Abstract
Juvenile polyposis syndrome (JPS; OMIM 174900) is a rare disorder which is characterized by the presence of hamartomatous polyps throughout the gastrointestinal tract and an increased risk of gastrointestinal malignancy. Mutations of the SMAD4 gene on chromosome 18q21.1 have been shown to cause a subset of JPS cases, with estimates ranging from 20% to >50%. Characterization of the genes that cause the remainder of JPS cases relies on the certainty that SMAD4 is not the causative gene. We have undertaken a comprehensive analysis of germline SMAD4 mutations in a cohort of JPS patients to define the spectrum of mutations that cause JPS. We have analyzed a series of polyps from these patients for SMAD4 protein expression. We have also performed a blinded assessment of polyp material to look for morphological differences between polyps from patients with and without a germline SMAD4 mutation. The results indicate that almost all germline SMAD4 mutations are readily detectable by screening genomic DNA using polymerase chain reaction-based methods; SMAD4 can be excluded as the causative gene in the majority of our JPS cohort. Loss of SMAD4 expression occurs in most polyps from SMAD4 mutation carriers, even those with missense germline mutations. SMAD4 loss in polyps is, however, not a feature of cases that are not caused by SMAD4 mutations, indicating that these polyps develop along a SMAD4-independent pathway. The morphology of polyps from SMAD4 mutation carriers is subtly different from other JPS polyps, notably including a more prominent epithelial component in the former.
Juvenile polyposis syndrome (JPS; MIM 174900) is a genetically heterogeneous disorder with a proportion of cases accounted for by mutations in the SMAD4/DPC4 gene on chromosome 18q21.1. 1,2 The main features of JPS are characteristic hamartomatous polyps throughout the gastrointestinal tract, and an increased risk of developing a gastrointestinal malignancy. Juvenile polyps range from a few millimeters to a few centimeters in size and are classically described as rounded with a hypercellular stroma, large mucin-filled cysts, lack of a smooth muscle core, and a flattened epithelium with no sign of hyperplasia. 3 Juvenile polyps also occur as part of other diseases such as Cowden, Bannayan-Zonana, and Gorlin syndromes, in which they occur with other syndrome-specific features. The gene that causes Cowden and Bannayan-Zonana syndromes has been shown to be PTEN (10q23.3), and germline PTCH (9q31) mutations cause Gorlin syndrome. PTEN and PTCH mutations have been excluded as the causative mutations in almost all JPS patients. 4,5
The SMAD4 gene has been shown to act as a tumor suppressor in JPS cases in which a germline mutation of SMAD4 has previously been demonstrated, with loss of the second copy leading to growth of the polyp. 6 Mutations and homozygous deletions of SMAD4, as well as allelic loss around 18q21.1, have been shown in sporadic cancers of the pancreas and colon, indicating the gene’s importance in the development of these tumors. 7,8 The SMAD4 protein acts as a cytoplasmic mediator in the transforming growth factor-β signaling pathway, by forming complexes with the phosphorylated receptor-regulated SMADs (SMAD2 and SMAD3). These complexes translocate from the cytoplasm to the nucleus, where association with DNA-binding proteins helps to regulate the transcription of genes involved in cell cycle and transcriptional regulation. 9 Despite being good candidates as they too are involved in the transforming growth factor-β signaling pathway, mutations in the other SMAD family members have not been found. 10
Trying to identify the genes which cause the remaining JPS cases unexplained by the SMAD4 gene relies on the certainty that it is indeed not the causative gene. Previously, an exon-by-exon mutation screen of SMAD4 in our JPS cohort has been performed using conformation-specific gel electrophoresis (CSGE) with 5 of 21 cases shown to have a SMAD4 mutation. 2,5 Such a technique, however, may only be at best 90% sensitive for base substitutions and small frameshifting changes, and would not detect large deletions. In addition, linkage analysis of 18q markers in informative pedigrees found four of eight families who provided good evidence against linkage, but four of eight families in whom linkage could not be excluded. 2
We have therefore undertaken a comprehensive analysis of the SMAD4 gene in our cohort of 26 familial and 18 sporadic JPS cases using a variety of techniques. In addition to the previously used methods, we have used several extra techniques. We have screened for germline deletions of whole or part of the SMAD4 gene by Southern blotting. Reverse transcriptase-polymerase chain reaction has been performed and used in the protein truncation test (PTT) that identifies nonsense germline mutations potentially missed by CSGE. F-SSCP (fluorescent single-stranded conformational polymorphism) analysis has been used to screen polymerase chain reaction products of all exons of SMAD4. We have used Western blotting to search for altered or reduced protein in the germline of those cases in which a lymphoblastoid cell line was available. Immunohistochemistry has been performed on all polyp and cancer material available from our cohort using a SMAD4 antibody in which detection levels have been shown to accurately mirror mutation status in pancreatic carcinomas. 11 Finally, to try and segregate the polyps by morphology according to SMAD4 mutation status, we performed a blinded analysis of hematoxylin and eosin (H&E)-stained sections from all available polyps and cancers.
