Abstract
F10 and BL6 cells of B16 mouse melanoma cells are metastatic after intravenous injection, but only BL6 cells can metastasize to lungs after subcutaneous injection. Differences in gene expression between the two cell lines were examined, and a greater expression of the Sik-similar protein (Sik-SP) gene was found in BL6 cells. Structurally, Sik-SP belongs to the nucleolar Nop5/Sik family whose members play central roles in ribosome biogenesis; however, the function of Sik-SP has not been examined. Cytology with green fluorescent protein-fused proteins showed that Sik-SP was localized to the nucleolus. To examine whether Sik-SP is involved in ribosome biogenesis, two parameters were measured: magnitude of ribosomal RNA synthesis per nucleus and magnitude of protein production from the same amount of mRNA of an exogenous luciferase gene. Both values and, in addition, nucleolar size were larger in COS-7 monkey kidney cells overexpressing Sik-SP and BL6 cells than in mock-transfected COS-7 and F10 cells, respectively. Sik-SP seemed to promote ribosome biogenesis in the nucleolus. Furthermore, the expression of Sik-SP seemed to confer a greater cell growth response to serum, because such a response was greater in BL6 cells and F10 cells overexpressing Sik-SP than in untreated and mock-transfected F10 cells. Sik-SP may render melanoma cells more competent to survive through augmenting the activity of nucleolus.
It has long been known by pathologists that hypertrophy of the nucleolus is one of the most consistent cytological features of cancer cells, 1,2 and is closely related to the rapidity of cancer cell proliferation. 3,4 The nucleolus is a structural and functional unit in which ribosomal genes are located and transcribed, and where ribosomal RNA (rRNA) and ribosome synthesis occur. 5 Therefore, hypertrophy of the nucleolus is regarded as a state in which rRNA and ribosome synthesis has increased. A dividing cell is required to duplicate its protein complement before division. It is likely that the nucleolar function is continuously up-regulated in progressively proliferating cancer cells to respond to the increased demand for ribosome biogenesis. Recently, molecular basis for the nucleolar hypertrophy has become gradually known. In cancer cells possessing larger nucleoli and a shorter cell-doubling time, several nucleolar proteins are expressed more abundantly, including the RNA polymerase I upstream-binding factor, 3 topoisomerase I, 3 fibrillarin, 3 nucleolin, 6 and protein B23. 6 These proteins have important functions in rRNA processing and synthesis. 3,6 Concomitantly, rRNA synthetic activity is elevated in cancer cells with larger nucleoli. 3 In addition to tumor growth rate, nucleolar size can predict the metastatic behaviors of a tumor. In cutaneous 7,8 and uveal 9,10 melanomas, the mean nucleolar area in patients who died of metastasis was significantly larger than the mean in patients who did not develop metastasis. The increase in nucleolar size is believed to reflect the increase in nuclear and nucleolar activity during the process of malignant progression from poorly to highly metastatic melanoma cells. 7-11 However, the genetic alterations that might be implicated in the changes in nucleolar size and function during such processes have not been studied intensively.
To study the molecular mechanisms of melanoma metastasis, various types of mouse models have been developed. 12,13 Among these, the C57BL/6 mouse model using B16 melanoma cells is one of the most useful. 14 From B16 cells, various sublines have been established that display distinct metastatic phenotypes. 15,16 The F10 and BL6 sublines heavily metastasize to the lungs after intravenous injection. 15,16 In contrast, although BL6 cells are also metastatic after subcutaneous injection, F10 cells are only poorly so. 15,17 We have attempted to reveal the difference in gene expression between the two sublines by using the library subtraction method, and were able to isolate several genes that are up- or down-regulated in BL6 cells. 18-22 The differentially expressed genes we analyzed had relevance to the clinical phenotypes of invasion and metastasis. The connexin 26 19 and MLZE 22 genes were up-regulated in BL6 cells, and were intensively expressed in the invasive descent of skin melanoma lesions. The annexin VII gene was down-regulated in BL6 cells, and its expression in human melanoma was related to a better prognosis. 20 Genetic differences between F10 and BL6 cells seemed to reflect the process by which human melanomas progress to more malignant phenotypes. If there is a difference in nucleolar function between the two sublines, the comparative approach we used could provide genes that are responsible for changes in the nucleolus during the malignant progression process.
In the present study, we first compared the nucleolar size in F10 and BL6 cells, and found that BL6 cells contain nucleoli that are larger than those of F10 cells. We therefore assumed that BL6 cells express particular genes involved in nucleolar sizing at higher levels than F10 cells do. With this assumption in mind, we screened the cDNA clone pool obtained after the subtraction of a BL6 cDNA library with F10-derived mRNA. We isolated the Sik-similar protein (Sik-SP) gene, a member of the Nop5/Sik family that consists of highly conserved nucleolar proteins. 23 The Nop5/Sik family members bind box C/D small nucleolar RNA. 24-26 The resulting complexes, named small nucleolar ribonucleoproteins, play a pivotal role in rRNA processing necessary for ribosome assembly. 27,28 In yeast, at least two members, Nop56 and Nop58, are present, 29 and yeast deficient in either undergoes growth arrest because of defects in protein synthesis. 29-31 In mammalian cells, the presence of counterparts to the yeast Nop5/Sik family members has already been confirmed, 24,25,32 but their roles in nucleolar function are not yet well understood. Such is the case with Sik-SP: the cDNA sequence of Sik-SP is in the GenBank database, but no functional analysis has been done on this molecule yet. We assumed that Sik-SP might have a role similar to that of the yeast Nop5/Sik family members. In this study, we used transient expression experiments to show that Sik-SP is located primarily at the nucleolus, and increased the area of the nucleolus recognized by toluidine blue stain. Functional analysis indicated that Sik-SP increases rRNA synthesis and the translational efficiency of the exogenous luciferase (Luc) gene. In addition, we found that expression of a human Nop5/Sik family member correlated well with the metastatic phenotypes of clinical melanomas. The Nop5/Sik family members may therefore promote nucleolar function in metastatic melanoma cells.
Materials and Methods
Cell Lines and Culture
Mouse melanoma (B16, F10, BL6, and K1735-P 12 ) and melanocytic (melan-a 33 ) cells were kindly provided by Dr. I. J. Fidler (The University of Texas, Houston, TX) and Dr. D. C. Bennett (St. Georges Hospital Medical School, London, UK), respectively. NIH3T3 and Balb3T3 fibroblastic cells, MC/9 and P-815 mastocytoma cells, and COS-7 kidney cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA). All cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal calf serum (FCS). Two hundred nmol/L of 12-O-tetradecanoylphorbol-13-acetate was added to grow melan-a cells.
For the experiment of serum starvation and stimulation, F10 and BL6 cells (2 × 104) were plated onto 35- or 100-mm culture dishes with DMEM/10% FCS (day 0). On the next day (day 1), some dishes of F10 cells were used for transfection. On day 2, the medium was replaced with DMEM, and on day 4, the medium was replaced with DMEM/10% FCS. Cells were harvested by trypsin-ethylenediaminetetraacetic acid solution (Sigma, St. Louis, MO) from 35-mm dishes on every day from days 1 to 6, and from 100-mm dishes on each of days 7 and 8, and the cell number was counted. A portion of the cells was stained with trypan blue (Life Technologies, Inc., Gaithersburg, MD), and the proportion of dead cells was determined. It was always <0.1%. Experiments were performed in triplicate, and repeated three times. The cell-doubling time was determined by counting cells in triplicate samples at 24-, 48-, and 72-hour intervals according to the method described by Patterson. 34
Isolation of cDNA Clones
The method for constructing a subtracted cDNA library has been described previously. 19,21,35 After four rounds of subtraction, ∼1400 clones were rescued and subjected to Northern blot analysis with the RNA of F10 and BL6 cells. Clones of interest were sequenced. The DNA sequences were used to search the National Center for Biotechnology Information database using the BLASTN algorithm.
In Vitro Transcription and Translation
The TNT T7 quick-coupled transcription/translation system (Promega, Madison, WI) was used according to the manufacturer’s instructions.
