::q24.1->qter)*† |
| 3 | 46,XY,t(8;12)(q12;q24) |
| 4 | 46,XY,r(8)* |
| 5 | 46,XY,del(6)(q11),der(7)t(6;7)(q11;p22),t(7;8)(q22;q12),del(8)(q12)* |
| 6 | 47,XX,del(3)(q12),-8,+der(?)t(8;?)(q12;?),+mar1 |
| 7 | 46,XY,t(1;8)(p22;q12) |
| 8 | 46,XX,t(8;9)(q12;p22)‡ |
| 9 | 46,XX |
| 10 | 46,XY |
| 11 | 46,XX |
| 12 | 46,XY,t(3;8)(q12;q12) |
| 13 | 49,XY,+8,+8,+8 |
| 14 | 46,XX,del(2)(q23q31),der(5)t(5;8)(p11;q12),der(8)(8pter->8q12::?::5p11->5pter) |
| 15 | 46,XX |
| 16 | Not done |
Figure 2.
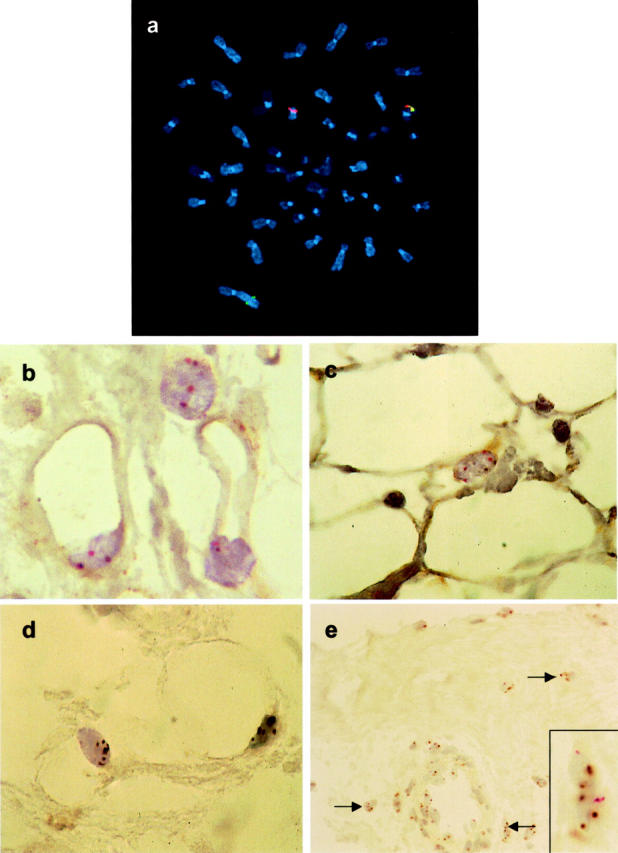
a: Fluorescence in situ hybridization on metaphase chromosomes with centromeric (red) and telomeric (green) PLAG1 probes demonstrates PLAG1 rearrangement in lipoblastoma case 2. b: CISH with PLAG1 probes in paraffin section from lipoblastoma case 2: splitting apart of red and brown probe signals is seen in adipocytes. c: CISH with chromosome 8 (red) and chromosome 10 (brown) probes in paraffin section from lipoblastoma case 9: polysomy 8 is evident in the adipocytic component. d and e: CISH with chromosome 8 (brown) and chromosome 11 (red) probes in paraffin section from lipoblastoma case 13: polysomy 8 is evident in both adipocytic (d) and fibrous (e) components but not in vascular element (e).