The results of this study define the spectrum of germline changes in SMAD4 associated with juvenile polyposis, analyze their effects on protein expression in juvenile polyps and associated tumors, and demonstrate that juvenile polyps in SMAD4 mutation carriers have different features from those in JPS caused by other, unknown genes.
Materials and Methods
Patients from 26 different JPS families, and 18 sporadic cases were selected (families: 1, 5, 6, 10, 12, 14, 15, 16, 17, 18, 19, 20, 21, 22, MD, FT, KS, YC, GP, DM, WN, WH, JP1, JP2, JP7, and JP8; sporadics: BN, CV, 1204, CN, 1868, SM106, HG, SM397, MTW, SM524, 1262, BW, RV, 1469, LB, CR1, FD, and HR). Patients had five or more juvenile polyps, or any number of juvenile polyps and a known family history, and none had any phenotypic features of Cowden syndrome, Bannayan-Zonana syndrome, or Gorlin syndrome. Germline PTEN and PTCH mutations had been excluded from all patients. Peripheral blood samples were used to provide a source of DNA and, in a subset of cases, permanent lymphoblastoid cell lines were made or snap-frozen normal tissue was available. DNA was extracted from peripheral blood lymphocytes and cell lines using standard methods. Archival, paraffin-embedded tissue from polyps and/or cancers was obtained from as many cases as possible. Twenty-one patients had previously been screened for germline SMAD4 mutations by CSGE, 12 with mutations found in families 17, 20, AC/CF, and BL and sporadic SV (the latter three not rescreened in this study except for AC/CF which was included in the immunohistochemistry and morphological review).
For F-SSCP, exon-by-exon amplification of SMAD4, covering all coding sequence and intron/exon boundaries, was performed using previously reported primers 2 with added fluorescent 5′ and 3′ labels (FAM, HEX, or TET). Polymerase chain reactions were then diluted 1:50 with distilled water and combined with an internal size standard (Tamra 500; PE Applied Biosystems, Warrington, UK) and formamide. F-SSCP analysis at 20°C was performed using an ABI310 sequencer (PE Applied Biosystems). Fragments showing aberrant migration were re-amplified alongside normal samples using nonfluorescently labeled primers, purified using Qiaquick columns (Qiagen, Hilden, Germany) and then sequenced in both forward and reverse orientations using the ABI Big Dye terminator kit (PE Applied Biosystems).
For the PTT, RNA was extracted from fresh-frozen tissue using Tri-reagent (Sigma, Poole, UK) and from lymphoblastoid cell lines using either Tri-reagent or Fast-track RNA extraction kit (Invitrogen, Grodingen, The Netherlands). cDNA was synthesized using the First Strand Synthesis kit (Promega, Madison, WI) and polymerase chain reactions performed using the iF/iiiR or iF/vR primer pairs used for the Southern analysis, with the forward primer tagged with a T7 RNA-polymerase binding site and an in-frame start codon. In vitro coupled transcription-translation was performed on the tagged polymerase chain reaction products using the TNT rabbit reticulocyte lysate kit (Promega) incorporating α35-S methionine and the resulting proteins separated according to size on a 12.5% polyacrylamide resolving gel. Once fixed and dried, gels were exposed to film overnight and developed.
