Abstract
Epidermal clones of p53-mutated keratinocytes are abundant in chronically sun-exposed skin and may play an important role in early development of skin cancer. Advanced laser capture microdissection enables genetic analysis of targeted cells from tissue sections without contamination from neighboring cells. In this study p53 gene mutations were characterized in single cells from normal, chronically sun-exposed skin. Biopsies were obtained from skin subjected to daily summer sun and skin totally protected from the sun by blue denim fabric. Using laser capture microdissection, 172 single-cell samples were retrieved from four biopsies and analyzed using single-cell polymerase chain reaction and direct DNA sequencing. A total of 14 different mutations were identified in 26 of 99 keratinocytes from which the p53 gene could be amplified. Mutations displayed a typical UV signature and were detected in both scattered keratinocytes and in a small cluster of p53-immunoreactive keratinocytes. This minute epidermal p53 clone had a diameter of 10 to 15 basal cells. Two missense mutations were found in all layers of epidermis within the p53 clone. The presented data show that p53 mutations are common in normal skin and that a clone of keratinocytes with a mutated p53 gene prevailed despite 2 months of total protection from ultraviolet light.
Nonmelanoma skin cancer [ie, squamous cell cancer (SCC) and basal cell cancer (BCC)] is the most common form of human cancer. 1 Ultraviolet (UV) radiation from the sun is accepted as a major risk factor and tumor cells exhibit mutations with typical UV signature in cancer-related genes. 2 Skin carcinogenesis is a multistep process, in which the early clandestine events, preceding malignant transformation are primarily unknown. In chronically sun-damaged skin SCC develops through stages of actinic keratosis and SCC in situ, whereas no such precursor lesions are known for BCC. Although several genes and pathways are important for development of skin cancer, the genetic events underlying the different steps from a normal cell to SCC or BCC are virtually unknown. Mutation of the p53 gene is one frequent, known genetic alteration found in SCC and BCC. 3 In addition, activation of the sonic hedgehog/patched signaling pathway seems essential for development of BCC. 4,5
In human skin there exists a multitude of p53-immunoreactive clusters of morphologically normal epidermal keratinocytes. 6-8 These p53 clones are predominantly found in chronically sun-exposed skin. Microdissection followed by polymerase chain reaction (PCR) and direct DNA sequencing has shown an underlying p53 mutation in at least 70% of analyzed cases. Epidermal p53 clones and adjacent cancers have never been shown to share the same p53 mutation and thus there is no solid evidence of a genetic link between p53 clones and any specific type of skin cancer. 9,10 However, the incidence and location of p53 clones suggest a role for p53 mutations in skin cancer. Mutations in the p53 gene have also been detected in UV-irradiated mouse skin months before the gross appearance of skin tumors, suggesting that p53 mutations are an early event for the development of skin cancer. 11,12 Clusters of keratinocytes with strong p53 immunoreactivity can furthermore be induced in mice subjected to UV irradiation. 13 Clonal expansion of keratinocytes with a mutated p53 gene is most likely facilitated because of a relative resistance to UV-induced apoptosis. 14 After DNA damage subsequent to normal sun-exposure, nonmutated keratinocytes will enter into apoptosis more easily and thus allow for expansion of p53-mutated keratinocytes.
Advanced laser capture microdissection techniques have made it possible to exploit targeted cells from histologically stained sections without contamination from neighboring cells and thereby reduced the obstacle of tissue complexity. 15-17 When combined with optimized techniques for gene amplification and sequencing, 18 detailed questions relating morphology to genetic background can be addressed. Recent studies using laser-assisted microdissection from tissue sections have provided insights into the biology of different diseases, eg, breast cancer, 19 malignant lymphoma, 20 thyroid cancer, 21 Barett’s adenocarcinoma. 22 In a previous study we have analyzed single tumor cells from a case of BCC. Irrespective of p53 immunoreactivity, we found the same p53 mutations in widely spread areas of the tumor as well as rare additional mutations not resulting in a clonal expansion. 23
It is known that a single dose of UVA, UVB, or γ irradiation will induce overexpression of p53 protein in human keratinocytes. 24 The genetic background underlying rare scattered p53-immunoreactive keratinocytes commonly found in normal skin is unclear. We have previously shown that the amount of p53-immunoreactive keratinocytes decreased 66% in skin covered by blue denim fabric compared to nonprotected skin after 2 months of natural sun exposure. 25 The nature of remaining p53-immunoreactive keratinocytes is unclear. In the present study immunohistochemistry, laser-assisted microdissection, gene amplification, and direct DNA sequencing were used to analyze biopsies from skin that had been subjected to ordinary daily summer sun and from adjacent skin that had been totally protected from solar radiation by blue denim fabric (SPF 1700). 26 The study reveals numerous persistent p53 mutations in normal human skin. By analyzing individual cells we found p53-mutated keratinocytes in both a clonal pattern and a dispersed pattern throughout the epidermis.
