Abstract
The histological diagnosis of hepatocellular carcinoma (HCC) can be complicated by difficulty in differentiation from cholangiocarcinoma and metastatic carcinoma. Immunohistochemical stains currently in use are suboptimal in terms of specificity and sensitivity. Using cDNA array analysis for differential gene expression, we demonstrated a significant increase in mRNA expression level of CD10/CALLA, a type 2 cell-surface metalloproteinase, in HCC, which was subsequently confirmed by reverse transcriptase-polymerase chain reaction and Western blotting analysis. To test the possibility of using CD10/CALLA as a diagnostic marker for HCC, various intrahepatic tumors were studied immunohistochemically using a monoclonal antibody for CD10. A characteristic canalicular-staining pattern was observed in normal hepatocytes and at the apical surface of bile duct epithelial cells. The canalicular expression of CD10 was identified in 9 of 15 HCCs examined (60%), whereas 10 cholangiocarcinomas and 8 of 9 metastatic carcinomas lacked this staining. In three of the six HCCs negative for CD10, the surrounding nonneoplastic liver tissue was also negative, suggesting fixation-associated loss of immunoreactivity. Six HCCs had stronger CD10 staining in tumor cells when compared to the surrounding nonneoplastic tissue. Three cases of benign bile duct adenomas also expressed CD10 at the luminal aspect. One of the MCs showed a diffuse, cytoplasmic staining for CD10, a pattern readily distinguishable from that of HCC. A panel of other immunohistochemical markers were also studied for comparison, including polyclonal anti-carcinoembryonic antigen, cytokeratin (CK) 7, CK20, and α-fetoprotein. Our results demonstrate that cDNA arrays can be effectively used to identify new diagnostic markers, and that CD10 is a reliable marker for identifying HCC, particularly when used in conjunction with a panel of immunohistochemical markers (polyclonal anti-carcinoembryonic antigen, CK7, CK20, and α-fetoprotein) and in the distinction from cholangiocarcinoma.
Hepatocellular carcinoma (HCC) is the most common primary malignancy of the liver in adults. The incidence in 1995 was reported as 2.1 per 100,000 population in the United States. 1 Its diagnosis requires histological examination of biopsied tissue from a clinically suspected liver mass. An accurate diagnosis, particularly of poorly differentiated ones, requires differentiation from cholangiocarcinoma (CC), metastatic carcinoma (MC), and other malignant tumors, and is critical in guiding appropriate clinical management. Well-differentiated HCCs exhibit readily recognizable features of hepatocytic differentiation and thus may cause little difficulty in microscopic diagnosis. For poorly differentiated lesions, on another hand, proper diagnosis can be problematic.
In conjunction with relevant clinical and laboratory information, such as the presence or absence of cirrhosis and the serum α-fetoprotein (AFP) level, a panel of immunohistochemical stains of biopsied or resected liver specimens is often helpful in histological differential diagnosis. Commonly used are a polyclonal antibody against carcinoembryonic antigen (CEA-p), monoclonal antibodies specific for cytokeratin (CK) 7 and CK20, and AFP. HCCs usually exhibit a characteristic membranous staining pattern with CEA-p. The antibody cross-reacts with the biliary glycoprotein I located at the bile canalicular aspect of the hepatocyte surface. 2 Most HCCs do not express CK7 or CK20, but most primary and metastatic adenocarcinomas express either or both antigens. 3 When performed in a well controlled setting, staining for AFP is relatively specific for HCC, but with a low sensitivity, with only ∼20% of HCCs being positive for this protein immunohistochemically.
The recent accumulation of genetic data derived from the human genome project has led to the development of new technologies and approaches to study gene expression profiles in a global setting. One such technology, cDNA arrays, allows large-scale gene expression profiling of human tissue to discover target genes that may be useful in the identification of potential therapeutic interventions and/or diagnostic probes. This technology has been used successfully in identifying many novel genes associated with neoplastic transformation, 4-10 and more recently as an investigative tool in molecular pathology. 11-13 Although genetic alterations in malignancies such as breast and colon cancers are relatively well explored, this is not the case with HCC. Furthermore, currently available diagnostic markers for HCC are not optimal, and thus more accurate immunohistochemical markers for HCC are needed.
In this study, we used cDNA array analysis to identify novel gene products that may serve as immunohistochemical markers for HCC. One such candidate identified is CD10, a cell surface protein. The potential usefulness of CD10 as a diagnostic marker in HCC was then examined immunohistochemically in archival tissue with hepatic tumors, which include HCC, CC, MC, and benign bile duct adenoma (BDA), using a monoclonal antibody for CD10. Several other antibodies were also included in this study for comparison. The results show that, used in combination with AFP, immunostaining for CD10 adds a significant sensitivity to the histological identification of HCC, while maintaining the specificity. In addition, the results presented in this report emphasize the effectiveness of using cDNA array technique to examine gene expression profiles in human tumors to identify potential diagnostic markers that are useful in surgical pathology.