Plasmid Construction
We previously described two modified pEGFP-C1 (Novagen, Madison, WI) vectors, termed pEGFP3B and pEGFP3. 36 Using clone 3-30 as a template, the polymerase chain reaction (PCR) was done with two sense primers (F-Asc-full and F-Asc-ΔN) containing an AscI site and two antisense primers (R-Not-full and R-Not-ΔC) containing a NotI site: F-Asc-full, 5′-CATGGCGCGCCGATGGAAGGCAAAATCAATAAACAG-3′; F-Asc-ΔN, 5′-CATGGCGCGCCGGAACAGGTAGTAGAAGAGGAGCCG-3′; R-Not-full, 5′-ATAGCGGCCGCTCACTAGCTCCCAACTGAATCGTAAGC-ATG-3′; R-Not-ΔC, 5′-ATAGCGGCCGCTCACTATTCCTCCGCCTCCTCCTCCTCCAT-3′. Primer sets and the amplified cDNA regions of Sik-SP are: F-Asc-full and R-Not-full, the full-length coding region (amino acid residues 1 to 474, Sik-SP); F-Asc-full and R-Not-ΔC, the C-terminal deletion form (amino acid residues 1 to 418, Sik-SPΔC); F-Asc-ΔN and R-Not-full, the N-terminal deletion form (amino acid residues 419 to 474, Sik-SPΔN). The PCR-amplified DNA fragments were digested with AscI and NotI, and then ligated into the pEGFP3B vector by way of the AscI-NotI sites (pEGFP3B-Sik-SP, pEGFP3B-Sik-SPΔC, and pEGFP3B-Sik-SPΔN). The plasmid constructs encode various forms of Sik-SP fused to the 3′ end of the green fluorescent protein (GFP). The pEGFP3 vector contains an in-frame stop codon between the GFP sequence and the AscI site, and was used as a control for the pEGFP3B vector. The pcDNA3 expression vector (Invitrogen, San Diego, CA) was modified to contain an AscI site as described previously, 21 and the AscI-NotI cDNA fragment of wild-type Sik-SP was subcloned from the pEGFP3B vector into the modified pcDNA3 vector (pcDNA3-Sik-SP).
Plasmid Transfection
The Fugene 6 transfection reagent (Roche Diagnostics Co., Indianapolis, IN) was used according to the manufacturer’s instructions. A 100-μl DMEM mixture containing 3 μl of Fugene 6 and 1 μg of plasmid DNA was added to COS-7 or F10 cells that were grown in 35-mm culture dishes at a confluency of 70%, unless otherwise indicated. For co-transfection, 1 μg of each of plasmid constructs was added to the mixture. When 100-mm culture dishes were used, the quantity of each component was scaled up. Subsequently, cells were cultured in DMEM/10% FCS.
Histocytological Examination
Fibronectin (Sigma)-coated coverslips (25 ng/cm2) were placed at the bottom of 35-mm culture dishes, and cells were plated onto them. The next day, cells were transfected with pEGFP3B, pEGFP3B-Sik-SP, pEGFP3B-Sik-SPΔC, or pEGFP-3B-Sik-SPΔN. After the cells were cultured for 24 hours, the coverslips were fixed in 3% glutaraldehyde, and mounted in an aqueous medium. Cells were observed through an Axioplan2 microscope (Carl Zeiss, OberKochen, Germany) equipped with epifluorescence and a charge-coupled device camera (Power HAD; Sony, Tokyo, Japan), and images were created by IPLab Spectrum (Scanalytics Inc., Fairfax, VA) software on a Macintosh computer. For subsequent cell staining, the mounting medium was washed out with distilled water.
To stain with toluidine blue, F10 and BL6 cells were cultured on fibronectin-coated coverslips (25 ng/cm2) for 3 hours, and fixed in 3% glutaraldehyde. Sections of paraffin-embedded tissues were deparaffinized with xylene, and hydrated in ethanol series. Cells and sections were stained for 10 minutes with 0.05% toluidine blue in a buffer (pH 4.1) containing 60 mmol/L citric acid and 80 mmol/L sodium phosphate. The samples were washed in water, dehydrated in ethanol, and mounted in Permount medium (Fisher Scientific, Fair Lawn, NJ). Stained cells were observed with an image processing system, which consisted of a charge-coupled device camera (HC-2500; Fuji Photo Film Co., Tokyo, Japan), a microscope (AX80; Olympus, Tokyo, Japan), a color monitor, a Macintosh computer (Power Mac G4), image processing software (Adobe Photoshop Ver 5.0; Adobe System Inc., Mountain View, CA), and a digital cursor. Images were created on the computer monitor screen at a magnification of ×7250. This magnification was achieved by an intermediate zoom magnification system (Provis, Olympus). On the monitor, the nucleolar diameters were between 7 and 38 mm. A cursor was used manually to delineate the margins of the toluidine blue-stained nucleoli in at least 30 fields for each sample. The area occupied by the delineation was expressed in a pixel unit, and converted into a μm 2 unit by the magnification rate on the monitor. The mean nucleolar area was calculated by observing 200 nucleoli for F10 and BL6 cells, and 50 nucleoli for COS-7 and human melanoma cells.
Luciferase Assay
The pRL-SV40, pRL-CMV, and pRL-null vectors were purchased from Promega. The XhoI-EcoRI DNA fragment including the modified chicken β-actin promoter was subcloned from the pAG-2 vector 37,38 into the pRL-null by way of the XhoI-EcoRI sites (pRL-actin). The three vectors express the Renilla Luc gene under the control of the simian virus 40 enhancer/early promoter, cytomegalovirus enhancer/immediate early promoter, and chicken β-actin promoter, respectively. Cells were transfected with each of the three vectors. In addition, cells were co-transfected with pcDNA3 or pcDNA3-Sik-SP. After the cells were cultured for 24 hours, the Renilla Luc activity was measured by using the Dual luciferase assay system (Promega) according to the manufacturer’s instructions. The level of Luc activity was normalized against the amount of Luc plasmid DNA introduced by transfection, as determined by Southern blot analysis. After the cells were extracted in the passive lysis buffer of the Luc assay kit, a half volume of the lysate was subjected to phenol/chloroform extraction, followed by ethanol precipitation. The DNA obtained was blotted with an [α-32P] dCTP-labeled NheI-XbaI DNA fragment of the pRL-null vector, which contained the Renilla Luc gene. Hybridized bands were quantified by measuring the peak area of the densitometric tracing (BAS 2000 system, Fuji Photo Film Co.). Values of Luc activity were divided by values of the band intensity, and expressed as relative Luc activity in arbitrary units. Experiments were performed in triplicate, and repeated three times.
Evaluation of in Vitro rRNA Synthesis
In vitro rRNA synthetic activity was evaluated essentially as described previously. 39,40 On the day before the assay, some cells were transfected in 100-mm culture dishes with the empty pcDNA3 vector or with pcDNA3 encoding the Sik-SP gene. On the next day, the cells (5 × 107) were washed with phosphate-buffered saline and resuspended in 0.25 mol/L sucrose containing 19 mmol/L Tris-HCl (pH 7.4), 10 mmol/L MgCl2, and 0.5% Triton X-100. After homogenization, cells were centrifuged at 2500 × g for 10 minutes. Pellets were washed twice in 0.25 mol/L sucrose containing 10 mmol/L Tris-HCl (pH 7.4) and 10 mmol/L MgCl2, and resuspended in 1.0 ml of 0.25 mol/L sucrose containing 1 mmol/L MgCl2. This suspension was considered to contain purified nuclei at concentration of 5 × 10 7 nuclei/ml, given no leakage. RNA synthetic activity in purified nuclei was measured in the absence or presence (5 μg/ml) of α-amanitin (Roche Diagnostics Co.), an inhibitor of RNA polymerases II and III. 41,42 Three hundred μl of the assay mixture contained 50 μmol of Tris-HCl buffer (pH 8.0); 0.2 μmol of MnCl2; 100 μmol of (NH4)2SO4; 0.9 μmol each of CTP, GTP, and ATP; 18 μmol of nonradioactive UTP; 0.05 μmol of [3H]UTP; and 100 μl of nuclear suspension including 5 × 10 6 nuclei. The mixture was incubated at 37°C for 10 minutes. An equal volume of 10% (w/v) trichloroacetic acid was added and the precipitate was collected on glass-fiber filters (Whatman GF/C; Whatman, Clifton, NJ). The filters were washed twice with 5% trichloroacetic acid, twice with 0.1 mol/L HCl, and once with 100% ethanol. After baking at 80°C for 3 minutes, the radioactivity of the filters was measured in a liquid scintillation counter. Experiments were performed in triplicate, and repeated three times.