Materials and Methods
Volunteers and Sampling of Keratinocytes
Skin biopsies for analysis of human keratinocytes were obtained from the skin of volunteers previously characterized in the study by Berne and colleagues. 25 This study was approved by the local ethics committee and involved 11 healthy volunteers. Throughout 5 to 10 weeks in the summer, volunteers covered one area of 9 mm 2 on the dorsal side of one forearm with blue denim fabric (SPF1700). Twenty-four hours after the last exposure to sunlight punch biopsies were taken from both covered skin and an equivalent area of sun-exposed skin. In the present study we analyzed keratinocytes from sun-exposed (V1) and shielded skin (V2) from one 41-year-old male volunteer and shielded skin from two additional female volunteers (V3 and V4), age 36 and 66 years, respectively.
Biopsies were embedded in OCT (Miles Inc, USA) and snap-frozen at −70°C. Consecutive cryosections were cut with a thickness of 16 μm. Sections were mounted on thin glass slides and immediately covered with 10 mmol/L ethylenediaminetetraacetic acid, before incubation at 50°C for 30 minutes. Sections were stored at −20°C before immunohistochemistry.
Immunohistochemistry
Immunohistochemistry was performed essentially as previously described. 9 Expression of p53 protein was visualized using a monoclonal antibody recognizing nuclear protein p53 (DO-7, code M7001; DAKO, Glostrup, Denmark). Sections were rinsed in phosphate-buffered saline (PBS) for 10 minutes before incubation in 0.3% hydrogen peroxide for 30 minutes to exhaust endogenous peroxidase. After preincubation in 1% bovine serum albumin in PBS, monoclonal antibody DO-7 was applied at room temperature (dilution 1:200; incubation time 30 minutes). Biotinylated rabbit anti-mouse antibody (code E354, dilution 1:200, incubation time 30 minutes; DAKO) was used as secondary antibody. The immunoreaction was visualized using the avidin/biotin system, (code K355; dilution 1:200; incubation time 30 minutes; DAKO) with 0.004% hydrogen peroxide as substrate and diaminobenzidine as chromogen. Mayer’s hematoxylin was used for counterstaining. All solutions contained 1 mmol/L of ethylenediaminetetraacetic acid to inhibit endogenous nucleases. Stained slides were kept at −20°C before microdissection.
Microdissection
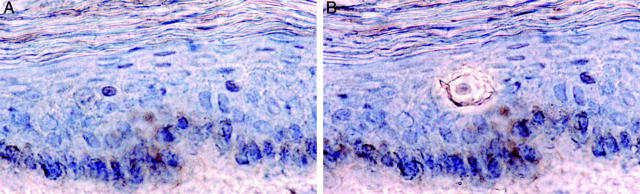
The dissections were performed using a PALM Robot-Microbeam laser microdissection system (P.A.L.M GmbH, Bernried, Germany) as previously described. 23 This system, which depends on a fine-focused laser beam, allows dissection of single cells from a tissue section mounted on a glass slide. 27 Single cells were isolated by eradication of surrounding cells with the laser (Figure 1) ▶ and then detached from the glass with the aid of a small glass capillary (Femtotips; Eppendorf) attached to the micromanipulator. The tip of the capillary, with the attached cell, was broken off against the bottom of a PCR tube containing 10 μl of PCR buffer (10 mmol/L Tris-HCl, pH 8.3, 50 mmol/L KCl) and subsequently covered with 50 μl of mineral oil. Capillaries were re-examined under the microscope to ensure that the cells had been transferred to the tubes.
Figure 1.
Immunohistochemically stained epidermis from skin protected from UV radiation during 2 summer months. Note rare p53-immunoreactive keratinocyte in epidermis before microdissection (A) and after isolation using laser-assisted microdissection (B). Original magnifications, ×400.