Southern blotting was performed using standard protocols. Briefly, overlapping cDNA probes were designed to cover the SMAD4 gene (GenBank accession number U44378) using Primer3 (http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi). Primer sequences are shown in Table 1 ▶ . The probes were gel-purified from a low-melting point gel using Geneclean II (Bio101, Anachem, UK), and labeled for the Southern blotting using α32-dCTP and Ready to Go beads (Amersham, UK). Ten μg of DNA was digested using the restriction enzymes HindIII, EcoRV (both four base cutters) and Sau3A1 (six base cutter), run on a 1% agarose gel for 18 to 20 hours and transferred to Hybond (Amersham, UK) membrane. Hybridization and subsequent washes were performed at 65°C.
Table 1.
Primers and Nucleotide Positions (Based on Genbank U44378 Sequence) Used for Southern Blotting and PTT
| Primer | Position | Sequence 5′-3′ |
|---|---|---|
| if | 1 | atggacaatatgtctattacga |
| iR | 317 | ttgtgaagatcaggccacct |
| ii F | 256 | ggtcggaaaggatttcctca |
| iiR | 601 | acagagctggggtgctgtat |
| iii F | 547 | cagcatccaccaagtaatcg |
| iiiR | 931 | ggaatgcaagctcattgtga |
| ivf | 895 | ggacattactggcctgttca |
| ivR | 1260 | acgccagcttctctgtcta |
| vF | 1207 | agtgaccacgcggtctttg |
| vR | 1659 | aaggttgtgggtctgcaatc |
For Western blotting, lymphoblastoid cell line pellets were lysed in 0.1 mol/L dithiothreitol/bromophenol blue and separated on a 15% resolving gel. After transfer to the polyvinylidene difluoride membrane (Millipore, Milton Keynes, UK) and blocking in 5% Marvel (Premier Brands, Stafford, UK), the primary antibodies were exposed to the membrane for 2 hours at room temperature. The anti-SMAD4 mouse monoclonal antibody (B8; Santa Cruz Biotechnology, Santa Cruz, CA) was diluted 1/100, and the control anti-β-actin mouse monoclonal antibody (Sigma) was diluted 1/1000, and exposed to the membrane both simultaneously and separately. A further control, anti-MLH1 (MLH1AB1; CN Biosciences, Luton, UK), was also used (there being no evidence of any changes in MLH1 expression in the normal tissues of any JPS patient). After detection of the proteins with enhanced chemiluminescence reagents (Amersham, UK) according to manufacturer’s instructions, the membrane was exposed to film for 1 and 5 minutes.
Immunohistochemistry was performed on 5-μm sections from all polyp and cancer tissue using the B8 nuclear staining antibody at 1/100 dilution, after baking of the sections for 20 minutes. After counterstaining with hematoxylin, the slides were examined for SMAD4 expression, with scoring of absent, weak, or strong.
In the morphological review, 6-μm paraffin sections of all polyps and cancers were H&E stained and examined by a histopathologist (NAW) with no previous knowledge of the SMAD4 mutation status of the material. The slides were scored for several categories including: 1) whether they resembled the classic juvenile polyp, particularly the predominance of epithelium or stroma; 2) the amount of inflammation; 3) whether dysplastic features (for example, adenomatous regions) or hyperplastic features were present in any region; 4) site of the polyp; and 5) any extra features such as colitis or cryptitis.
For statistical tests of association, Fisher’s exact and two-tailed t-tests were used.