RNA and DNA Analysis
Northern and Southern blotting and hybridization were performed using standard methods. Relative signal intensity was calculated with the BAS 2000 system (Fuji Photo Film Co.). Total RNA was prepared from cultured cells and mouse tissues using 3 mol/L lithium chloride and 6 mol/L urea. 43 To extract total RNA from paraffin-embedded tissues, the Paraffin block RNA isolation kit (Ambion, Austin, TX) was used. One sheet of section was cut at a thickness of 20 μm and processed according to the manufacturer’s instruction. Reverse transcriptase (RT)-PCR was performed as described, 35 using a half volume of the RNA yields and two sets of primers: F-actin, 5′-AACCGCGAGAAGATGACCCAGAT-3′; R-actin, 5′-GAAGGAAGGTTGGAAGAGAGCCT-3′; and F-hNop58, 5′-GTTATGATGCTTTTGGTGAG-3′; R-hNop58, 5′-TGCTGGTACATGGTTCTTCCTCAG-3′. The PCR profile with the β-actin primers consisted of 20 cycles of denaturation at 94°C for 30 seconds, reannealing at 51°C for 30 seconds, and polymerization at 72°C for 1 minute. The PCR profile with the hNop58 primers consisted of 33 cycles of the same settings. For probe preparation, PCR-amplified DNA fragments derived from mRNA of BL6 cells were subcloned into the pBluescript vector (Stratagene, La Jolla, CA). After amplification, DNA inserts of clone 3-30 (Sik-SP), β-actin, and hNop58 were excised from the vectors, and labeled with [α-32P] dCTP by the random labeling method.
Human Surgical Samples
Melanoma samples were obtained from 15 patients with skin melanoma. The patients underwent surgical removal of the melanoma at the time of diagnosis and were followed-up for more than 5 years at the Osaka University Hospital. During the follow-up, lymph nodes with or without metastasis were excised from the patients. Several lesions with intradermal nevus, a benign melanocytic disease, were excised from individual patients in the same hospital. All patients were informed of and consented to this study. Surgical samples were fixed in 4% paraformaldehyde, and embedded in paraffin. The sections were used for diagnosis, nucleolar size measurement, and RT-PCR analysis.
Statistical Analysis
The t-test was performed for analysis of nucleolar area and Luc activity data, and the chi-square test was performed for analysis of positivity ratio data of gene expression using the StatView (Abacus Concepts Inc., Cary, NC) software on a Macintosh computer. Data are given as mean ± SD. A P value <0.05 was considered to be significant.
Results
Nucleoar Size of F10 and BL6 Cells
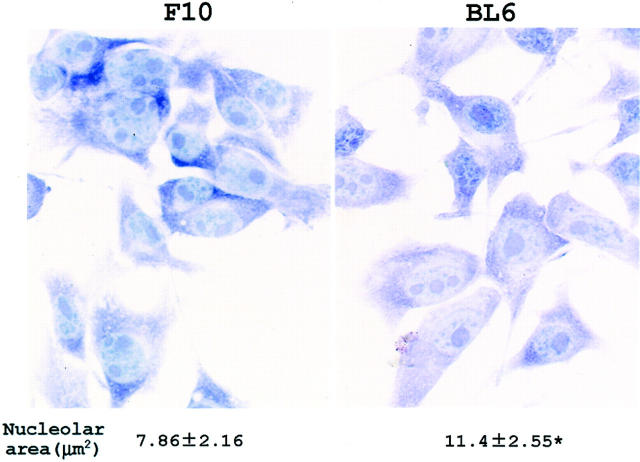
To evaluate the nucleolar size, F10 and BL6 cells were stained with toluidine blue (Figure 1) ▶ . Nucleoli were often detected as separate objects in single nuclei, but the margin of each nucleolus was clearly defined. In cells with multiple nucleoli, the nucleolar area was measured by summing the entire toluidine blue-stained area present in a single nucleus. The mean nucleolar area was 1.5-fold larger in BL6 cells than in F10 cells (Figure 1) ▶ .
Figure 1.
Nucleolar size of F10 and BL6 cells. Cells were plated onto coverslips, incubated for 3 hours, and stained with toluidine blue. The mean nucleolar size is shown under the cytological images. *, P < 0.01 by t-test when compared with the value of F10 cells. Original magnification, ×400.
Sik-SP Is Transcriptionally Up-Regulated in BL6 Cells
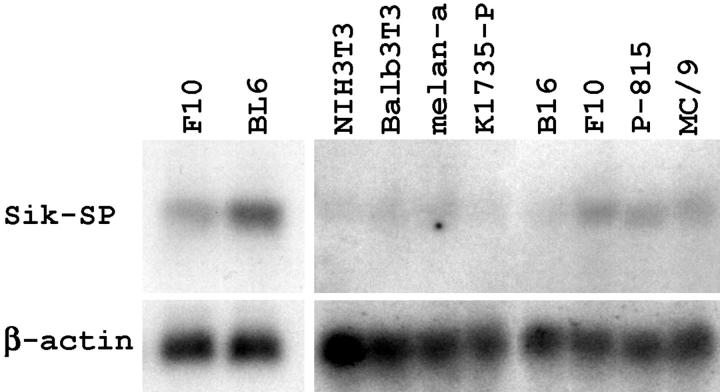
We previously constructed a subtracted cDNA library by removing clones complementary to the mRNA of F10 cells from a BL6 cDNA library. 18-22 After four rounds of subtraction, ∼1400 clones were recovered. Blots of total RNA from F10 and BL6 cells were hybridized with each clone sequentially. One clone, 3-30, was found to hybridize to a single 2.5-kb transcript. As shown in Figure 2 ▶ , this transcript was expressed at low levels in F10 cells, but its levels were fivefold higher in BL6 cells. Clone 3-30 was sequenced and found to carry a 1798-bp-long cDNA insert. Searches of the National Center for Biotechnology Information database using the BLASTN program revealed that clone 3-30 encompassed a large part of the sequence designated Sik-SP (accession no. AF053232). Structurally, Sik-SP belongs to the Nop5/Sik family, 23 but no functional analysis has been reported yet. We screened a BL6 cDNA library by using clone 3-30 as a probe, and obtained five positive clones that contained cDNA inserts as long as clone 3-30. Sequencing revealed that all five clones encoded Sik-SP. When clone 3-30 was transcribed and translated in the cell lysate of reticulocytes, the translation product was detected as a band of 55 kd (data not shown), consistent with the length of the open reading frame of the Sik-SP gene.
Figure 2.
Expression of Sik-SP mRNA in various mouse cells. Ten μg of total RNA from the indicated cells was blotted with the clone 3-30 probe (top). After stripping, the blots were hybridized with the β-actin probe to indicate the amount of RNA loaded per lane.
Northern blots of mouse normal tissues were probed with clone 3-30. For comparison, the amount of total RNA loaded per lane was matched with that in the blot of F10 and BL6 cells used in Figure 2 ▶ . The 3-30 probe did not hybridize to any transcripts from normal mouse tissues, including lung, intestine, liver, kidney, spleen, thymus, skeletal muscle, heart, and brain (data not shown). In contrast, hybridized bands were detected faintly in all mouse cell lines examined (Figure 2) ▶ . The band intensity in nontumor cells, such as NIH3T3, Balb3T3, and melan-a, and the intensity in K1735-P and B16 melanoma cells were weaker than that in F10 cells. By contrast, the band intensity in P-815 and MC/9 mastocytoma cells was similar to that in F10 cells. The four types of melanoma and mastocytoma cells examined are much less metastatic than BL6 cells. 12,13 Expression levels for the Sik-SP gene seemed to increase in some tumor cells, especially in highly metastatic BL6 cells.