Amplification
PCR amplification of chromosomal DNA was performed essentially as previously described. 18,23 In brief, exons 4 to 9 of the human p53 gene were amplified in a multiplex/nested configuration. The outer multiplex amplification was performed in one tube with 12 primers located in intronic sequences flanking the six exons. The PCR mixture (20 μl) contained 20 mmol/L Tris-HCl, pH 8.75, 10 mmol/L (NH4) SO4, 2 mmol/L MgCl2, 10 mmol/L KCl, 0.1% Triton 20 X-100, 0.1 mg/ml bovine serum albumin, 0.2 mmol/L dNTPs, 0.25 μmol/L of each primer, and 1.8 U Pfu Turbo DNA polymerase (Stratagene, La Jolla, CA). The first four cycles in this outer PCR consisted of denaturation at 98°C for 0.25 minute; annealing at 50°C for 4 minutes, and extension at 72°C for 30 minutes. The following 26 cycles were performed with denaturation at 98°C for 0.25 minute, annealing at 50°C for 0.5 minute, and extension at 72°C for 2 minutes. After dilution (25-fold for exons 4, 5, 7, 8; and 100-fold for exons 6 and 9), inner region-specific amplifications for exons 4 to 9 were performed as previously described. 18 Each PCR was initiated by a 2-minute denaturation at 98°C and the final cycle was followed by a 10-minute extension at 72°C. For each set of 10 samples at least three negative controls without DNA were included.
DNA Sequencing
The sequence analysis was performed using the BigDye Terminator Cycle Sequencing kit (Perkin-Elmer Applied Biosystems, Inc., Foster City, CA). The DNA sequence was then determined by direct sequencing on the ABI 377 DNA sequencer (Perkin-Elmer-Applied Biosystems Inc.). Each sample was sequenced in both directions at least once and all exons containing mutations were resequenced using the product from a new inner PCR. All mutations displayed either 50 or 100% mutation signal in comparison to the wild-type sequence.
Results
In the previous study, 25 in which positive and negative cells were counted under the microscope there were 23 (V1), 20 (V2), 14 (V3), and 44 (V4) p53-immunoreactive keratinocytes per mm epidermis. The PALM laser microscope was used to retrieve a total of 172 single cells from immunohistochemically stained sections from four different biopsies. In 99 of 172 (58%) it was possible to amplify one or more p53 exons, suggesting loss of the single cell because of handling in 42% of the cases. In the 99 samples that contained a template for p53 amplification, a total of 494 of 594 (82%) possible exons were amplified (Table 1) ▶ . Twenty-six morphologically normal keratinocytes, of which 15 overexpressed p53 protein determined by immunohistochemistry, showed one or more mutations in the p53 gene. Eighteen of these mutated keratinocytes were clustered as an epidermal p53 clone in V2 (Figures 2 and 3) ▶ ▶ . A total of 14 different mutations were found (Table 2) ▶ . In shielded skin (V1) there were four mutations in 64 amplified exons (6.3%) and in nonshielded skin (V2, V3 and V4) there were five mutations in 134 exons (3.7%), two mutations in 102 exons (2%), and three mutations in 194 exons (1.5%).
Table 1.
Summary of Single Cells Collected from Four Biopsies Using Laser-Assisted Microdissection
| Biopsy | No. of cells collected | No. of cells with PCR product | No. of amplified exons (% of possible) | No. cells with p53 mutation | No. different mutations |
|---|---|---|---|---|---|
| V1 | 14 | 11 | 64 (97%) | 2 | 4 |
| V2 | 48 | 26 | 134 (83%) | 18 | 5 |
| V3 | 30 | 19 | 102 (89%) | 2 | 2 |
| V4 | 80 | 43 | 194 (75%) | 4 | 3 |
| Total | 172 | 99 | 494 (82%) | 26 | 14 |
Number of cells yielding PCR products and amount of amplified exons are shown. Percentage amplified exons in samples yielding a PCR product. The frequency of keratinocytes with a mutated p53 gene as well as number of different p53 gene mutations is also included in the table.
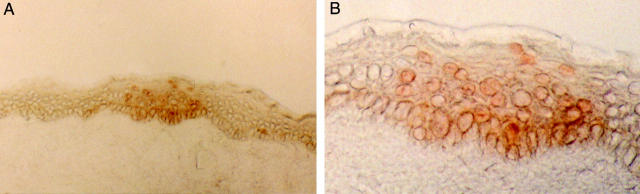
Figure 2.
Immunohistochemically stained (no counter stain) epidermis from B, revealing a small cluster of p53-immunoreactive keratinocytes found in normal skin after 2 months of total protection from UV radiation. Original low magnification, ×100 (A) and original high magnification ×400 (B). The epidermal p53 clone measures 10 to 15 cells in diameter and is enclosed within an area of 0.05 mm2.