Results
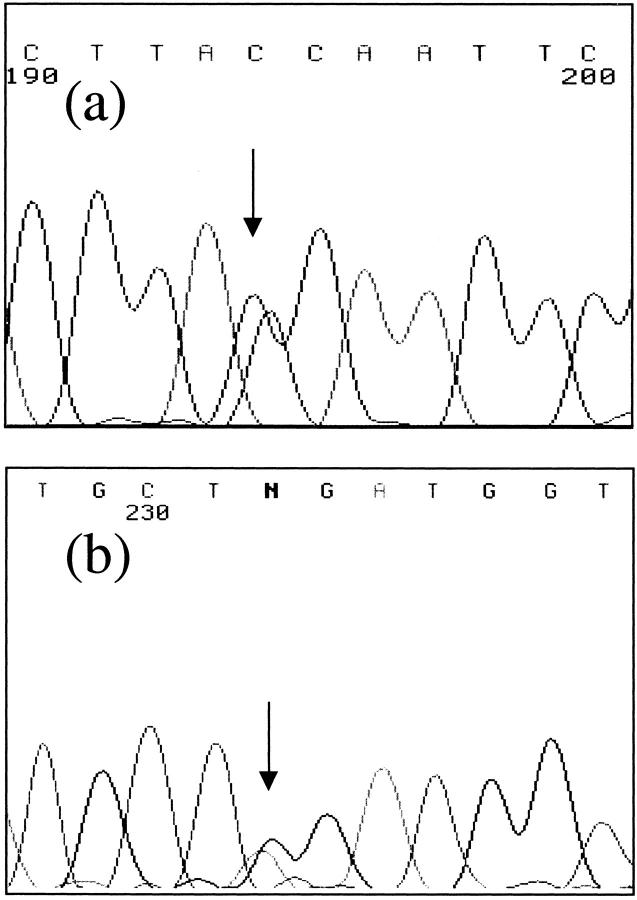
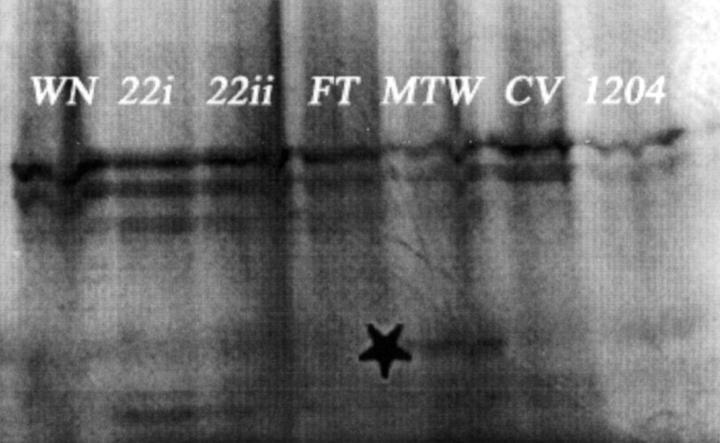
A summary of the germline SMAD4 mutations detected and the method used is shown in Table 2 ▶ . Only those cases in which a mutation was found or where polyps were assessed by immunohistochemistry are included in Table 2 ▶ . In addition to the mutations previously detected by CSGE, two further germline changes were found, both in patients previously analyzed. DNAs from all 26 families and 18 sporadic cases were used for the F-SSCP. In family 21, a G to A heterozygous change at the +1 splice donor site of intron 2 was found (Figure 1) ▶ . This mutation is predicted to abrogate the correct splicing of exons 2 and 3 of SMAD4 (although no mRNA was available to determine the precise effects). The mutation was seen in two affected sisters and their affected maternal aunt, but not in their unaffected father or 50 controls. The PTT was performed on 19 JPS individuals from whom cDNA was available (families 5, 6, 19, 22, MD, FT, GP, WN, JP1, JP2, JP7, and JP8; and sporadic cases CV, 1204, 1868, MTW, 1262, 1469, and HR), plus controls. No sample that had a known SMAD4 mutation was included in the PTT. One patient (MTW) had an extra PTT band corresponding to a truncated protein (Figure 2) ▶ . Sequencing of new products of MTW from exons 1 to 7 revealed a nonsense change Q180X in exon 4 (Figure 1) ▶ .
Table 2.
Summary of Germline SMAD4 Mutations and Immunohistochemistry
| Family/ID | F,S | Mutation (Nucleotide no.) | Predicted effect | Method | SMAD4 expression |
|---|---|---|---|---|---|
| 20 | f | 189–197dellins44† | stop codon 70 | CSGE* | 0/16 polyps |
| 17 | f | 1564–1565del | stop codon 525 | CSGE* | 0/37 polyps |
| SV‡ | s | 516–527del | stop codon 187 | CSGE* | − |
| BL‡ | s | c → a 1333 | R445X | CSGE* | − |
| AC/AF‡ | f | c → t 1083 | R361C | CSGE* | 1/6 polyps, 0/1 cancer |
| 21 | f | +1 splice donor intron 2 g → a | Abrogation of splicing | F-SSCP | 0/5 polyps |
| MTW | s | c → t 541 | Q180X | PTT | − |
| MD | f | No | 3/3 polyps | ||
| LB | s | No | 7/8 polyps | ||
| 12 | f | No | 3/3 polyps, 6/7 cancers | ||
| 15 | f | No | 19/19 polyps | ||
| 6 | f | No | 2/2 polyps 2/2 cancers | ||
| Wh | f | No | 3/3 polyps |
F,S denotes familial or sporadic case.