Nucleolar Localization and Sizing of Sik-SP
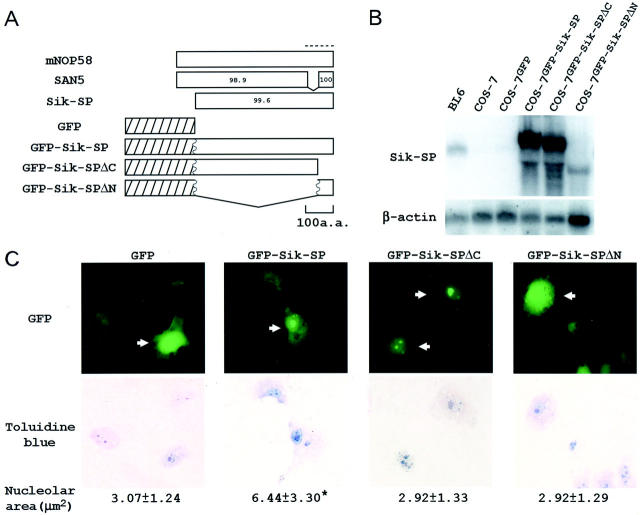
Recently, SAN5, another mouse member of the Nop5/Sik family, was shown to locate to the nucleolus. 44 To learn the subcellular localization of Sik-SP, COS-7 cells were transfected with a plasmid encoding the GFP-tagged form of Sik-SP. Because an abundance of lysine residues in the C-terminal region is characteristic of the Nop5/Sik family members, 23-25,29,30 we wished to know whether the C-terminal region is important for the localization of Sik-SP. Therefore, a similar transfection was done with plasmids encoding GFP-tagged forms of the two deletion mutants (Sik-SPΔC and Sik-SPΔN). Sik-SPΔN consists of 55 C-terminal amino acid residues of Sik-SP, while these same 55 amino acid residues are deleted in Sik-SPΔC (Figure 3A) ▶ . Expression of the introduced genes was confirmed by the Northern blots with the clone 3-30 probe (Figure 3B) ▶ . No transcript was detected in untreated COS-7 cells or in COS-7 cells transfected with the GFP gene alone. On the other hand, one transcript hybridized heavily in GFP-tagged Sik-SP-transfected cells. The larger mobility size of this transcript as compared to the Sik-SP transcript of BL6 cells is consistent with the message length for the gene transfected. Similarly, in GFP-tagged Sik-SPΔC and Sik-SPΔN-transfected cells, two transcripts of different length were detected at the expected mobility sizes. GFP-tagged forms of Sik-SP seemed to be transcribed efficiently in COS-7 cells.
Figure 3.
Nucleolar localization of GFP-tagged Sik-SP. A: Schematic representation of the GFP-tagged forms of wild-type Sik-SP and two deletion mutants. Top: The three open bars show the amino acid structure of mNop58, SAN5, and Sik-SP. The numbers in the bars indicate the similarity (%) to the corresponding regions of mNop58. Bottom: The three bars show the amino acid structure of GFP-tagged Sik-SP, Sik-SPΔC, and Sik-SPΔN. Shaded and open bars correspond to the regions of GFP and Sik-SP, respectively. Wavy lines indicate the fusion site of GFP to various forms of Sik-SP. A dotted line indicates the C-terminal lysine-rich region. B: Detection of the transcripts for various forms of GFP-tagged Sik-SP in COS-7 cells. COS-7 cells were transfected with pEGFP3B (COS-7GFP), pEGFP3B-Sik-SP (COS-7GFP-Sik-SP), pEGFP3B-Sik-SPΔC (COS-7GFP-Sik-SPΔC), and pEGFP3B-Sik-SPΔN (COS-7GFP-Sik-SPΔN). After the cells were cultured for 24 hours, total RNA (10 μg) was extracted from each cell, and blotted with clone 3-30. Total RNA (10 μg) of BL6 and untreated COS-7 cells were blotted as a control. After stripping, the blot was hybridized with the β-actin probe to indicate the amount of RNA loaded per lane. C: Nucleolar size of COS-7 cells expressing various forms of GFP-tagged Sik-SP in COS-7 cells. COS-7 cells were transfected as described in B, and observed with a microscope equipped with epifluorescence (top). It was easy to distinguish COS-7 cells containing GFP fluorescence (indicated by arrows) from COS-7 cells not containing it. After images were created, the cells were stained with toluidine blue (bottom), and their nucleolar area was measured (at the bottom). *, P < 0.01 by t-test when compared with the values of other types of COS-7 cells. Original magnification, ×400.
In COS-7 cells, GFP alone accumulated evenly in the cytoplasm and nucleus (Figure 3C) ▶ . GFP-tagged Sik-SP and GFP-tagged Sik-SPΔC showed a well-concentrated localization within the nucleus (Figure 3C) ▶ . We stained the cells with toluidine blue, and found that both GFP-tagged proteins localized to the nucleolus. In contrast, GFP-tagged Sik-SPΔN was not concentrated at the nucleolus, but was found in both the cytoplasm and nucleus (Figure 3C) ▶ . A similar subcellular localization was observed in F10 and BL6 cells after transfection with the GFP-tagged forms (data not shown). These results indicated that Sik-SP is localized to the nucleolus, and that the N-terminal region is required for its nucleolar localization, but the C-terminal region is not.
While observing cells, we noticed that nucleoli expressing GFP-tagged Sik-SP were larger than nucleoli not expressing it. We compared the nucleolar area in COS-7 cells expressing various GFP-tagged forms of Sik-SP. After the transfection with each plasmid construct, COS-7 cells containing GFP fluorescence were stored in an image processing system. Then, the cells were stained with toluidine blue, and the stored cells were identified on the computer monitor screen. The nucleolar area of cells expressing GFP-tagged Sik-SP was larger than that of cells expressing GFP alone and GFP-tagged Sik-SPΔN (Figure 3C) ▶ . In addition, GFP-tagged Sik-SPΔC did not increase the nucleolar area despite its nucleolar localization. Sik-SP seemed to directly affect the nucleolar size when overexpressed.
Increased Nucleolar Function in Sik-SP-Transfected COS-7 Cells
The Nop5/Sik family members are essential for ribosome biogenesis in yeast, 29,30 whereas their roles are not defined precisely in mammalian cells. We examined whether Sik-SP was involved in ribosome biogenesis in COS-7 cells by the following two experiments. For fear that GFP might cause nonspecific effects, COS-7 cells were transfected with the empty pcDNA3 vector (COS-7pcDNA3), or pcDNA3 encoding the Sik-SP gene (COS-7Sik-SP).
Increased rRNA Biogenesis
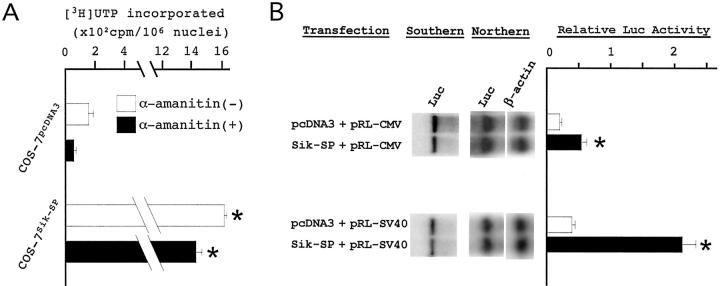
Twenty-four hours after the transfection, we purified nuclei from COS-7pcDNA3 and COS-7Sik-SP cells, and examined the amounts of transcript that was newly synthesized in the nuclei in vitro during a given time period. The amount was expressed as the value of [3H]UTP incorporated into already initiated and elongating transcripts. COS-7Sik-SP cells showed a remarkably higher value of [3H]UTP incorporation than COS-7pcDNA3 cells in the absence of α-amanitin (Figure 4A) ▶ . The presence of α-amanitin reduced the value of [3H]UTP incorporation comparably in both cells. Because α-amanitin is an inhibitor for RNA polymerases II and III, 41,42 the results indicated that COS-7Sik-SP cells have higher levels of rRNA biogenesis than COS-7pcDNA3 cells, while the synthesis of other types of RNA was comparable in both cells.
Figure 4.
Sik-SP up-regulates nucleolar function. A: In vitro RNA synthetic activity. Nuclei were purified from COS-7pcDNA3 and COS-7Sik-SP cells, and examined for RNA synthetic activity. The amount of [3H]UTP incorporated into already initiated and elongating transcripts was measured in the absence (−) or presence (+) of α-amanitin. The data represent the mean ± SD of two experiments. *, P < 0.01 by t-test when compared with the two values of COS-7pcDNA3 cells. B: Evaluation of translational efficiency of the exogenous Luc gene. COS-7 cells were transfected with pcDNA3 or pcDNA3-Sik-SP together with pRL-CMV or pRL-SV40. After the cells were cultured for 24 hours, they were examined for Luc gene dosage (Southern), Luc gene expression (Northern), and Luc activity (bar graph). Relative Luc activity was calculated as described in Materials and Methods. *, P < 0.01 by t-test when compared with the value of COS-7pcDNA3 cells in each Luc construct. To quantify the Luc gene expression, total RNA (5 μg) was blotted with the Luc probe, and after stripping, hybridized again with the β-actin probe (Northern). Signal intensity was quantified by the densitometric method.