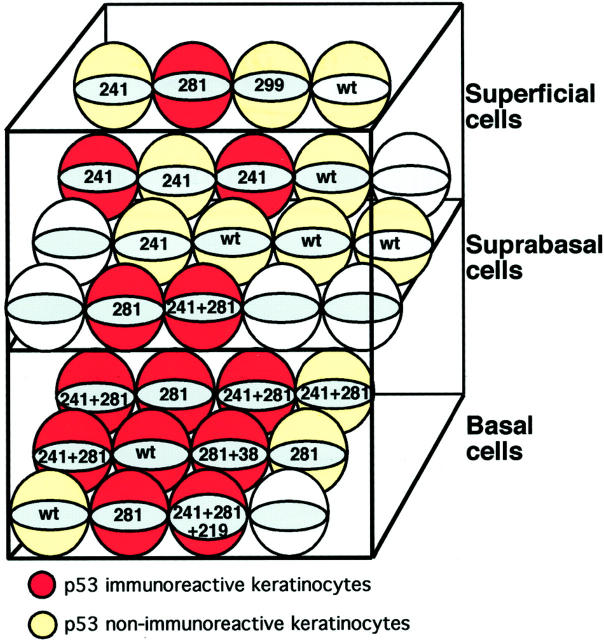
Figure 3.
Topography of the detected p53 mutations identified in the small cluster of p53-immunoreactive keratinocytes from Figure 2B ▶ is illustrated. Two frequent missense mutations (codon 241 and codon 218) were present in all three layers of epidermis. Both p53-immunoreactive (red) and nonimmunoreactive (yellow) keratinocytes show mutations.
Table 2.
Summary of the 14 Different Mutations Found in Normal Human Skin
| Biopsy | Mutated exon (codon) | Base change | Amino acid change | Consequence | Mutated cell type | p53 immuno-reactivity | Frequency of cells with mutation |
|---|---|---|---|---|---|---|---|
| V1 | 5 (148) | GAT-TAT | Asp-Tyr | Uncertain | b | + | 1 |
| 4 (82) | CCG-CTG* | Pro-Leu | Apoptosis | b | + | 1 | |
| 4 (104) | CAG-TAG* | Gln-stop | Uncertain | b | + | 1 | |
| 6 (213) | CGA-TGA* | Arg-stop | Uncertain | b | + | 1 | |
| V2 | 8 (281) | GAC-AAC* | Asp-Asn | DNA contact | b(9)/sb(2)/sf(1) | +(10)/−(2) | 12 |
| 7 (241) | TCC-TTC* | Ser-Phe | DNA contact | b(5)/sb(5)/sf(1) | +(7)/−(4) | 11 | |
| 6 (219) | CCC-TCC* | Pro-Ser | Phosphorylation | b | + | 1 | |
| 4 (38) | CAA-TAA* | Gln-stop | Transactivation | b | + | 1 | |
| 8 (299) | CTG-CTT | Leu-Leu | Silent | sf | − | 1 | |
| V3 | 4 (92) | CCC-CAC | Pro-His | Apoptosis | sb | − | 1 |
| 5 (153) | CCC-CAC | Pro-His | Backbone and side chain | b | + | 1 | |
| V4 | 4 (36) | CCG-CTG* | Pro-Leu | Transactivation | b | − | 1 |
| 4 (81) | ACA-ATA | Thr-Ile | Apoptosis | b | − | 1 | |
| 8 (275) | TGT-TAT | Cys-Tyr | DNA contact | sb/sf | − | 2 |
Mutated p53 exon, codon, base change, amino acid change, and possible functional consequence is displayed. *, represents mutations with typical UV signature (C-T or G-A transition at dipyrimidine site). Cell type (b, basal cell; sb, suprabasal cell; and sf, superficial cell). p53 immunoreactivity and frequency of cells containing one or more of the 14 mutations is also included.
Chronically Sun-Exposed Skin
Nonshielded Skin
Biopsy V1: In the sun-exposed skin, 64 of 66 possible exons were amplified from 11 of 14 collected keratinocytes. Four different mutations were found in two keratinocytes. One p53-immunoreactive, basal keratinocyte showed three different mutations (one missense and two stop mutations). All three mutations were C-T transitions at dipyrimidine sites. The fourth, missense mutation (G-T transversion) was also found in a p53-immunoreactive basal keratinocyte.
Shielded Skin
Biopsies V2, V3, and V4: Immunohistochemical staining revealed a small cluster of p53-immunoreactive keratinocytes suggestive of an epidermal p53 clone in V2 (Figure 2) ▶ . Forty-eight single cells were isolated from 12 consecutive immunostained cryosections. Immunoreactive and nonimmunoreactive basal, suprabasal, and superficial keratinocytes from this area were analyzed. Amplification yielded a product in 134 of 162 exons from 27 keratinocytes. Five different mutations were found. All p53-mutated cells were clustered within an area of 0.05 mm2. Mutations were found in both immunoreactive and nonimmunoreactive cells in all layers of epidermis (Figure 3) ▶ . Mutations were found in 12 of 13 p53-immunoreactive and 6 of 12 nonimmunoreactive keratinocytes. Two missense mutations were dominating (codon 241 and 281) and found in all but one of the mutated keratinocytes. Both mutations are within conserved regions of the p53 gene and code for amino acids involved in sequence-specific DNA binding. The remaining three mutations (one missense, one stop, and one silent mutation) were found only in solitary keratinocytes. Of the five different mutations found three were C-T transitions at dipyrimidine sites, one was a G-A at a dipyrimidine site and the odd, silent mutation in codon 299 was a G-T transversion.