*Mutation previously reported 2,3 and confirmed in this study.
†Mutation previously reported as 189–197 deletion only. 2,3
‡Mutation previously reported 2,3 and not reanalyzed in this study.
Those families not shown had both no mutation detected and no tumours analysed by immunohistochemistry.
−, not done.
Association between loss of SMAD4 expression and SMAD4 mutation is highly significant (Fisher’s exact test, P ∼ 0.0).
Figure 1.
Sequence changes in patients 21 (a) and MTW (b). a: SMAD4 exon 2 reverse sequence of family 21 is shown with the +1 splice donor intron 2 c->t (g->a in forward) change arrowed. b: SMAD4 exon 4 reverse sequence of MTW is shown with the change arrowed, g->a (c->t in forward).
Figure 2.
Protein truncation test. Shown are the PTT results using primers iF and iiiR (Table 1) ▶ covering exons 1 to 7 from seven patients. The truncated protein in patient MTW is shown by an asterisk.
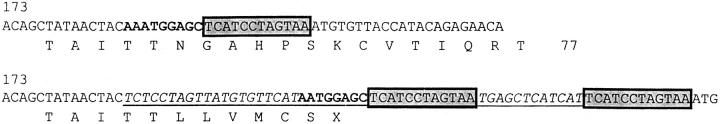
We reclassified one mutation previously detected using CSGE. The mutation of family 20 has been previously reported as 189-197del, an in-frame deletion of nine bases in exon 1. 12 This mutation has been further characterized as a most unusual and complex change, which comprises a net 1-bp deletion and 44-bp insertion, resulting in a stop at codon 70 (Figure 3) ▶ .
Figure 3.
Details of sequence change in germline of patient 20. Wild-type cDNA sequence is shown above and the mutant below (based on GenBank U44378). The insertion is underlined. The sequence in bold shows deletion of the A. The boxed sequences show a region duplicated in the mutant. The flanking sequences of the insertion are shown in italics and have no known similarity to any gene or Alu sequence.
In an attempt to detect large germline changes, Southern blotting was performed on 24 individuals from whom sufficient DNA was available (families: 1, 5, 6, 10, 12, 14, 16, 17, 19, 21, 22, MD, FT, KS, DM, and WN; sporadics: BN, CV, SM106, HG, MTW, SM524, 1469, and HR). Only one aberrant band was observed in one individual, under HindIII digestion using probe iv (details not shown). This change was not observed with any other restriction endonuclease or in the patient’s affected brother and is therefore most unlikely to be pathogenic, but may be a polymorphism changing a restriction site.
Western analysis was used as a further method of detecting truncating germline SMAD4 mutations. Previous analyses had demonstrated the anti-SMAD4 B8 antibody used to recognize an epitope in exon 5 (codons 68 to 108) (data not shown). A total of 12 lymphoblastoid cell lines from JPS patients were available for Western blotting (from families 19, 22, MD, FT, WN, JP1, JP2, JP7, and JP8; and sporadics CV, MTW, and HR). Detection of SMAD4 (64 kd) and β-actin (42 kd) was performed both simultaneously and separately. No bands of aberrant size were seen.