Materials and Methods
Patients and Specimens
For molecular analysis, fresh tissue samples of HCC and surrounding nonneoplastic liver were obtained at the time of resection. They were snap-frozen in liquid nitrogen and stored at −80°C before RNA extraction and protein analysis. For retrospective immunohistochemical studies, samples were retrieved from the surgical pathology archives at the University of Texas Medical Branch Hospitals and University of Chicago Hospitals. Multiple sections were examined microscopically to confirm the tumor type and the degree of differentiation. One representative block was then selected for immunohistochemical studies. The demographics of the patients and histological classification of the tumors are summarized in Table 1 ▶ . Among the nine cases of MCs, four had colorectal primaries (including one neuroendocrine carcinoma), two had pancreatic head primaries, and three had unknown primaries.
Table 1.
Demographic Data of Patients Studied and Tumor Types
| Tumor type | Number of cases | Age range (yrs) | Male | Female |
|---|---|---|---|---|
| HCC | 15 | 43–72 | 12 | 3 |
| BDA | 3 | 25–63 | 2 | 1 |
| CC | 10 | 56–80 | 7 | 3 |
| MC | 9 | 35–74 | 6 | 3 |
HCC, Hepatocellular carcinoma; BDA, bile duct adenoma; CC, cholangiocarcinoma; MC, metastatic carcinoma.
RNA Isolation and cDNA Array Analysis
Total cellular RNA was isolated from HCC and the surrounding nonneoplastic liver tissue, using the RNAqueous-4-PCR RNA extraction kit (Ambion, Austin, TX), according to the manufacturer’s instructions. Briefly, 50 mg of liver tissue was pulverized under liquid nitrogen using a mortar and pestle and the powder used for extraction of RNA. After removal of contaminating DNA by treatment with DNase 1 (Ambion), the RNA was precipitated using 5 mol/L of ammonium acetate and absolute ethanol. The quality of RNA was monitored by agarose gel electrophoresis, as demonstrated by the presence of intact ribosomal RNAs (28S and 18S bands).
cDNA probes for array analysis were synthesized following the manufacturer’s directions (Clontech, Palo Alto, CA). Briefly, 4 to 5 μg of total cellular RNA was used to generate cDNA probes in the presence of [α-32P]dATP, 10× dNTP mix (5 mmol/L each of dCTP, dGTP, and dTTP), Human 1.2II 10× CDS primer mix, 5× reaction buffer, Moloney-murine leukemia virus reverse transcriptase (100 U/μl), and dithiothreitol (100 mmol/L). Probes were then hybridized to Atlas Human Array 1.2II nylon membranes (Clontech, Palo Alto, CA) that contain 1176 spotted human cDNA’s (10 ng/spot), overnight at 68°C. Arrays were washed using the manufacturer’s protocols and differential gene expression patterns were detected by phosphorimaging. Data analysis was performed using the AtlasImage software (Clontech). Arrays were performed in duplicate and gene expression was normalized to overall global gene expression of each array, as recommended by the manufacturer.
Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR)
Total cellular RNA was used in a RT-PCR reaction to confirm array data. cDNA synthesis was performed using the cDNA synthesis kit (Clontech) according to the manufacturer’s protocol. Briefly, 1 μg of total RNA was used in a 20-μl reaction mixture containing 5× reaction buffer (250 mmol/L Tris-HCl, pH 8.3, 375 mmol/L KCl, 15 mmol/L MgCl2), 10 mmol/L each dNTP, Moloney-murine leukemia virus reverse transcriptase (40 U/μl), RNase inhibitor (1 U), and an oligo (dT)18 primer. The reaction mix was incubated at 42°C for 1 hour to synthesize the first strand cDNA, and the reaction was then terminated at 94°C for 5 minutes and diluted to 100 μl with 80 μl of diethyl pyrocarbonate H2O. Subsequently, 5 to 10 μl of diluted reaction mixtures were subjected to 35 PCR cycles, using the AdvanTaq PCR kit (Clontech) according to the manufacturer’s suggested protocol. The primers used were as follows: CD10: sense, 5′-CTGGAGTTCATAATGGATCTTGTAAGC-3′ and antisense 5′-CATCCAAGTGAGGTCATCTAAAGTCTG-3′; GAPDH: sense 5′-GGCTCTCCAGAACATCATCCCTGC-3′ and antisense 5′-GGGTGTCGCTGTTGAAGTCAGAGG-3′.