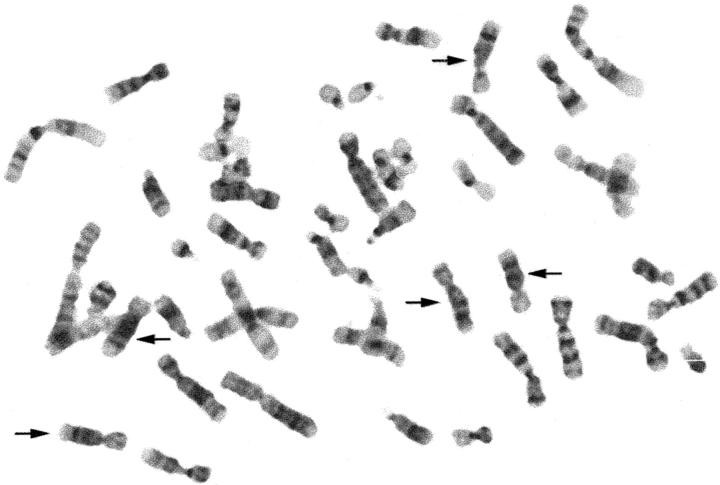
Figure 3.
Metaphase cell from lipoblastoma case 13, stained by trypsin-Giemsa. Arrows indicate the five copies of chromosome 8.
Notably, PLAG1 rearrangement or copy number gain was found in lipoblastomas irrespective of histological subtype (Table 1) ▶ . In particular, PLAG1 alterations were demonstrated in each of two cases, which were composed predominantly of mature adipocytic cells (lipoma-like lipoblastoma). Six lipoblastomas, including three with PLAG1 rearrangement (cases 1,7, and 12) and three with PLAG1 copy number gain (cases 9, 13, and 16), were evaluated to determine which cell types contained the genomic alterations. All six tumors contained monovacuolated and multivacuolated lipoblasts as well as more mature adipocyte-like cells. PLAG1 rearrangement was demonstrated in each of the mesenchymal cell components—lipoblastic, mature adipocytic, stellate primitive mesenchymal cell, and fibroblast-like cell—in these tumors. However, PLAG1 was not rearranged in the vascular elements, including endothelial cells and leukocytes, within the tumor parenchyma. Similarly, the three lipoblastomas with polysomy 8 contained additional PLAG1 signals in both the lipogenic cells and fibroblast-like cells, including those in fibrous septae (Figure 2, d and e) ▶ , but not in the vessels (Figure 2e) ▶ . The size of the polysomic cell populations varied in these tumors. Case 13 had three to six copies of chromosome 8 in all cells of the tumor, except for the vessels, whereas cases 9 and 16 had localized cell populations with three or four copies of chromosome 8.
Discussion
Most lipogenic tumors exhibit characteristic chromosomal rearrangements, which are associated with rearrangement and/or altered expression of specific oncoproteins. Myxoid liposarcomas typically have chromosomal abnormalities involving band 12q13, resulting in rearrangement of the CHOP gene with either FUS in 16p11 or EWS in 22q12. 9,10 The FUS-CHOP and EWS-CHOP fusion oncoproteins are transcriptional regulators, affecting the expression of various genes associated with adipocytic differentiation. 11,12 Well-differentiated liposarcomas, on the other hand, are characterized by supernumerary ring and giant marker chromosomes, containing amplified sequences from the long arm of chromosome 12. 13,14 These amplicons generally include the MDM2 oncogene, the product of which inhibits normal TP53 function and thereby promotes cell cycle progression. 15,16 Nonrandom chromosomal alterations are also found in benign adipocytic tumors. Approximately 60% of typical, mature, lipomas have rearrangements of the HMGIC locus at chromosome band 12q15. 17-19 HMGIC encodes a DNA-binding protein, expressed primarily in embryogenic tissues, the functions of which include regulation of mesenchymal cell growth and differentiation. 20 A similar situation exists in lipoblastoma, where 8q12 chromosomal rearrangements result in up-regulated expression of the PLAG1 transcription factor. 5,21
In the present study, we found PLAG1 rearrangements in 11 of 16 lipoblastomas, and we also showed that the PLAG1 nonrearranged tumors may have novel oncogenic mechanisms involving PLAG1 copy number gains. These data suggest an either/or relationship between PLAG1 rearrangement and PLAG1 amplification in lipoblastoma tumorigenesis. An analogous situation has been observed in uterine leiomyomas, where ∼25% (80 of 325) of tumors contain rearrangements of the HMGIC region in 12q15, whereas another 10% (27 of 325) are trisomic for chromosome 12 (Mitelman F, Johansson B, Mertens F: Mitelman Database of Chromosome Aberrations in Cancer. http://cgap.nci.nih.gov/Chromosomes/Mitelman. 2001). Although PLAG1 expression has not been compared in lipoblastomas containing PLAG1 rearrangements versus amplifications, we hypothesize that PLAG1 transcriptional up-regulation will be a consistent feature in both groups. Moreover, it is possible that the low-level amplified PLAG1 loci are not wild type. Rather, these loci might contain point mutations within binding sites of PLAG1 transcriptional repressors, which would thereby contribute to PLAG1 transcriptional up-regulation. There is precedent for promoter-region point mutations in association with nonfusion MYC and BCL6 oncogenes in Burkitt lymphomas 22 and in follicular center cell lymphomas, 23 respectively. However, it is yet known whether analogous genomic mechanisms contribute to transcriptional up-regulation in nonlymphoid tumors.