Thirty single keratinocytes were microdissected and analyzed from V3. Exons (102 of 114) were successfully amplified in 19 cells. Two keratinocytes, one p53-immunoreactive basal cell and one nonimmunoreactive suprabasal cell showed a p53 mutation in codon 153 and codon 92, respectively. Both were missense mutations and C-A transversions. V3 also displayed heterozygosity at codon 72.
From 43 of 80 single-cell samples that contained DNA, 194 of 258 exons were amplified from V4. Four nonimmunoreactive keratinocytes were found to contain a missense mutation. Two basal cells showed each one mutation (codon 81 and codon 36). One suprabasal and one superficial keratinocyte located in close proximity shared a common mutation in codon 275. The types of mutations were one C-T transition at a dipyrimidine site and two transitions (C-T and G-A) at nondipyrimidine sites.
Discussion
The power of microdissection from tissue sections combined with fine tuned techniques for gene amplification and sequencing, can successfully be used to scrutinize links between gene function and morphology. The multistep theory of carcinogenesis has been proposed as a general model for environmental carcinogenesis, starting from clonal expansion of target cells through stages of precancerous lesions to eventual invasive and metastasizing cancer. Many studies have been designed to dissect the cellular and molecular mechanisms involved in this process. One obstacle has been cellular heterogeneity, a hallmark of cancer. The complexity of tumor tissue may result in masking of important genetic alterations in certain cells because of the existence of the variety of cell types present within a tumor. Laser-assisted isolation, which allows for micrometer-sized precision, and subsequent molecular characterization of defined individual cells provides a useful strategy. In this study we have used the refined technique to investigate the minimal template acquired from single cells isolated from normal skin. A major finding in this study was that p53 mutations are frequent in normal skin. Keratinocytes with mutations in the p53 gene were found dispersed in the epidermis, however, one cluster representing a clandestine clone of p53-mutated keratinocytes was also found (Figure 2) ▶ . The detected mutations, of which a majority displayed a typical UV signature, persisted despite 2 months of total sun protection.
The central role of the p53 pathway in human carcinogenesis is well accepted. 28 Although mutations in the p53 gene are up to date the most common alteration found in human cancer, 29,30 large differences exist depending on tumor type. In certain forms of cancer p53 alterations seem to be a rather late event during progression to a higher grade of malignancy, eg, colon cancer, 31 whereas in other forms of cancer p53 mutations seem to be early events, eg, skin cancer. 32 Most frequent alterations include point mutations resulting in amino acid substitutions and deletions that may lead to abrogation of p53-dependent pathways involved in important cellular functions, eg, cell-cycle control, DNA repair, differentiation, genomic plasticity, and apoptosis.
The consequence of a mutation in the p53 gene will depend not only on type of genetic alteration, but also on the target cell, which has been mutated. Mutations in terminally differentiating cells or cells that have lost their capacity to re-enter a more stem-cell-like phenotype, are probably less detrimental than mutations in stem cells. Such cells would be shed as a result of the constant turnover of epidermis regardless of defects in cell cycle control, apoptosis, and so forth. The turnover time in viable, normal epidermis is 26 to 42 days. 33 The type of mutations detected in morphologically normal keratinocytes from sun-protected skin, suggested that these were induced by UV radiation (Table 2) ▶ . If indeed these mutations were caused by UV irradiation, they were induced at least 2 months before biopsy. Alternatively, blue denim fabric (SPF 1700) is permeable for enough UV irradiation to induce mutations. We interpret our findings as consistent with mutations occurring in epidermal stem cells. The scattered p53-mutated keratinocytes found in different cell layers would thus represent offspring from one mutated stem cell. This becomes even more evident in the minute p53 clone found in one of the volunteers (V2), where a cluster of p53-immunoreactive keratinocytes appears to share the same typical UV signature mutations (Figure 3) ▶ . The findings here are well in concert with a mutated epidermal stem cell, which continues to give rise to transient amplifying and terminally differentiating cells within an area of approximately the size of one epidermal proliferative unit. 34,35