Detection of CD10 by Western Immunoblotting Analysis
Soluble cell lysates from HCC and surrounding nonneoplastic liver were prepared by lysis in 200 μl of a buffer containing 50 mmol/L Tris-Cl (pH 7.5), 150 mmol/L NaCl, 1% Nonidet P-40, 0.1% sodium deoxycholate, and 25 mg/ml of aprotinin. Equal concentrations of protein were subjected to electrophoresis in a 12% sodium dodecyl sulfate-polyacrylamide gel, followed by transfer onto Hybond-N membranes (Amersham, Arlington Heights, IL). Nonspecific binding was blocked by incubation in phosphate-buffered saline (PBS) containing 0.1% Tween 20 (PBS-T) and 5% nonfat milk for 1 hour at room temperature. The membrane was incubated overnight at 4°C with anti-CD10/CALLA (clone 56C6; Neomarkers, Freemont, CA) at a 1:200 dilution in PBS-T containing 1% bovine serum albumin (PBS-T-BSA). After four washes in PBS-T, the membrane was incubated with anti-mouse IgG conjugated to horseradish peroxidase (Sigma Chemical Co. St. Louis, MO) diluted in PBS-T-BSA for 1 hour at room temperature. After four additional washes with PBS-T, the signals were visualized by enhanced chemiluminescence (ECL Plus; Amersham).
Antibodies and Immunohistochemical Staining
Multiple 3- to 4-μm sections were cut from a formalin-fixed, paraffin-embedded tissue block, and mounted on positively charged slides. Deparaffinization and quenching in 3% methanol H2O2 were performed as described previously. 14
Primary antibodies used were purchased from various vendors. Most of these antibodies had been previously titrated and used routinely in our lab. These included rabbit anti-human CEA (1:1600), monoclonal mouse anti-human CK7 (isotype IgG1, kappa, 1:200), monoclonal mouse anti-human CK20 (IgG2a, kappa, 1:100), and rabbit anti-human AFP (1:1600), purchased from DAKO (Carpinteria, CA). CD10/CALLA (neutral endopeptidase) Ab-2 antibody (clone 56C6) is a mouse monoclonal antibody purchased from NeoMarkers. The Anti-CD10 was titrated and a dilution of 1:80 was chosen in our study.
Incubation condition for each primary antibody was 30 minutes at room temperature. Appropriate positive and negative controls were used for all of the stainings. For detection of the bound primary antibodies, the LSAB2 kit with horseradish peroxidase and diaminobenzidine as chromogen was used (DAKO), following the manufacturer’s instructions. All of the slides were counterstained with Harris hematoxylin (Fisher Scientific, Pittsburgh, PA).
Microscopic Examination
Immunohistochemical staining was evaluated microscopically and a complete absence of staining is recorded as negative. Presence of staining was further specified as membranous, luminal, or cytoplasmic. The intensity of staining was recorded at a subjective spectrum of 1 to 3, with 1 being minimal and 3 being strong. Staining of at least 5% of the tumor cells was considered to be positive for the antibody.
Results
Differential Gene Expression in HCC
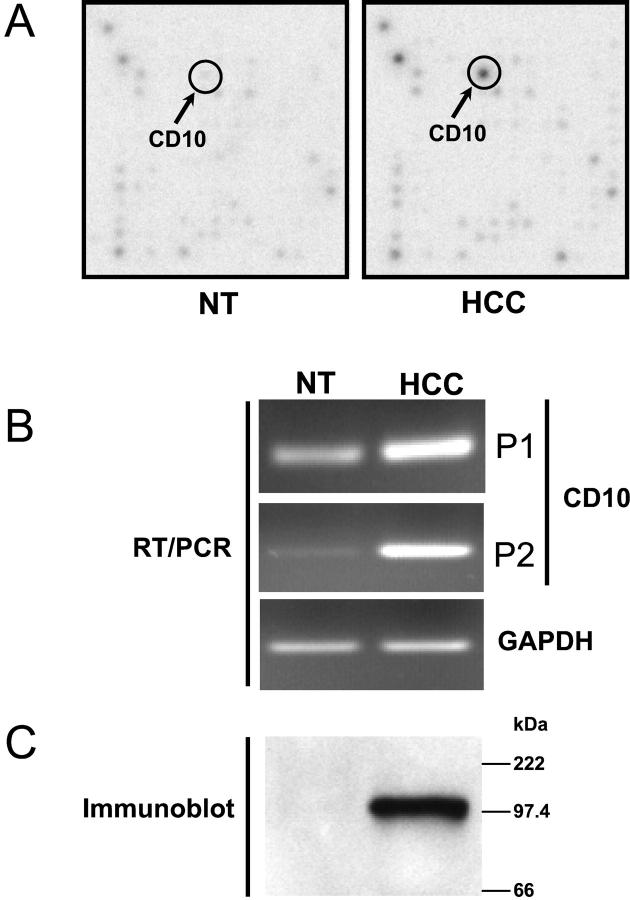
To search for potential diagnostic markers for HCC and to better understand genetic changes involved in HCC, we used cDNA expression arrays to simultaneously assess the expression patterns of 1176 human genes. We have identified a number of genes that were differentially expressed in HCC when compared with nonneoplastic liver tissues (MR Beard, S-Y Xiao, D Fleming, manuscript in preparation). Among these, CD10/CALLA mRNA level was consistently up-regulated in two of four HCC samples examined. Compared to other identified genes, CD10 was one of the most highly differentially regulated genes, which showed ∼40-fold increase in mRNA abundance in HCC, as compared to the surrounding nontumor tissue (Figure 1A) ▶ . The increase in its mRNA level noted in arrays was confirmed by RT-PCR using specific primers to CD10 (Figure 1B) ▶ . Furthermore, Western blotting analysis demonstrated that the protein expression of CD10 was also markedly increased in HCC (Figure 1C) ▶ , in concordance with the array and RT-PCR data.