Translational Rather than Transcriptional Up-Regulation of the Exogenous Luc Gene
Next, we compared translational levels of exogenous Luc gene in COS-7pcDNA3 and COS-7Sik-SP cells. The pRL-CMV or pRL-SV40 vector was transfected into COS-7 cells along with pcDNA3-Sik-SP or the empty pcDNA3 vector. Twenty-four hours after the transfection, the cells were harvested and divided into three aliquots to examine gene dosage, transcriptional levels, and translational levels of the Luc gene. To measure gene dosage and transcriptional levels of the Luc gene, DNA and total RNA were extracted from the cell pellets and hybridized with the Luc probe (Figure 4B) ▶ . Translational levels of the Luc gene were evaluated by measuring Luc activity. For the comparison between COS-7pcDNA3 and COS-7Sik-SP cells, the values for Luc activity were normalized to the amount of Luc plasmid DNA introduced into each cell. The normalized values were expressed as relative Luc activity. COS-7Sik-SP cells showed significantly higher levels of relative Luc activity than COS-7pcDNA3 cells (Figure 4B) ▶ . On the other hand, mRNA expression levels for the Luc gene were comparable in both cells (Figure 4B ▶ , Northern). Sik-SP seemed to up-regulate translation rather than transcription of the exogenous Luc gene in COS-7 cells.
Nucleolar Function in F10 and BL6 Cells
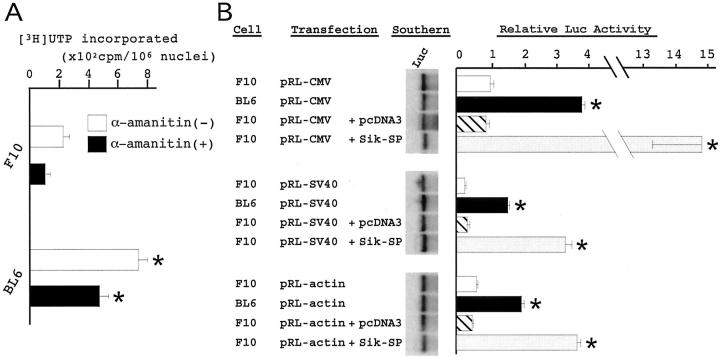
RNA synthetic activity was evaluated in the nuclei isolated from F10 and BL6 cells (Figure 5A) ▶ . BL6 cells showed threefold higher levels of RNA synthesis than F10 cells in the absence of α-amanitin. The presence of α-amanitin reduced the levels of RNA synthesis in both cells. Although the magnitude of reduction by α-amanitin was slightly larger in BL6 cells, BL6 cells showed fivefold higher levels for RNA synthesis than F10 cells even in the presence of α-amanitin. This result indicated that BL6 cells synthesize rRNA at higher levels, in preference to the other types of RNA.
Figure 5.
Nucleolar function of F10 and BL6 cells. A: In vitro RNA synthetic activity. Nuclei were purified from F10 and BL6 cells, and examined for RNA synthetic activity. The amount of [3H]UTP incorporated into already initiated and elongating transcripts was measured in the absence (−) or presence (+) of α-amanitin. The data represent the mean ± SD of two experiments. *, P < 0.01 by t-test when compared with the value of F10 cells in each assay condition. B: Evaluation of the translational efficiency of the exogenous Luc gene. F10 and BL6 cells were transfected with the pRL-CMV, pRL-SV40, or pRL-actin. F10 cells were also transfected with pcDNA3 or pcDNA3-Sik-SP together with the pRL-CMV, pRL-SV40, or pRL-actin. After the cells were cultured for 24 hours, they were examined for Luc gene dosage (Southern), and Luc activity (bar graph). Relative Luc activity was calculated as described in the Materials and Methods. *, P < 0.01 by t-test when compared with the values of F10 and pcDNA3-co-transfected F10 cells in each Luc construct.
The pRL-CMV, pRL-SV40, and pRL-actin vectors express the Luc gene under the control of distinct cis-acting elements, and were transfected into F10 and BL6 cells. In addition to the Luc constructs, the empty pcDNA3 vector or pcDNA3-Sik-SP was co-expressed in F10 cells. Twenty-four hours after the transfection, cells were harvested and examined for gene dosage, transcriptional levels, and translational levels of the Luc gene according to the same procedure as used for COS-7 cells. In every Luc plasmid construct, the four types of cells exhibited similar profiles of relative Luc activity (Figure 5B) ▶ : the Luc activity of pcDNA3-co-transfected F10 cells was comparable to that of F10 cells, whereas BL6 and Sik-SP-co-transfected F10 cells showed several-fold to 10-fold higher Luc activity than the other two types of F10 cells. Because there was no difference in the mRNA levels of the Luc gene among the four types of cells (data not shown), these results indicate that the Luc message was translated more efficiently in BL6 and Sik-SP-co-transfected F10 cells. In addition, they suggested that the difference in the translational efficiency between F10 and BL6 cells is attributable, at least in part, to the expression level of Sik-SP.
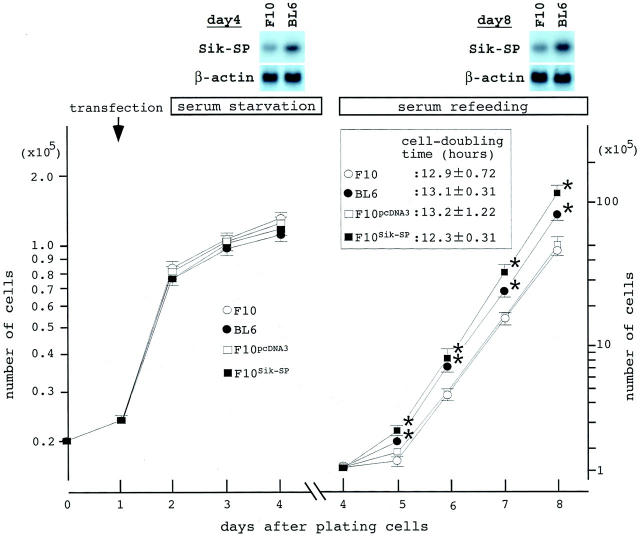
Previous reports indicated a clear relationship between nucleolar size and cell proliferation rate. 3,4 However, there was no difference in the mean cell-doubling time between F10 and BL6 cells (Figure 6) ▶ . We examined the effects of Sik-SP on cell growth recovery after serum-starved culture conditions. F10 and BL6 cells were plated onto culture dishes (day 0), serum-starved for 2 days from day 2, and refed with serum on and after day 4. Some dishes of F10 cells were used for transfection with an empty pcDNA3 vector (F10pcDNA3), or pcDNA3-Sik-SP (F10Sik-SP) on day 1. F10 and BL6 cells ceased to grow actively toward the end of starvation, as did F10pcDNA3 and F10Sik-SP cells. After 2 days of the serum refeeding, all of the cell types started to grow exponentially at a similar rate of cell-doubling. The number of BL6 and F10Sik-SP cells was noticeably larger than that of F10 and F10pcDNA3 cells on every day after cells were refed with serum. This was primarily attributable to the difference in the cell number on day 5, the day after the serum refeeding. BL6 and F10Sik-SP cells seemed to have a greater growth response to serum stimulation than F10 and F10pcDNA3 cells. Expression of Sik-SP mRNA was examined in F10 and BL6 cells on days 4 and 8 (Figure 6 ▶ , top). There was no significant change in the expression levels on either day when compared with the levels under the standard culture conditions (see Figure 2 ▶ ).
Figure 6.
Growth recovery of melanoma cells from serum-starvation. F10 and BL6 cells (2 × 104) were plated onto 35- or 100-mm culture dishes with DMEM/10% FCS (day 0). On the next day (day 1), some dishes of F10 cells were used for transfection. On day 2, the medium was replaced with DMEM, and on day 4, the medium was replaced with DMEM/10% FCS. The number of cells was counted every day, and dotted in a semilogarithmic graph. A representative result is shown. The cell-doubling time was calculated from the cell numbers on days 5, 6, 7, and 8. *, P < 0.01 by t-test when compared with the values of F10 and F10pcDNA3 cells. Top: Representative results of Northern blots of F10 and BL6 cells on days 4 and 8. RNA blots (10 μg) from each cell were hybridized with the clone 3-30 probe, and after stripping, hybridized again with the β-actin probe.