Epidermal p53 clones are frequently found in morphologically normal epidermis from chronically sun-exposed skin. 6,10,36,37 Confocal microscopy revealed that epidermal p53 clones originated from putative stem cell compartments, and it was hypothesized that these mutated keratinocytes were awaiting further genetic change before developing a malignant phenotype. 7 Although one study suggests a role for p53 clones as early precursors for BCC, 38 no genetic link has been detected between p53 clones and skin cancer. In addition, experimental studies have shown the early onset of epidermal p53 clones after chronic UV irradiation in histologically normal appearing mouse skin. 13 In a recent study using mice, it was also shown that p53 clones are indicators of tumor risk. 11 The clustering of cells in V2 with the same mutations strongly suggests that these cells belong to one clone as opposed to the scattered cells with different p53 mutations found elsewhere. Two missense mutations (codons 241and 281), found in all layers of the epidermis were comprised in the keratinocytes within the minute p53 clone. Both mutations show a typical UV signature and result in amino acid changes affecting p53 residues involved in direct DNA binding. Thus, prerequisites for a selective growth advantage of these keratinocytes exist. Because of lack of solar radiation the selective force may have been lost so that this p53 clone rather represents a regressed clone with a size of a normal epidermal proliferative unit. 34
In shielded skin containing scattered p53-immunoreactive cells, five different missense mutations were detected: codon 36, 81, 92, 153, and 275. The mutation at codon 275 was found in both one suprabasal and one superficial cell that were located close to each other, suggesting a common progenitor. It is unclear why some p53 mutations found in single, dispersed keratinocytes do not result in clonal expansion. Several of these mutations also showed a typical UV signature and also affected potentially important regions of the p53 gene. Although residual mutations are not infrequent in skin that has been shielded from the sun, nonshielded skin shows a notably higher mutation frequency [4 mutations in 64 amplified exons (6.3%) as compared to 10 mutations in 430 exons (2.3%)]. This data are well in agreement with the disappearance of mutated cells because of normal epidermal turnover.
Single-cell PCR has been used in various applications, 39 however, the role of allelic drop out (ADO) remains unclear. In a previous study the average ADO rate was 50% in single cells. 18 Interestingly, the epidermal p53 clone analyzed in the present study shows that the ADO rate in the basal cells (two of eight) is significantly lower compared to suprabasal (five of six) and superficial cells (three of three). Technical artifacts because of suboptimal amplification do not easily explain such a finding. Perhaps ADO is part of the terminal differentiation pathway so that cells continuously lose alleles during transit from a basal-proliferating cell to a highly differentiated keratinocyte in the upper, granular cell layer of the epidermis. Our data concerning clonal arrangement, p53 gene mutations, and ADO during differentiation in keratinocytes from normal human skin provide novel insights into the complexity of a self-renewing tissue. The presented strategy has a wide potential to dissect unique features in the different cell populations that are present in normal as well as diseased tissue.
Footnotes
Address reprint requests to Fredrik Pontén, Department of Genetics and Pathology, Rudbeck Laboratory, Uppsala University, S-75185 Sweden. E-mail: fredrik.ponten@genpat.uu.se.
Supported by the Swedish Cancer Foundation (grant no. 99 0373) and The Foundation for Strategic Research.
G. L. and A. P. contributed equally to this article.
References
- 1.Leigh IM, Newton Bishop JA, Kripke ML: Skin cancer. Tooze J eds. Cancer Surveys 1996, vol 26.:p 361 Cold Spring Harbor Laboratory Press, Cold Spring Harbor [Google Scholar]