Figure 1.
A: Differential mRNA expression profiles between HCC and adjacent nonneoplastic liver tissue (NT) as monitored using an Atlas Human cDNA 1.2II microarray. A representative section of the array is shown in which differential mRNA expression of CD10 is indicated by arrows. B: Up-regulation of CD10 mRNA expression as confirmed by RT-PCR. Semiquantitative RT-PCR was performed using specific primers for CD10 and total cellular RNA isolated from tissue surrounding the HCC (NT) and the HCC from two patients (P1 and P2). Co-amplification of human GAPDH was used as an internal control. C: Increased CD10 protein expression in HCC. Total cellular extracts from tissue surrounding the HCC (NT) and HCC were prepared from patient P1, and separated by sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis and CD10 was identified by immunoblotting with anti-CD10 antibody. A single species at ∼100 kd is seen, consistent with the reported molecular weight of CD10.
Expression of CD10 and Several Other Markers in HCC, CC, BDA, and Metastatic Carcinoma
The above results clearly demonstrate that in a subset of HCC cases, CD10 is highly up-regulated and thus may be potentially a useful diagnostic marker for HCC. To examine this possibility, immunohistochemical analysis was performed using a monoclonal antibody against CD10. The results showed that nonneoplastic hepatocytes and bile duct epithelial cells both expressed CD10. In normal hepatocytes, however, the expression was limited to the canalicular aspects of the cell membrane (Figure 2A) ▶ ; whereas in bile duct epithelium, it was present on the luminal surface of the membranes (Figure 2A) ▶ . Neoplastic hepatocytes in HCCs also exhibited CD10 positivity on the canalicular aspect (Figure 2B) ▶ . Benign BDAs showed a CD10 expression pattern similar to that seen in normal bile duct epithelium (Figure 2C) ▶ , but their malignant counterpart, CCs, were negative for CD10 expression (Figure 2D) ▶ .
Figure 2.
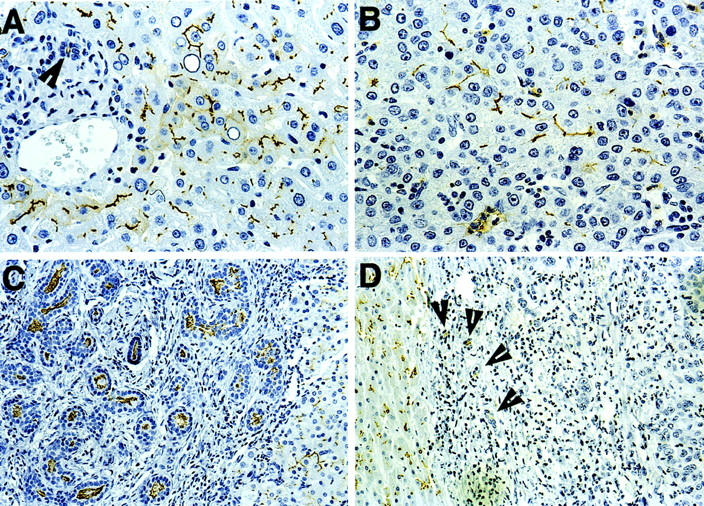
Immunohistochemical staining using anti-CD10 monoclonal antibody. A: Normal liver showing membranous staining of hepatocytes at the bile canalicular aspects, and luminal staining of a bile duct at the portal tract at left (arrowhead). B: Membranous staining of tumor cells in an HCC. C: Luminal staining of proliferating bile ductules in a BDA (left). The neighboring normal hepatocytes exhibit normal membranous staining. D: No staining for CD10 in tumor cells of a CC. Note membranous staining of neighboring normal hepatocytes (left), and luminal staining of a few residual benign bile ductules (arrowhead). Original magnifications: ×125 (C and D), ×250 (A and B).