Nop58 Expression in Human Malignant Melanoma
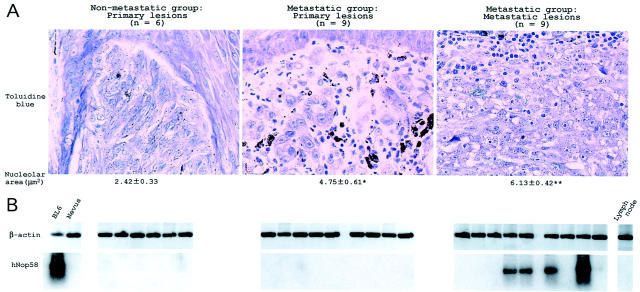
To assess the clinical relevance of the Nop5/Sik family members, we examined gene expression for human Nop58 (hNop58), a human member of the family, in human melanoma lesions. Tumor samples were obtained from 15 patients who were retrospectively divided into two groups, nonmetastatic (n = 6) and metastatic (n = 9). The primary lesions of all of the patients had been excised at the time of diagnosis. Patients of the nonmetastatic group survived at least 5 years after that time without developing lymph node metastasis. Patients of the metastatic group developed lymph node metastasis within 5 years. The nucleolar area of melanoma cells increased in the following order: primary lesions of the nonmetastatic group < primary lesions of the metastatic group < metastatic lesions of the metastatic group. This result is consistent with the previous report that nucleolar size could be a marker for poor prognosis of melanomas. 7-10
Total RNA was extracted from sections of melanoma tissues, nevus lesions, and lymph node tissues. Because melanoma lesions occupied more than half of the area in each of the adjacent sections of melanoma tissues (data not shown), melanoma cells were considered to be the major source of RNA in the melanoma samples. The yield of total RNA for each sample was <1.0 μg. All of the RNA samples were reverse-transcribed, PCR-amplified with β-actin-specific primers, and subjected to Southern blot analysis. As a positive control, total RNA (5 ng) of BL6 cells was used. The β-actin probe hybridized to a band of 456 bp, indicating that β-actin mRNA was clearly and equally amplified in all RNA samples (Figure 7B ▶ , top). After the RT reaction, PCR was performed with hNop58-specific primers, and the products were blotted with the hNop58 probe (Figure 7B ▶ , bottom). The message levels for hNop58 were below the limit of detection in all of the primary lesions. On the other hand, a 417-bp-long band was clearly detected in four of nine of the metastatic lesion samples, indicating a marked amplification of hNop58 mRNA. No hNop58-specific bands were detectable in the RNA samples from either nevus lesions or lymph node tissues, indicating a specific expression of hNop58 mRNA in melanoma cells. When total RNA samples were directly used as PCR templates in a negative control experiment, no hybridized bands were detected with any probes (data not shown), indicating little amount of genomic contamination. The ratio of hNop58-positive to -negative samples was significantly larger in the metastatic lesions (4:5) than in the primary lesions (0:15) (P < 0.01 by the chi-square test). We compared the nucleolar size between metastatic melanoma cells expressing hNop58 (5.71 ± 1.31 μm2; n = 4) and cells not expressing hNop58 (6.20 ± 0.953 μm2; n = 5), but found no significant difference. The hNop58 gene expression seemed to be closely linked to metastasis and nucleolar size, but seemed not to be the only determinant for either phenotype.
Figure 7.
Nucleolar size and hNop58 expression in human melanoma lesions. A: Sections were cut from paraformaldehyde-fixed, paraffin-embedded tissues, and stained with toluidine blue. A representative case for each group indicated above was shown. The nucleolar area was calculated by analyzing all of the samples belonging to each group, and appeared under the image. *, P < 0.01 by t-test when compared with the value of the nonmetastatic group. **, P < 0.05 by t-test when compared with the value of the primary lesions of the metastatic group. Epidermal structure is seen in the primary lesions, and a lot of lymphocytes are seen in the metastatic lesions (metastasis to the lymph nodes). Melanin deposition is seen in both lesions. Original magnification, ×400. B: Expression of hNop58 mRNA in human melanoma lesions. Total RNA was extracted from paraffin-embedded sections of melanoma lesions, nevus lesions, and metastasis-free lymph node tissues, and subjected to the RT-PCR reaction, followed by Southern blot analysis. Total RNA of BL6 cells was used as a positive control. RNA was analyzed with β-actin-specific primers and probe (top), or with hNop58-specific primers and probe (bottom). Each sample was blotted in a row of the groups indicated above in A. Nevus lesions and lymph node tissues served as negative controls for hNop58 expression in the primary and metastatic lesions, respectively.
Discussion
Although F10 and BL6 cells are originated from B16 melanoma cells, there is an apparent difference in the metastatic phenotypes of the two sublines. When injected subcutaneously into the mouse footpad, BL6 cells colonize in lungs much more heavily than do F10 cells. 16 We also found that BL6 cells contained larger nucleoli than F10 cells. While screening the BL6 cDNA library after subtraction with F10-derived mRNA, we isolated Sik-SP as a gene transcriptionally up-regulated in BL6 cells. Expression profiles of the Sik-SP gene in normal mouse tissues and cell lines indicated an especial abundance of the Sik-SP message in BL6 cells. Structurally, Sik-SP belongs to the conserved nucleolar Nop5/Sik family that is postulated to play a central role in ribosome biogenesis. 24,25,29,30 Thus, we hypothesized that BL6 cells might possess higher levels of nucleolar function than F10 cells because of an increased expression of Sik-SP.
An abundance of lysine residues in a region of the C-terminus including 100 amino acid residues is characteristic of Nop5/Sik family members. 23-25,29,30 However, the significance of this region is still unclear. Phylogenetic analysis divides the Nop5/Sik family members into two subgroups, Nop56 and Nop58. 24 Recently, Newman and colleagues 24 reported the amino acid sequence for mNop58 by fusing two EST clones (ATCC no. 1416459 and ATCC no. 971387). The sequence is the most probable mouse counterpart for yeast Nop58 (yNop58), because there is 47% identity and 71% similarity between mNop58 and yNop58. On the other hand, Nelson and colleagues 44 isolated a novel mouse member of the Nop58 subgroup from a NIH3T3 cDNA library, and termed it SAN5. In the present study, we isolated clones encoding Sik-SP from a BL6 cDNA library. The genetic basis for the diversity of the mouse Nop58 subgroup remains to be determined. Both SAN5 and Sik-SP are deletion forms of mNop58: Sik-SP lacks 63 amino acid residues at the N-terminus, and SAN5 lacks 36 amino acid residues near the C-terminus (Figure 3A) ▶ . The C-terminal lysine-rich region is entirely conserved in Sik-SP, and is partially deleted in SAN5, yet both Sik-SP (Figure 3C) ▶ and SAN5 44 localize to the nucleolus. Moreover, deletion of the part of the C-terminal region that is conserved in SAN5 did not change the nucleolar localization of Sik-SP. The C-terminal lysine-rich region thus does not seem to be necessary for the correct targeting of the Nop58 subgroup members to the nucleolus, as it is in yNop58. 29 However, our results point to a possible role of the C-terminal region in the nucleolar function of Sik-SP. Nucleolar localization of GFP-tagged Sik-SP increased the nucleolar area stained with toluidine blue, whereas nucleolar localization of GFP-tagged Sik-SPΔC did not. Deletion of the C-terminal region likely resulted in loss of Sik-SP function, suggesting a close relation between nucleolar size and the function of Sik-SP in the transfected COS-7 cells. Further analysis is necessary to determine the functional importance of the C-terminal region with more confidence.
Yeast deficient in yNop58 undergoes growth arrest because of defects in early pre-rRNA processing events necessary for ribosome assembly. 29-31 This lethal defect in yeast ribosome biogenesis was complemented by SAN5. 44 Here we obtained consistent results for Sik-SP in mammalian cells. Nucleolar size was increased in COS-7 cells that expressed GFP-tagged Sik-SP, suggesting a role for Sik-SP in the nucleolus. Transient expression of Sik-SP increased rRNA synthesis and translational levels of an exogenous Luc gene in COS-7 cells (Figure 4) ▶ . The two effects of Sik-SP were reproducible in F10 and BL6 cells (Figure 5) ▶ . Higher levels of rRNA synthesis and exogenous Luc gene translation were detected in BL6 cells. Transient expression of Sik-SP in F10 cells compensated for the inefficiency in translation. These results support the idea that Sik-SP promotes ribosome biogenesis in mammalian cells. In addition, BL6 cells appeared superior to F10 cells with respect to nucleolar function including rRNA and protein synthesis, because of the increased expression of Sik-SP.