Most lipoblastomas contain primitive mesenchymal cells and adipocytic cells at various stages of maturation. 1 Typically, these cells are arranged in lobules showing a pattern of gradual differentiation, with the immature mesenchymal cells and multivacuolated lipoblasts at the periphery and the monovacuolated lipoblasts and adipocytes toward the center. This organization recapitulates the architecture of embryonal fat and it has been shown that some lipoblastomas eventually evolve into mature fat, presumably by terminal differentiation. 3,23-26 If this is the case, one would anticipate a lipoma-like morphology in some lipoblastomas. Indeed, in the present study, two lipoma-like lipoblastomas were shown to contain PLAG1 rearrangements. However, it is unlikely that any substantial number of adult lipomas result from lipoblastoma maturation. Adult lipomas are vastly more common than lipoblastoma, and they are characterized by a cytogenetic profile very different from that in lipoblastoma. Previous cytogenetic data have shown that only 1% (2 of 216) of lipomas from patients aged 15 years or older contain 8q11-13 rearrangements (Mitelman F, Johansson B, Mertens F: Mitelman Database of Chromosome Aberrations in Cancer. http://cgap.nci.nih.gov/Chromosomes/Mitelman. 2001). The frequency of trisomy 8 in these tumors is 2% (4 of 216). This paucity of lipoma PLAG1-region rearrangements is striking, particularly because—as discussed below—we demonstrated PLAG1 rearrangements in the lipoma components of lipoblastomas. It follows that lipoblastomas and lipomas generally arise by distinct tumorigenic mechanisms, and possibly from different adipocytic progenitor cells.
CISH analysis of paraffin sections in six lipoblastomas revealed that the PLAG1 alterations were present in lipoblasts, mature adipocytes, stellate primitive mesenchymal cells, and fibroblast-like cells. The presence of PLAG1 rearrangements in variably differentiated mesenchymal cells suggests an origin from a transformed mesenchymal progenitor that proliferates and differentiates in a manner similar to white fat. Because lipoblastomas occur in children primarily younger than 10 years of age, we expect that such progenitors are only present, or only susceptible to PLAG1-mediated neoplastic transformation, during the first few years of life. Such a window of vulnerability in cellular development has been suggested in connection with other neoplasms, in particular hematological malignancies. However, the difficulties in determining histological origin of tumor cell populations have left this hypothesis primarily unsubstantiated in solid neoplasms. 27 Lipoblastomas provide a compelling example of such a scenario, and in contrast to leukemias, in which expression of fusion transcription factor oncogenes can block cell maturation, 28 lipoblastomas follow a remarkably well-ordered sequence of differentiation and remain benign. Much might be learned by characterizing the biological impact of abnormally expressed developmental factors, such as PLAG1, in benign tumors. In particular, it would be important to determine whether the benign nature of lipoblastomas is dictated by cell context (the specific progenitor cell) or whether this is a general aspect of PLAG1-mediated transformation.