Table 2 ▶ summarizes the data derived from immunohistochemical studies and showed that 9 of 15 HCCs examined expressed CD10 on the cell membrane of the tumor cells, but accentuated at the bile canalicular aspects, similar to normal, nontumor hepatocytes (Figure 2, A and B ▶ ). Among these nine positive cases, six exhibited stronger staining intensity in tumor cells when compared to the surrounding nontumor tissue (Figure 3) ▶ . In contrast, CCs were all negative for this marker (Figure 2D) ▶ . All MCs in the liver we examined were also negative, except for one adenocarcinoma from an unknown origin, where a diffuse cytoplasmic staining in tumor cells was observed (data not shown).
Table 2.
Immunohistochemical Staining of Different Intrahepatic Tumors
| Types of tumors | Number of cases | CEA | CK7 | CK20 | AFP | CD10 | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| No. positive | % | No. positive | % | No. positive | % | No. positive | % | No. positive | % | ||
| HCC | 15 | 15 | 100 | 1 | 6.7 | 0 | 0 | 3 | 20 | 9 | 60 |
| BDA | 3 | 3 | 100 | 2 | 100 | 0 | 0 | 0 | 0 | 3* | 100 |
| CC | 10 | 10 | 100 | 9 | 90 | 2 | 20.0 | 0 | 0 | 0 | 0 |
| MC | 9 | 9 | 100 | 3 | 33.3 | 5 | 55.6 | 0 | 0 | 1† | 11.1 |
HCC, Hepatocellular carcinoma; BDA, bile duct adenoma; CC, cholangiocarcinoma; MC, metastatic carcinoma.
*Luminal staining.
†Diffuse cytoplasmic staining.
Figure 3.
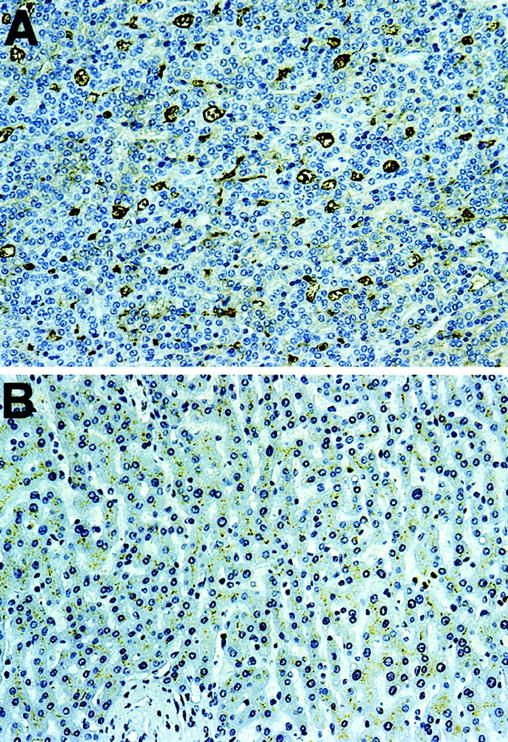
An example of HCC with increased expression of CD10. A: Strong membranous-positive reaction with anti-CD10. B: Weaker reactivity in normal area of the same liver. A and B were from the same section. Original magnifications, ×125.
Table 2 ▶ also shows that all HCCs examined expressed CEA-p in a membranous pattern (100%), with accentuation at the bile canalicular aspects. One hundred percent of the CCs and 100% of the MCs were also positive for this antigen, but with both cytoplasmic and membranous staining (data not shown). All HCCs were negative for CK20, and only one was positive for CK7. CCs were positive for CK7 and CK20 in 90% and 20% of cases, respectively. The one CC case that was negative for CK7 was poorly differentiated and expressed only weak (1+) staining for CEA-p. As a group, the MCs expressed CK7 and CK20 in 33.3% and 44.4% of cases, respectively. All three metastatic adenocarcinomas with colorectal primary were CK7−CK20+. The two adenocarcinomas with pancreatic primary were CK7+/CK20-. The two adenocarcinomas with unknown primary were both CK7+/CK20+. Two metastatic tumors (a carcinoid tumor of unknown primary, and a neuroendocrine carcinoma from the rectum) were negative for both CK7 and CK20. AFP was found to be expressed in only 20% of the HCCs. Only one HCC showed concurrent staining for both AFP and CD10. All other tumors were negative for AFP.