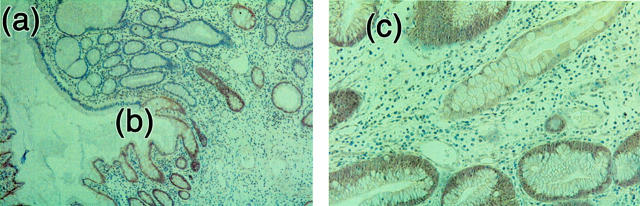
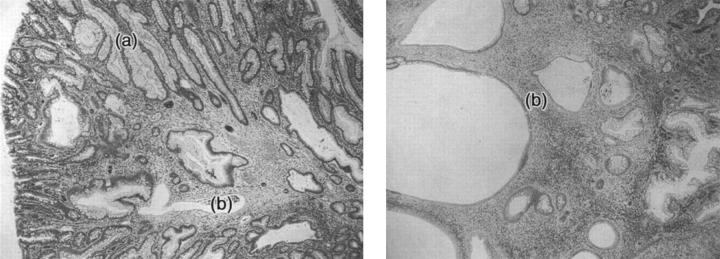
A total of 102 polyps and 10 cancers (from families 17, 20, 21, AC/CF, MD, 6, 12, and 15; and sporadics LB and Wh) were assessed for SMAD4 expression using immunohistochemistry with the B8 antibody. The results of the immunohistochemistry are summarized in Table 2 ▶ . In total, 37 of 38 juvenile polyps and 8 of 9 cancers from six SMAD4 wild-type families were positive for B8 staining, reflecting retention of SMAD4 expression. In stark contrast, only 1 of 64 polyps and 0 of 1 cancers from four SMAD4-mutant families were positive for B8, reflecting loss of SMAD4 expression in the great majority of tumors (Figure 4) ▶ . Thus, there was excellent concordance between our mutation screening and the immunohistochemistry. The results strongly suggest that disease in families without SMAD4 mutations develops along a SMAD4-independent pathway, whereas the families who have a SMAD4 germline mutation have lost the second copy of SMAD4, leading to growth of the polyp. These data corroborate results showing that SMAD4 acts as a tumor suppressor gene in JPS 6 and suggest that even missense changes are associated with loss of protein expression.
Figure 4.
Immunohistochemistry for SMAD4 using B8 antibody. Areas of expression of SMAD4 are brown and loss of SMAD4 expression areas are blue. Left: A juvenile polyp (original magnification, ×5) from a SMAD4 germline mutation carrier (family 20). Note the absence of staining in the polyp (a), and the border between the normal/polyp tissue where loss of expression begins (b). Right: A juvenile polyp (original magnification, ×20) from the SMAD4 mutation-negative family 12. Note the strong expression of SMAD4 indicated by brown staining (c).
A total of 101 H&E-stained polyp sections were reviewed to look for potential differences between polyps derived from patients who possess a germline SMAD4 mutation and polyps from patients who do not harbor SMAD4 mutations. A summary of the findings is shown in Table 3 ▶ . Polyps from patients without SMAD4 mutations were generally of the classical morphology, with expanded cysts, predominant stroma, and large numbers of inflammatory cells. 13 Although many polyps from SMAD4 mutation carriers had features of juvenile polyps, that is, expanded cysts and high levels of inflammation, polyps from mutation carriers were much more epithelial/nonclassical, with many long elongated crypts replacing the round cysts (Figure 5) ▶ . Polyps from both mutation carriers and nonmutation carriers had similar frequencies of hyperplasia/dysplasia (Table 3) ▶ . It was evident that there are morphological differences between polyps arising as a result of SMAD4 loss, and those arising via a SMAD4-independent pathway, making it possible to segregate tumors according to SMAD4 mutation status, as long as a large enough sample set from any family was available.
Table 3.
Summary of Morphology Results
| Patient | SMAD4 mutation? | Classical JP polyps | Hyperplastic/dysplastic/ adenomatous areas | Nonclassical JP polyps | Hyperplastic/dysplastic/ adenomatous areas | Notes |
|---|---|---|---|---|---|---|
| AF/CF | Yes | 3 /6 | All 3 with dysplasia and hyperplasia | 3 /6 | 2 with hyperplasia, 1 without | Cryptitis in 3. All very epithelial. |
| 20 | Yes | 5 /14 | All 5 with areas of hyperplasia | 9 /14 | All complex. 7/9 very dysplastic and/or adenomatous, 4/9 hyperplastic (inc. 2 which were not dysplastic) | Very elongated, dense crypts. Larger polyps very epithelial. Smooth muscle in 7. |
| 17 | Yes | 6 /37 | All 6 with hyperplasia | 31 /37 | All very hyperplastic. 12/31 with dysplasia | Very elongated, dense crypts. Larger polyps very epithelial |
| 21 | Yes | 5 /5 | 4/5 classical with hyperplasia; 1 with dysplasia and adenomatous region. | 0 /5 | N/A | Small |
| MD | No | 3 /3 | 2/3 had region of hyperplasia, one of these with small adenomatous region | 0 /3 | N/A | Very inflamed, prominent stroma |
| LB | No | 6 /8 | 5/6 with region of hyperplasia, 2/6 with region of dysplasia | 2 /8 | Note: both small bowel. 2/2 with region of hyperplasia | Very inflamed, prominent stroma, large cysts. Smooth muscle in 4. |