- 2.Brash DE, Rudolph JA, Simon JA, Lin A, Mckenna GJ, Baden HP, Halperin AJ, Pontén J: A role for sunlight in skin cancer: UV-induced p53 mutations in squamous cell carcinoma. Proc Natl Acad Sci USA 1991, 88:10124-10128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ziegler A, Jonason AS, Simon J, Leffell D, Brash DE: Tumor suppressor gene mutations and photocarcinogenesis. Photochem Photobiol 1996, 63:432-435 [DOI] [PubMed] [Google Scholar]
- 4.Gailani MR, Stahle Backdahl M, Leffell DJ, Glynn M, Zaphiropoulos PG, Pressman C, Undén AB, Dean M, Brash DE, Bale AE, Toftgard R: The role of the human homologue of Drosophila patched in sporadic basal cell carcinomas. Nat Genet 1996, 14:78-81 [DOI] [PubMed] [Google Scholar]
- 5.Nilsson M, Unden AB, Krause D, Malmqwist U, Raza K, Zaphiropoulos PG, Toftgard R: Induction of basal cell carcinomas and trichoepitheliomas in mice overexpressing GLI-1. Proc Natl Acad Sci USA 2000, 97:3438-3443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pontén F, Berne B, Ren ZP, Nistér M, Pontén J: Ultraviolet light induces expression of p53 and p21 in human skin; effect of sunscreen and constitutive p21 expression in skin appendages. J Invest Dermatol 1995, 105:402-406 [DOI] [PubMed] [Google Scholar]
- 7.Jonason AS, Kunala S, Price GJ, Restifo RJ, Spinelli HM, Persing JA, Leffell DJ, Tarone RE, Brash DE: Frequent clones of p53-mutated keratinocytes in normal human skin. Proc Natl Acad Sci USA 1996, 93:14025-14029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ren ZP, Pontén F, Nistér M, Pontén J: Two distinct p53 immunohistochemical patterns in human squamous cell skin cancer, precursors and normal epidermis. Int J Cancer 1996, 66:174-182 [DOI] [PubMed] [Google Scholar]
- 9.Pontén F, Berg C, Ahmadian A, Ren ZP, Nistér M, Lundeberg J, Uhlén M, Pontén J: Molecular pathology in basal cell cancer with p53 as a genetic marker. Oncogene 1997, 15:1059-1067 [DOI] [PubMed] [Google Scholar]
- 10.Ren ZP, Hedrum A, Pontén F, Nistér M, Ahmadian A, Lundeberg J, Uhlén M, Pontén J: Human epidermal cancer and accompanying precursors have identical p53 mutations different from p53 mutations in adjacent areas of clonally expanded non-neoplastic keratinocytes. Oncogene 1996, 12:765-773 [PubMed] [Google Scholar]
- 11.Rebel H, Mosnier LO, Berg RJ, Westerman-de Vries A, van Steeg H, van Kranen HJ, de Gruijl FR: Early p53-positive foci as indicators of tumor risk in ultraviolet-exposed hairless mice: kinetics of induction, effects of DNA repair deficiency, and p53 heterozygosity. Cancer Res 2001, 61:977-983 [PubMed] [Google Scholar]
- 12.Ananthaswamy HN, Loughlin SM, Cox P, Evans RL, Ullrich SE, Kripke ML: Sunlight and skin cancer: inhibition of p53 mutations in UV-irradiated mouse skin by sunscreens. Nat Med 1997, 3:510-514 [DOI] [PubMed] [Google Scholar]
- 13.Berg RJ, van Kranen HJ, Rebel HG, de Vries A, van Vloten WA, Van Kreijl CF, van der Leun JC, de Gruijl FR: Early p53 alterations in mouse skin carcinogenesis by UVB radiation: immunohistochemical detection of mutant p53 protein in clusters of preneoplastic epidermal cells. Proc Natl Acad Sci USA 1996, 93:274-278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brash DE, Ziegler A, Jonason AS, Simon JA, Kunala S, Leffell DJ: Sunlight and sunburn in human skin cancer: p53, apoptosis, and tumor promotion. J Invest Dermatol Symp Proc 1996, 1:136-142 [PubMed] [Google Scholar]
- 15.Becker I, Becker KF, Rohrl MH, Minkus G, Schutze K, Hofler H: Single-cell mutation analysis of tumors from stained histologic slides. Lab Invest 1996, 75:801-807 [PubMed] [Google Scholar]
- 16.Bernsen MR, Dijkman HB, de Vries E, Figdor CG, Ruiter DJ, Adema GJ, Muijen GN: Identification of multiple mRNA and DNA sequences from small tissue samples isolated by laser-assisted microdissection. Lab Invest 1998, 78:1267-1273 [PubMed] [Google Scholar]
- 17.Schutze K, Lahr G: Identification of expressed genes by laser-mediated manipulation of single cells. Nature Biotechnol 1998, 16:737-742 [DOI] [PubMed] [Google Scholar]
- 18.Persson AE, Gao L, Williams C, Backvall H, Ponten J, Ponten F, Lundeberg J: Analysis of p53 mutations in single cells obtained from histological tissue sections. Anal Biochem 2000, 287:25-31 [DOI] [PubMed] [Google Scholar]