Discussion
CD10 is a 100-kd cell surface glycoprotein. It was originally identified on tumor cells of acute lymphoblastic leukemia, 15,16 and thus named common acute lymphoblastic leukemia antigen (CALLA). Besides lymphoid progenitor cells and tumors cells, CD10 was also found to be expressed on fetal liver cells, 17 and normal liver cells. 18 Biochemically, CD10 is a type II integral membrane protein known as neutral endopeptidase 24.11 (NEP 24.11), which functions to cleave small biologically active peptide at the amino terminus to hydrophobic residues within the peptide sequences. 19 Because it is differentially expressed on lymphoid cells, CD10 has been used routinely in the differential diagnosis of B cell lymphomas.
Microarray analysis of gene expression profiles in normal and neoplastic tissues is rapidly becoming a useful tool to identify genetic alterations and potential diagnostic markers in cancer. In this study we have used cDNA arrays containing 1176 genes to explore the gene expression profiles in HCC and surrounding liver tissue to identify novel genes differentially expressed in HCC, and to explore their potential roles in clinical diagnosis. We identified CD10 to be up-regulated at the mRNA level in two HCCs as compared to nonneoplastic liver tissue. The up-regulation of CD10 mRNA level was among the highest differentially regulated genes we found, with ∼40-fold increase in mRNA abundance in both HCC samples. The reliability of our array data was validated by semiquantitative RT-PCR that demonstrated an increase in CD10 mRNA level in tumor cells. Further analysis by Western blotting corroborated these findings by demonstrating an increase in CD10 protein expression level in HCC, and thus suggested the potential use of this antigen as a diagnostic marker for HCC.
By studying archival cases of HCC, CC, and MC immunohistochemically, we demonstrated the usefulness of CD10 in differential diagnosis among these tumors. Although 60% of HCCs in this series were positive for CD10 in a canalicular pattern, all of the CCs lacked the expression of this antigen, despite the fact that CD10 is normally expressed in bile duct epithelial cells and benign BDAs.
One of nine MCs examined in our study was positive for CD10, but in a diffuse cytoplasmic staining pattern. When interpreting the result of CD10-positive staining, one must be strict about the particular canalicular staining pattern in HCCs. CD10 may be expressed in various other types of adenocarcinomas, including clear cell or papillary renal cell carcinomas, and non-small cell carcinoma of the lung. 20,21 It has been reported that in some well-differentiated adenocarcinomas, CD10 positivity is restricted to the apical surface of the malignant glandular cells, which may be confused with the canalicular pattern seen in HCC. However, because these tumors are well-differentiated adenocarcinomas, they do not pose the difficulty in differential diagnosis from HCCs by routine hematoxylin and eosin stain. 22 On the other hand, poorly differentiated carcinomas may shows a diffuse cytoplasmic staining pattern 22 in cases that express CD10. This should be readily distinguishable from the canalicular pattern seen in HCC. Nevertheless, caution should be exercised in interpreting the result of CD10 staining, and other immunomarkers should always be considered in conjunction.
There are several other immunohistochemical markers and special mucin stains currently available to aid in the differential diagnosis of intrahepatic tumors. The CEA-p cross-reacts with the biliary glycoprotein I at the bile canalicular aspects of the hepatocyte surface. 2 Although it shows a characteristic canalicular staining pattern, the interpretation of the staining with this antibody is not always straightforward, because weak cell membrane staining and cytoplasmic staining can hinder interpretation. Staining for AFP produces a cytoplasmic pattern in HCC, but is present in a small proportion of cases 23 (20% of HCCs in this series). Immunostains for CKs are sometimes helpful, because HCCs are consistently negative for CK20, and usually for CK7 as well. In contrast, most CCs are CK7+/CK20−. Another recently developed immunohistochemical marker, hepatocyte paraffin-1 (HepPar1), has a relatively higher sensitivity and specificity compared to those mentioned previously. 23,24 However, a significant proportion of CCs can also be positive for this marker. 23 Histochemical stains for apomucin expression, such as MUC3, MUC5AC, MUC6, and MUC7, have also been described. 25
Among the 15 cases of HCCs examined in this study only 1 case concurrently expresses CD10 and AFP. However, 11 cases are positive for either AFP or CD10, resulting in a combined sensitivity of 73.3%. Therefore, if used in combination in routine practice, immunostaining for AFP and CD10 alone will give rise to an acceptable sensitivity and specificity in identifying HCC. The sensitivity and specificity will be further increased if other markers are also included (ie, CEA-p, CK7, CK20).