Footnotes
Address reprint requests to Dr. Jonathan A. Fletcher, Department of Pathology, Brigham and Women’s Hospital, 75 Francis St., Boston, MA 02115. E-mail: jfletcher@partners.org.
Supported by the Blanceflor Boncompagni-Ludovisi, née Bildt Foundation, the John and Augusta Persson Foundation for Scientific Medical Research, the Swedish Medical Society, and a Lund Family American Cancer Society Grant.
References
- 1.Enzinger FM, Weiss SW: Soft Tissue Tumors, ed 3 1994, Mosby-Year Book Inc., St. Louis
- 2.Collins MH, Chatten J: Lipoblastoma/lipoblastomatosis: a clinicopathologic study of 25 tumors. Am J Surg Pathol 1997, 21:1131-1137 [DOI] [PubMed] [Google Scholar]
- 3.Van Meurs DP: The transformation of an embryonic lipoma to a common lipoma. Br J Surg 1947, 34:282-284 [DOI] [PubMed] [Google Scholar]
- 4.Kas K, Voz ML, Röijer E, Åstrom AK, Meyen E, Stenman G, Van de Ven WJ: Promoter swapping between the genes for a novel zinc finger protein and beta-catenin in pleiomorphic adenomas with t(3;8)(p21;q12) translocations. Nat Genet 1997, 15:170-174 [DOI] [PubMed] [Google Scholar]
- 5.Hibbard MK, Kozakewich HP, Dal Cin P, Sciot R, Tan X, Xiao S, Fletcher JA: PLAG1 fusion oncogenes in lipoblastoma. Cancer Res 2000, 60:4869-4872 [PubMed] [Google Scholar]
- 6.Fletcher JA, Kozakewich HP, Schoenberg ML, Morton CC: Cytogenetic findings in pediatric adipose tumors: consistent rearrangement of chromosome 8 in lipoblastoma. Genes Chromosom Cancer 1993, 6:24-29 [DOI] [PubMed] [Google Scholar]
- 7.Dal Cin P, Sciot R, De Wever I, Van Damme B, Van den Berghe H: New discriminative chromosomal marker in adipose tissue tumors. The chromosome 8q11–q13 region in lipoblastoma. Cancer Genet Cytogenet 1994, 78:232-235 [DOI] [PubMed] [Google Scholar]
- 8.Xiao S, Renshaw A, Cibas ES, Hudson TJ, Fletcher JA: Novel fluorescence in situ hybridization approaches in solid tumors. Characterization of frozen specimens, touch preparations, and cytological preparations. Am J Pathol 1995, 147:896-904 [PMC free article] [PubMed] [Google Scholar]
- 9.Crozat A, Åman P, Mandahl N, Ron D: Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma. Nature 1993, 363:640-644 [DOI] [PubMed] [Google Scholar]
- 10.Panagopoulos I, Höglund M, Mertens F, Mandahl N, Mitelman F, Åman P: Fusion of the EWS and CHOP genes in myxoid liposarcoma. Oncogene 1996, 12:489-494 [PubMed] [Google Scholar]
- 11.Adelmant G, Gilbert JD, Freytag SO: Human translocation liposarcoma-CCAAT/enhancer binding protein (C/EBP) homologous protein (TLS-CHOP) oncoprotein prevents adipocyte differentiation by directly interfering with C/EBPbeta function. J Biol Chem 1998, 273:15574-15581 [DOI] [PubMed] [Google Scholar]
- 12.Kuroda M, Wang X, Sok J, Yin Y, Chung P, Giannotti JW, Jacobs KA, Fitz LJ, Murtha-Riel P, Turner KJ, Ron D: Induction of a secreted protein by the myxoid liposarcoma oncogene. Proc Natl Acad Sci USA 1999, 96:5025-5030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heim S, Mandahl N, Kristoffersson U, Mitelman F, Rööser B, Rydholm A, Willén H: Marker ring chromosome—a new cytogenetic abnormality characterizing lipogenic tumors? Cancer Genet Cytogenet 1987, 24:319-326 [DOI] [PubMed] [Google Scholar]