In F10 and BL6 cells, the nucleolar size correlated well with nucleolar function, but there was no difference in the mean cell-doubling time. In addition, transfection of Sik-SP did not significantly shorten the cell-doubling time of F10 cells (Figure 6) ▶ . This was also the case with SAN5, which did not significantly increase fibroblast proliferation. 44 Instead, we found that Sik-SP promoted the escape of melanoma cells from starved states, and their subsequent entrance into a logarithmic growth phase. Interestingly, SAN5 was recently characterized as an early response gene that is induced by multiple growth factors and links growth factor receptors to the cellular translational machinery. 44 Moreover, it has recently been shown that Sik-SP gene expression is markedly induced in endometrial epithelial cells by estrogen, 45 which also acts as a potent inducer of proliferation in these cells. Both Sik-SP and SAN5 are assumed to up-regulate ribosome biogenesis in response to an increased demand for protein synthesis. By virtue of the induction of either factor, cells could transition from a quiescent to an actively growing state. In BL6 cells, Sik-SP gene expression still remained high at the end of the serum-starved culture condition, and the cells appeared ready to start active growth immediately after serum stimulation.
Once metastasis has developed, melanoma is rarely curable and the median survival time drops to 6 months. 46 It is therefore important to diagnose the metastatic potentials of melanoma cells. As shown in Figure 7 ▶ , nucleolar size and hNop58 gene expression could be markers for metastatic potentials. Although nucleolar size is inversely related with cell-doubling time in many cancer cells, metastatic potentials are not explained only by the rapidity of cancer cell proliferation. In fact, there is no difference in either the in vivo or in vitro growth rate between F10 and BL6 cells. 15 The present results suggest that increased nucleolar function, but not rapid cell growth rate, might be involved in the acquisition of high metastatic potentials by BL6 cells. Cancer cells in tumors, especially in metastatic microfoci, 47,48 are often exposed to various stress conditions, such as mechanical stress, hypoxia, low pH, nutrient deprivation, and host immune defense. 49-52 Increased nucleolar function could help cancer cells grow promptly in response to environmental changes, such as tumor angiogenesis, which release cells from such stress conditions. It has been reported that transient exposure to and subsequent recovery from hypoxia, glucose starvation, and low pH enhance the metastatic ability of several types of cancer cells, including F10 cells. 53-55 This phenomenon may result from the preferential growth of cancer cells with increased nucleolar function. Whether nucleolar function plays a causative role in metastasis remains unknown. However, the present study has shown that the Nop5/Sik family members are among the factors that cause increased nucleolar size and function in metastatic melanoma cells.
Acknowledgments
We thank I. J. Fidler and D. C. Bennett for the mouse melanoma and melanocytic cells, respectively, and K. Morihana for technical assistance with the histocytological experiments.
Footnotes
Address reprint requests to Akihiko Ito, MD., Department of Pathology, Osaka University Medical School, 2-2 Yamada-oka, Suita, Osaka 565-0871, Japan. E-mail: aito@patho.med.osaka-u.ac.jp.
Supported by grants from the Ministry of Education, Science, and Culture; the Ministry of Health and Welfare; the Osaka Cancer Society; Public Trust Haraguchi Memorial Cancer Research Fund; and the Ichiro Kanehara Foundation.
K. N. and A. I. contributed equally to this work.
References
- 1.Busch H, Smetana K: Nucleoli of tumor cells. Busch H Smetana K eds. The Nucleolus. 1970, :pp 448-471 London, Academic Press, New York [Google Scholar]
- 2.Pianese G: Beitrag zur Histologie und Aetiologie der Carcinoma. Histologische und exprimentelle Untersuchungen. Beitr Pathol Anat Allgem Pathol 1896, 142 (suppl):S1-S193 [Google Scholar]
- 3.Derenzini M, Trere D, Pession A, Montanaro L, Sirri V, Ochs RL: Nucleolar function and size in cancer cells. Am J Pathol 1998, 152:1291-1297 [PMC free article] [PubMed] [Google Scholar]
- 4.Derenzini M, Trere D, Pession A, Govoni M, Sirri V, Chieco P: Nucleolar size indicates the rapidity of cell proliferation in cancer tissues. J Pathol 2000, 191:181-186 [DOI] [PubMed] [Google Scholar]
- 5.Olson MOJ, Dundr M, Szebeni A: The nucleolus: an old factory with unexpected capabilities. Trends Cell Biol 2000, 10:189-196 [DOI] [PubMed] [Google Scholar]
- 6.Olson MOJ: The role of proteins in nucleolar structure and function. Strauss PR Wilson SH eds. The Eukaryotic Nucleolus: Molecular Biochemistry and Molecular Assemblies. 1990, :pp 519-559 Telford Press, Inc., Cadwell [Google Scholar]
- 7.Gambini C, Casazza S, Borgiani L, Canepa M, Rovida S, Rongioletti F, Rebora A: Counting the nucleolar organizer region-associated proteins is a prognostic clue of malignant melanoma. Arch Dermatol 1992, 128:487-490 [PubMed] [Google Scholar]
- 8.Barzilai A, Goldberg I, Yulash M, Pavlotsky F, Zuckerman A, Trau H, Azizi E, Kopolovic J: Silver-stained nucleolar organizer regions (AgNORs) as a prognostic value in malignant melanoma. Am J Dermatopathol 1998, 20:473-477 [DOI] [PubMed] [Google Scholar]
- 9.McLean IW, Sibug ME, Becker RL, McCurdy JB: Uveal melanoma: the importance of large nucleoli in predicting patient outcome—an automated image analysis study. Cancer 1997, 79:982-988 [PubMed] [Google Scholar]
- 10.Bechrakis NE, Sehu KW, Lee WR, Damato BE, Forester MH: Transformation of cell type in uveal melanomas. Arch Ophthalmol 2000, 118:1406-1412 [DOI] [PubMed] [Google Scholar]
- 11.Crocker J: Nucleolar organizer regions. Underwood JC eds. Current Topics in Pathology. 1990, :pp 91-150 Springer-Verlag, Inc., New York [DOI] [PubMed] [Google Scholar]
- 12.Nicolson GL: Cancer metastasis: organ colonization and the cell-surface properties of malignant cells. Biochim Biophys Acta 1982, 695:113-176 [DOI] [PubMed] [Google Scholar]
- 13.Nicolson GL: Cancer metastasis: tumor cell and host organ properties important in metastasis to specific secondary sites. Biochem Biophys Acta 1988, 948:175-224 [DOI] [PubMed] [Google Scholar]
- 14.Poste G, Fidler IJ: The pathogenesis of cancer metastasis. Nature 1980, 283:139-146 [DOI] [PubMed] [Google Scholar]
- 15.Hart IR: The selection and characterization of an invasive variant of the B16 melanoma. Am J Pathol 1979, 97:587-600 [PMC free article] [PubMed] [Google Scholar]
- 16.Poste G, Doll J, Hart IR, Fidler IJ: In vitro selection of murine B16 melanoma variants with enhanced tissue-invasive properties. Cancer Res 1980, 40:1636-1644 [PubMed] [Google Scholar]
- 17.Fidler IJ: Selection of successive tumour lines for metastasis. Nat New Biol 1973, 242:148-149 [DOI] [PubMed] [Google Scholar]
- 18.Nakaji T, Kataoka TR, Watabe K, Nishiyama K, Nojima H, Shimada Y, Sato F, Matsushima H, Endo Y, Kuroda Y, Kitamura Y, Ito A, Maeda S: A new member of the GTPase superfamily that is upregulated in highly metastatic cells. Cancer Lett 1999, 147:139-147 [DOI] [PubMed] [Google Scholar]