It is not known at the present whether the increased expression of CD10 in HCCs has any role in carcinogenesis or simply represents aberrant expression in transformed cells. Biochemically, CD10 is a neutral endopeptidase that functions to reduce cellular responses to specific peptide hormones. 19 CD10 has also been found to be expressed on fetal liver cells 17 and hepatoblastoma. 26 Further studies using normal and neoplastic hepatocyte cell lines will be necessary to elucidate the roles of CD10 and its substrates, if any, in regulating hepatocyte growth, function, and neoplastic transformation.
In summary, the data presented in this report demonstrate for the first time that a canalicular staining pattern for CD10 expression can be used as a useful diagnostic marker for HCC. This new marker identification process is achieved by using the powerful cDNA array techniques in combination with immunohistochemical approaches.
Acknowledgments
We thank Y.-L. Zhang for help in retrieving some of the specimens, and Linda Haden for her assistance in immunohistochemical stains.
Footnotes
Address reprint requests to Shu-Yuan Xiao, Department of Pathology, Route 0588, University of Texas Medical Branch, 301 University Blvd., Galveston, TX 77555. E-mail: syxiao@utmb.edu.
Supported in part by an Institutional Research Grant from the American Cancer Society (to M. R. B.).
Presented in part at the 90th Annual Meeting of United States and Canadian Academy of Pathology, March 2001, Atlanta, GA.
References
- 1.Di Bisceglie AM, Carithers RL, Jr, Gores GJ: Hepatocellular carcinoma. Hepatology 1998, 28:1161-1165 [DOI] [PubMed] [Google Scholar]
- 2.Nap M, ten Hoor KA, Fleuren GJ: Cross-reactivity with normal antigens in commercial anti-CEA sera, used for immunohistology. The need for tissue controls and absorptions. Am J Clin Pathol 1983, 79:25-31 [DOI] [PubMed] [Google Scholar]
- 3.Chu P, Wu E, Weiss LM: Cytokeratin 7 and cytokeratin 20 expression in epithelial neoplasms: a survey of 435 cases. Mod Pathol 2000, 13:962-972 [DOI] [PubMed] [Google Scholar]
- 4.Cole KA, Krizman DB, Emmert-Buck MR: The genetics of cancer—a 3D model. Nat Genet 1999, 21:38-41 [DOI] [PubMed] [Google Scholar]
- 5.Moch H, Schraml P, Bubendorf L, Mirlacher M, Kononen J, Gasser T, Mihatsch MJ, Kallioniemi OP, Sauter G: High-throughput tissue microarray analysis to evaluate genes uncovered by cDNA microarray screening in renal cell carcinoma. Am J Pathol 1999, 154:981-986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Richter J, Wagner U, Kononen J, Fijan A, Bruderer J, Schmid U, Ackermann D, Maurer R, Alund G, Knonagel H, Rist M, Wilber K, Anabitarte M, Hering F, Hardmeier T, Schonenberger A, Flury R, Jager P, Fehr JL, Schraml P, Moch H, Mihatsch MJ, Gasser T, Kallioniemi OP, Sauter G: High-throughput tissue microarray analysis of cyclin E gene amplification and overexpression in urinary bladder cancer. Am J Pathol 2000, 157:787-794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rihn BH, Mohr S, McDowell SA, Binet S, Loubinoux J, Galateau F, Keith G, Leikauf GD: Differential gene expression in mesothelioma. FEBS Lett 2000, 480:95-100 [DOI] [PubMed] [Google Scholar]
- 8.Welsh JB, Zarrinkar PP, Sapinoso LM, Kern SG, Behling CA, Monk BJ, Lockhart DJ, Burger RA, Hampton GM: Analysis of gene expression profiles in normal and neoplastic ovarian tissue samples identifies candidate molecular markers of epithelial ovarian cancer. Proc Natl Acad Sci USA 2001, 98:1176-1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu J, Stolk JA, Zhang X, Silva SJ, Houghton RL, Matsumura M, Vedvick TS, Leslie KB, Badaro R, Reed SG: Identification of differentially expressed genes in human prostate cancer using subtraction and microarray. Cancer Res 2000, 60:1677-1682 [PubMed] [Google Scholar]
- 10.Graveel CR, Jatkoe T, Madore SJ, Holt AL, Farnham PJ: Expression profiling and identification of novel genes in hepatocellular carcinomas. Oncogene 2001, 20:2704-2712 [DOI] [PubMed] [Google Scholar]