- 19.Lehmann U, Glockner S, Kleeberger W, von Wasielewski HF, Kreipe H: Detection of gene amplification in archival breast cancer specimens by laser-assisted microdissection and quantitative real-time polymerase chain reaction. Am J Pathol 2000, 156:1855-1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cerroni L, Arzberger E, Putz B, Hofler G, Metze D, Sander CA, Rose C, Wolf P, Rutten A, McNiff JM, Kerl H: Primary cutaneous follicle center cell lymphoma with follicular growth pattern. Blood 2000, 95:3922-3928 [PubMed] [Google Scholar]
- 21.Volante M, Papotti M, Roth J, Saremaslani P, Speel EJ, Lloyd RV, Carney JA, Heitz PU, Bussolati G, Komminoth P: Mixed medullary-follicular thyroid carcinoma. Molecular evidence for a dual origin of tumor components. Am J Pathol 1999, 155:1499-1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walch AK, Zitzelsberger HF, Bruch J, Keller G, Angermeier D, Aubele MM, Mueller J, Stein H, Braselmann H, Siewert JR, Hofler H, Werner M: Chromosomal imbalances in Barrett’s adenocarcinoma and the metaplasia-dysplasia-carcinoma sequence. Am J Pathol 2000, 156:555-566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pontén F, Williams C, Gao L, Ahmadian A, Nistér M, Lundeberg J, Pontén J, Uhlén M: Genomic analysis of single cells from human basal cell using laser-assisted capture microscopy. Mutat Res Genomics 1997, 382:45-55 [DOI] [PubMed] [Google Scholar]
- 24.Ponten F, Lindman H, Bostrom A, Berne B, Bergh J: Induction of p53 expression in skin by radiotherapy and UV radiation: a randomized study. J Natl Cancer Inst 2001, 93:128-133 [DOI] [PubMed] [Google Scholar]
- 25.Berne B, Ponten J, Ponten F: Decreased p53 expression in chronically sun-exposed human skin after topical photoprotection. Photodermatol Photoimmunol Photomed 1998, 14:148-153 [DOI] [PubMed] [Google Scholar]
- 26.Berne B, Fischer T: Protective effects of various types of clothes against UV radiation. Acta Derm Venereol 1980, 60:459-460 [PubMed] [Google Scholar]
- 27.Schütze K, Clement-Sengewald A: Catch and move—cut or fuse. Nature 1994, 368:667-669 [DOI] [PubMed] [Google Scholar]
- 28.Hussain SP, Harris CC: Molecular epidemiology of human cancer: contribution of mutation spectra studies of tumor suppressor genes. Cancer Res 1998, 58:4023-4037 [PubMed] [Google Scholar]
- 29.Hainaut P, Hernandez T, Robinson A, Rodriguez-Tome P, Flores T, Hollstein M, Harris CC, Montesano R: IARC Database of p53 gene mutations in human tumors and cell lines: updated compilation, revised formats and new visualisation tools. Nucleic Acids Res 1998, 26:205-213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levine AJ: p53, the cellular gatekeeper for growth and division. Cell 1997, 88:323-331 [DOI] [PubMed] [Google Scholar]
- 31.Fearon ER, Vogelstein B: A genetic model for colorectal tumorigenesis. Cell 1990, 61:759-767 [DOI] [PubMed] [Google Scholar]
- 32.Ziegler A, Jonason AS, Leffel DJ, Simon JA, Sharma HW, Kimmelman J, Remington L, Jacks T, Brash DE: Sunburn and p53 in the onset of skin cancer. Nature 1994, 372:773-776 [DOI] [PubMed] [Google Scholar]
- 33.Wilkinson R, Eblling FJG: Textbook of Dermatology, ed 5, vol 2. Edited by R Champion, JL Burton, FJG Ebling. Oxford, Blackwell Scientific Publications, 1992
- 34.Asplund A, Guo ZM, Hu XR, Wassberg C, Ponten J, Ponten F: Mosaic pattern of maternal and paternal keratinocyte clones in normal human epidermis revealed by analysis of X-chromosome inactivation. J Invest Dermatol 2001, 117:128-131 [DOI] [PubMed] [Google Scholar]
- 35.Potten CS: The epidermal proliferative unit: the possible role of the central basal cell. Cell Tissue Kinet 1974, 7:77-88 [DOI] [PubMed] [Google Scholar]
- 36.Brash DE, Ponten J: Skin precancer. Tooze J eds. Cancer Surveys. 1998, :pp 69-113 Cold Spring Harbor Laboratory Press, Cold Spring Harbor [PubMed] [Google Scholar]
- 37.Ren ZP, Ahmadian A, Pontén F, Nistér M, Berg C, Lundeberg J, Uhlén M, Pontén J: Benign clonal keratinocyte patches with p53 mutations show no genetic link to synchronous squamous cell precancer or cancer in human skin. Am J Pathol 1997, 150:1791-1803 [PMC free article] [PubMed] [Google Scholar]
- 38.Tabata H, Nagano T, Ray A, Flanagan N, Birch-MacHin M, Rees J: Low frequency of genetic change in p53 immunopositive clones in human epidermis. J Invest Dermatol 1999, 113:972-976 [DOI] [PubMed] [Google Scholar]
- 39.Hahn S, Zhong XY, Troeger C, Burgemeister R, Gloning K, Holzgreve W: Current applications of single-cell PCR. Cell Mol Life Sci 2000, 57:96-105 [DOI] [PMC free article] [PubMed] [Google Scholar]