- 19.Ito A, Katoh F, Kataoka TR, Okada M, Tsubota N, Asada H, Yoshikawa K, Maeda S, Kitamura Y, Yamasaki H, Nojima H: A role for heterologous gap junctions between melanoma and endothelial cells in metastasis. J Clin Invest 2000, 105:1189-1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kataoka TR, Ito A, Asada H, Watabe K, Nishiyama K, Nakamoto K, Itami S, Yoshikawa K, Ito M, Nojima H, Kitamura Y: Annexin VII as a novel marker for invasive phenotype of malignant melanoma. Jpn J Cancer Res 2000, 91:75-83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ito A, Kataoka TR, Watanabe M, Nishiyama K, Mazaki Y, Sabe H, Kitamura Y, Nojima H: A truncated isoform of the PP2A B56 subunit promotes cell motility through paxillin phosphorylation. EMBO J 2000, 19:562-571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watabe K, Ito A, Asada H, Endo Y, Kobayashi T, Nakamoto K, Itami S, Takao S, Shinomura Y, Aikou T, Yoshikawa K, Matsuzawa Y, Kitamura Y, Nojima H: Structure, expression and chromosome mapping of MLZE, a novel gene which is preferentially expressed in metastatic melanoma cells. Jpn J Cancer Res 2001, 92:140-151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vorbruggen G, Onel1 S, Jackle H: Restricted expression and subnuclear localization of the Drosophila gene Dnop5, a member of the Nop/Sik family of the conserved rRNA processing factors. Mech Dev 2000, 90:305-308 [DOI] [PubMed] [Google Scholar]
- 24.Newman DR, Kuhn JF, Shanab GM, Maxwell ES: Box C/D snoRNA-associated proteins: two pairs of evolutionarily ancient proteins and possible links to replication and transcription. RNA 2000, 6:861-879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Y, Isaac C, Wang C, Dragon F, Meier UT: Conserved composition of mammalian box H/ACA and box C/D small nucleolar ribonucleoprotein particles and their interaction with the common factor Nopp140. Mol Biol Cell 2000, 11:567-577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lafontaine DL, Tollervey D: Synthesis and assembly of the box C+D small nucleolar RNPs. Mol Cell Biol 2000, 20:2650-2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tollervey D: Trans-acting factors in ribosome synthesis. Exp Cell Res 1996, 229:226-232 [DOI] [PubMed] [Google Scholar]
- 28.Maxwell ES, Fournier MJ: The small nucleolar RNAs. Annu Rev Biochem 1995, 64:897-934 [DOI] [PubMed] [Google Scholar]
- 29.Gautier T, Berges T, Tollervey D, Hurt E: Nucleolar KKE/D repeat proteins Nop56p and Nop58p interact with Nop1p and are required for ribosome biogenesis. Mol Cell Biol 1997, 17:7088-7098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu P, Brockenbrough JS, Metcalfe AC, Chen S, Aris JP: Nop5p is a small nucleolar ribonucleoprotein component required for pre-18 S rRNA processing in yeast. J Biol Chem 1998, 273:16453-16463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lafontaine DL, Tollervey D: Nop58p is a common component of the box C+D snoRNPs that is required for snoRNA stability. RNA 1999, 5:455-467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lyman SK, Gerace L, Baserga SJ: Human Nop5/Nop58 is a component common to the box C/D small nucleolar ribonucleoproteins. RNA 1999, 5:1597-1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bennett CD, Cooper JP, Hart RI: A line of nontumorigenic mouse melanocytes, syngeneic with the B16 melanoma and requiring a tumor promoter for growth. Int J Cancer 1987, 39:414-418 [DOI] [PubMed] [Google Scholar]
- 34.Patterson MK, Jr: Measurement of growth and viability of cells in culture. Jokoby WB Pastran IH eds. Methods in Enzymology. 1979, :pp 141-167 Academic Press, New York [DOI] [PubMed] [Google Scholar]
- 35.Ito A, Morii E, Maeyama K, Jippo T, Kim DK, Lee YM, Ogihara H, Hashimoto K, Kitamura Y, Nojima H: Systematic method to obtain novel genes that are regulated by mi transcription factor: impaired expression of granzyme B and tryptophan hydroxylase in mi/mi cultured mast cells. Blood 1998, 91:3210-3221 [PubMed] [Google Scholar]
- 36.Fujii T, Tamura K, Copeland NG, Gilbert DJ, Jenkins NA, Yomogida K, Tanaka H, Nishimune Y, Nojima H, Abiko Y: Sperizin is a murine RING zinc-finger protein specifically expressed in haploid germ cells. Genomics 1999, 57:94-101 [DOI] [PubMed] [Google Scholar]
- 37.Miyazaki J, Takaki S, Araki K, Tashiro F, Tominaga A, Takatsu K, Yamamura K: Expression vector system based on the chicken beta-actin promoter directs efficient production of interleukin-5. Gene 1989, 79:269-277 [DOI] [PubMed] [Google Scholar]
- 38.Niwa H, Yamamura K, Miyazaki J: Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 1991, 108:193-199 [DOI] [PubMed] [Google Scholar]
- 39.Stirpe F, Novello F: Factors affecting the activity of ribonucleic acid polymerase solubilized from rat liver nuclei: effect on ionic strength, spermine and divalent ions with native and denatured deoxyribonucleic acid. Eur J Biochem 1970, 15:505-512 [DOI] [PubMed] [Google Scholar]
- 40.Janne O, Bullock LP, Bardin CW, Jacob ST: Early androgen action in kidney of normal and androgen-insensitive (tfm/y) mice. Changes in RNA polymerase and chromatin template activities. Biochem Biophys Acta 1976, 418:330-343 [DOI] [PubMed] [Google Scholar]
- 41.Blair DG, Dommasch M: Nuclear DNA-dependent RNA polymerases of Ehrlich ascites tumor cells: two discrete-amanitin-sensitive forms. Biochem Biophys Res Commun 1972, 49:877-883 [DOI] [PubMed] [Google Scholar]
- 42.Capobianco G, Vescia A: DNA-dependent RNA polymerases: II) Effect of α-amanitin and denaturated DNA on multiple forms separated from rat liver nuclei. Ital J Biochem 1974, 23:245-256 [PubMed] [Google Scholar]
- 43.Uckert W, Walther W, Stein U: Large and small scale RNA preparations from eukaryotic cells. Methods Mol Biol 1998, 86:7-14 [DOI] [PubMed] [Google Scholar]
- 44.Nelson SA, Aris JP, Patel BK, LaRochelle WJ: Multiple growth factor induction of a murine early response gene that complements a lethal defect in yeast ribosome biogenesis. J Biol Chem 2000, 275:13835-13841 [DOI] [PubMed] [Google Scholar]
- 45.Das SK, Tan J, Raja S, Halder J, Paria BC, Dey SK: Estrogen targets genes involved in protein processing, calcium homeostasis, and Wnt signaling in the mouse uterus independent of estrogen receptor-α and -β. J Biol Chem 2000, 275:28834-28842 [DOI] [PubMed] [Google Scholar]
- 46.Barnhill RL, Mihm MC, Fitzpatrick TB, Sober EJ: Neoplasms: malignant melanoma. Fitzpatrick TB Eisen AZ Wolff K Freedberg IM Austen KF eds. Dermatology in General Medicine. 1993, :pp 1078-1116 McGraw-Hill, New York [Google Scholar]
- 47.Folkman J: Tumor angiogenesis: a possible control point in tumor growth. Ann Intern Med 1975, 82:96-100 [DOI] [PubMed] [Google Scholar]
- 48.Folkman J: What is the evidence that tumors are angiogenesis dependent? J Natl Cancer Inst 1990, 82:4-6 [DOI] [PubMed] [Google Scholar]
- 49.Wike-Hooley JL, Haveman J, Reinhold HS: The relevance of tumour pH to the treatment of malignant disease. Radiother Oncol 1984, 2:343-366 [DOI] [PubMed] [Google Scholar]
- 50.Vaupel P, Kallinowski F, Okunieff P: Blood flow, oxygen and nutrient supply, and metabolic microenvironment of human tumors: a review. Cancer Res 1989, 49:6449-6465 [PubMed] [Google Scholar]
- 51.Tannock IF, Rotin D: Acid pH in tumors and its potential for therapeutic exploitation. Cancer Res 1989, 49:4373-4384 [PubMed] [Google Scholar]
- 52.Cuvier C, Jang A, Hill RP: Exposure to hypoxia, glucose starvation and acidosis: effect on invasive capacity of murine tumor cells and correlation with cathepsin (L + B) secretion. Clin Exp Metastasis 1997, 15:19-25 [DOI] [PubMed] [Google Scholar]
- 53.Young SD, Hill RP: Effects of reoxygenation on cells from hypoxic regions of solid tumors: anticancer drug sensitivity and metastatic potential. J Natl Cancer Inst 1990, 82:371-380 [DOI] [PubMed] [Google Scholar]
- 54.Schlappack OK, Zimmermann A, Hill RP: Glucose starvation and acidosis: effect on experimental metastatic potential, DNA content and MTX resistance of murine tumour cells. Br J Cancer 1991, 64:663-670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Young SD, Marshall RS, Hill RP: Hypoxia induces DNA overreplication and enhances metastatic potential of murine tumor cells. Proc Natl Acad Sci USA 1988, 85:9533-9537 [DOI] [PMC free article] [PubMed] [Google Scholar]