- 11.Emmert-Buck MR, Strausberg RL, Krizman DB, Bonaldo MF, Bonner RF, Bostwick DG, Brown MR, Buetow KH, Chuaqui RF, Cole KA, Duray PH, Englert CR, Gillespie JW, Greenhut S, Grouse L, Hillier LW, Katz KS, Klausner RD, Kuznetzov V, Lash AE, Lennon G, Linehan WM, Liotta LA, Marra MA, Munson PJ, Ornstein DK, Prabhu VV, Prange C, Schuler GD, Soares MB, Tolstoshev CM, Vocke CD, Waterston RH: Molecular profiling of clinical tissue specimens: feasibility and applications. Am J Pathol 2000, 156:1109-1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moch H, Kononen T, Kallioniemi OP, Sauter G: Tissue microarrays: what will they bring to molecular and anatomic pathology? Adv Anat Pathol 2001, 8:14-20 [DOI] [PubMed] [Google Scholar]
- 13.Snijders AM, Meijer GA, Brakenhoff RH, van den Brule AJ, van Diest PJ: Microarray techniques in pathology: tool or toy? Mol Pathol 2000, 53:289-294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao S-Y, Zhang H, Yang Y, Tesh R: Pirital virus (Arenaviridae) infection in the Syrian golden hamsters, mesocricetus auratus: a new model for arenaviral hemorrhagic fever. Am J Trop Med Hyg 2001, 64:111-118 [DOI] [PubMed] [Google Scholar]
- 15.Greaves MF, Brown G, Rapson NT, Lister TA: Antisera to acute lymphoblastic leukemia cells. Clin Immunol Immunopathol 1975, 4:67-84 [DOI] [PubMed] [Google Scholar]
- 16.Ritz J, Pesando JM, Notis-McConarty J, Lazarus H, Schlossman SF: A monoclonal antibody to human acute lymphoblastic leukaemia antigen. Nature 1980, 283:583-585 [DOI] [PubMed] [Google Scholar]
- 17.Hokland P, Rosenthal P, Griffin JD, Nadler LM, Daley J, Hokland M, Schlossman SF, Ritz J: Purification and characterization of fetal hematopoietic cells that express the common acute lymphoblastic leukemia antigen (CALLA). J Exp Med 1983, 157:114-129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McIntosh GG, Lodge AJ, Watson P, Hall AG, Wood K, Anderson JJ, Angus B, Horne CH, Milton ID: NCL-CD10–270: a new monoclonal antibody recognizing CD10 in paraffin-embedded tissue. Am J Pathol 1999, 154:77-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shipp MA, Vijayaraghavan J, Schmidt EV, Masteller EL, D’Adamio L, Hersh LB, Reinherz EL: Common acute lymphoblastic leukemia antigen (CALLA) is active neutral endopeptidase 24.11 (“enkephalinase”): direct evidence by cDNA transfection analysis. Proc Natl Acad Sci USA 1989, 86:297-301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Avery AK, Beckstead J, Renshaw AA, Corless CL: Use of antibodies to RCC and CD10 in the differential diagnosis of renal neoplasms. Am J Surg Pathol 2000, 24:203-210 [DOI] [PubMed] [Google Scholar]
- 21.Ganju RK, Sunday M, Tsarwhas DG, Card A, Shipp MA: CD10/NEP in non-small cell lung carcinomas. Relationship to cellular proliferation. J Clin Invest 1994, 94:1784-1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chu P, Arber DA: Paraffin-section detection of CD10 in 505 nonhematopoietic neoplasms. Frequent expression in renal cell carcinoma and endometrial stromal sarcoma. Am J Clin Pathol 2000, 113:374-382 [DOI] [PubMed] [Google Scholar]
- 23.Minervini MI, Demetris AJ, Lee RG, Carr BI, Madariaga J, Nalesnik MA: Utilization of hepatocyte-specific antibody in the immunocytochemical evaluation of liver tumors. Mod Pathol 1997, 10:686-692 [PubMed] [Google Scholar]
- 24.Fasano M, Theise ND, Nalesnik M, Goswami S, Garcia de Davila MT, Finegold MJ, Greco MA: Immunohistochemical evaluation of hepatoblastomas with use of the hepatocyte-specific marker, hepatocyte paraffin 1, and the polyclonal anti-carcinoembryonic antigen. Mod Pathol 1998, 11:934-938 [PubMed] [Google Scholar]
- 25.Sasaki M, Nakanuma Y, Ho SB, Kim YS: Cholangiocarcinomas arising in cirrhosis and combined hepatocellular-cholangiocellular carcinomas share apomucin profiles. Am J Clin Pathol 1998, 109:302-308 [DOI] [PubMed] [Google Scholar]
- 26.von Schweinitz D, Leuschner I, Gluer S, Pietsch T: Expression of cell adhesion molecules and common acute lymphoblastic leukaemia antigen in hepatoblastoma. Virchows Arch 1996, 429:239-241 [DOI] [PubMed] [Google Scholar]