- 14.Dal Cin P, Kools P, Sciot R, De Wever I, Van Damme B, Van de Ven W, Van den Berghe H: Cytogenetic and fluorescence in situ hybridization investigation of ring chromosomes characterizing a specific pathologic subgroup of adipose tissue tumors. Cancer Genet Cytogenet 1993, 68:85-90 [DOI] [PubMed] [Google Scholar]
- 15.Oliner JD, Kinzler KW, Meltzer PS, George DL, Vogelstein B: Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature 1992, 358:80-83 [DOI] [PubMed] [Google Scholar]
- 16.Nilbert M, Rydholm A, Willén H, Mitelman F, Mandahl N: MDM2 gene amplification correlates with ring chromosome in soft tissue tumors. Genes Chromosom Cancer 1994, 9:261-265 [DOI] [PubMed] [Google Scholar]
- 17.Mandahl N, Heim S, Arheden K, Rydholm A, Willen H, Mitelman F: Three major cytogenetic subgroups can be identified among chromosomally abnormal solitary lipomas. Hum Genet 1988, 79:203-208 [DOI] [PubMed] [Google Scholar]
- 18.Ashar HR, Fejzo MS, Tkachenko A, Zhou X, Fletcher JA, Weremowicz S, Morton CC, Chada K: Disruption of the architectural factor HMGI-C: DNA-binding AT hook motifs fused in lipomas to distinct transcriptional regulatory domains. Cell 1995, 82:57-65 [DOI] [PubMed] [Google Scholar]
- 19.Schoenmakers EF, Wanschura S, Mols R, Bullerdiek J, Van den Berghe H, Van de Ven WJ: Recurrent rearrangements in the high mobility group protein gene, HMGI-C, in benign mesenchymal tumours. Nat Genet 1995, 10:436-444 [DOI] [PubMed] [Google Scholar]
- 20.Zhou X, Benson KF, Ashar HR, Chada K: Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGI-C. Nature 1995, 376:771-774 [DOI] [PubMed] [Google Scholar]
- 21.Åström A, D’Amore ES, Sainati L, Panarello C, Morerio C, Mark J, Stenman G: Evidence of involvement of the PLAG1 gene in lipoblastomas. Int J Oncol 2000, 16:1107-1110 [DOI] [PubMed] [Google Scholar]
- 22.Bhatia K, Huppi K, Spangler G, Siwarski D, Iyer R, Magrath I: Point mutations in the c-Myc transactivation domain are common in Burkitt’s lymphoma and mouse plasmacytomas. Nat Genet 1993, 5:56-61 [DOI] [PubMed] [Google Scholar]
- 23.Migliazza A, Martinotti S, Chen W, Fusco C, Ye BH, Knowles DM, Offit K, Chaganti RS, Dalla-Favera R: Frequent somatic hypermutation of the 5′ noncoding region of the BCL6 gene in B-cell lymphoma. Proc Natl Acad Sci USA 1995, 92:12520-12524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O’Donnell KA, Caty MG, Allen JE, Fisher JE: Lipoblastoma: better termed infantile lipoma? Pediatr Surg Int 2000, 16:458-461 [DOI] [PubMed] [Google Scholar]
- 25.Mentzel T, Calonje E, Fletcher CD: Lipoblastoma and lipoblastomatosis: a clinicopathological study of 14 cases. Histopathology 1993, 23:527-533 [DOI] [PubMed] [Google Scholar]
- 26.Coffin CM: Lipoblastoma: an embryonal tumor of soft tissue related to organogenesis. Semin Diagn Pathol 1994, 11:98-103 [PubMed] [Google Scholar]
- 27.Åman P: Fusion genes in solid tumors. Semin Cancer Biol 1999, 9:303-318 [DOI] [PubMed] [Google Scholar]
- 28.Olsson I, Bergh G, Ehinger M, Gullberg U: Cell differentiation in acute myeloid leukemia. Eur J Haematol 1996, 57:1-16 [DOI] [PubMed] [Google Scholar]