| WN | No | 6 /6 | 2 with hyperplasia and dysplasia, 3 with hyperplasia | 0 /6 | N/A | Very inflamed, prominent stroma, large cysts. Granuloma in 2 |
| 12 | No | 10 /10 | Some regions hyperplastic-like 6/10 with dysplasia | 0 /6 | N/A | Very inflamed, prominent stroma, large cysts. |
| 15 | No | 17 /19 | 11 with region of hyperplasia, 2 with dysplasia | 2 /19 | Very small polyps | Very inflamed, prominent stroma with large cysts. |
| 6 | No | 2 /2 | No hyperplasia | 0 /2 | N/A | Very inflamed, prominent stroma, large cysts |
| Wh | No | 3 /3 | No hyperplasia | 0 /3 | N/A | Very inflamed, prominent stroma, large cysts |
All polyps were from colorectum unless stated otherwise. The overall epithelial content was far more pronounced in the polyps of SMAD4 mutation carriers than those without mutations, and consequently the number of classical juvenile polyps was significantly lower in the mutation carriers than in the nonmutation carriers (Fisher’s exact test, P < 1 × 10−10). The size of polyps was significantly greater in SMAD4 mutation carriers versus noncarriers (mean 15.96 mm versus 9.83 mm, t = 4.98, v = 102, P < 0.001). There was only a borderline difference between the frequency of hyperplasia/dysplasia in polyps from mutation carriers and noncarriers (Fisher’s exact test, P = 0.06), although the spatial extent of hyperplasia and dysplasia appeared to be greater in SMAD4 mutation carriers.
Figure 5.
H&E-stained slides of juvenile polyps. Left: A juvenile polyp (original magnification, ×2.5) from a SMAD4 mutation carrier (family 20). Note areas that look hyperplastic (a) and areas of classical juvenile polyp morphology, with expanded cysts and normal epithelium (b). Right: A classical juvenile polyp (original magnification, ×2.5) from a SMAD4 mutation-negative patient (family MD) with morphology of type (b).
Discussion
We have performed a comprehensive analysis of SMAD4 mutation and expression in juvenile polyposis. Germline SMAD4 mutations undoubtedly account for a minority of JPS cases. Most germline mutations are detectable by F-SSCP analysis or CSGE, and sequencing. After an initial screen with CSGE detected five mutations, we used F-SSCP and detected one extra change, a novel splice site mutation at the +1 donor site of intron 2. One additional germline SMAD4 mutation, not detected by CSGE or F-SSCP under the conditions we used, was found using PTT. Southern analysis detected no large-scale mutations. Western analysis found no evidence of truncated proteins. None of our patients with known truncating germline mutations C-terminal to the B8 antibody epitope was analyzed by this technique.
Nevertheless, our results using immunohistochemistry suggest that cryptic SMAD4 mutations are very rare. Just 1 of 38 polyps from patients without a germline SMAD4 mutation showed loss of protein expression, confirming the results of our mutation detection and showing that these tumors grow along a genetic pathway that does not involve SMAD4, at least in the early stages. By contrast, almost all polyps and cancers from our known SMAD4 mutation carriers had absent protein expression. It seems, therefore, that if the wild-type SMAD4 allele is generally deleted 6,14 as the second hit that initiates the growth of JPS polyps, the remaining mutant protein is unstable. Although not unexpected for truncated proteins, it seems that even if the germline change is of a missense type, protein instability generally results.
A previous study 11 had found a strong association between SMAD4/DPC4 changes and absent protein expression in pancreatic cancer. However, the great majority of tumors studied had homozygous deletions of SMAD4; only three tumors with SMAD4 loss had small-scale mutations, and it was not clear whether these were of missense or truncating types. Our patient with a germline missense mutation carried an R361C change. This mutation maps to the loop/helix domain in the C-terminal of SMAD4 and has also been found in a sporadic colorectal. 8 The functional effects of R361C have been well evaluated 15 and it prevents both hetero- and homo-oligomerization of SMAD4. Our results also suggest that SMAD4 protein, such as R361C, which is not bound into a complex, is degraded or unstable in vivo. This is upheld by data showing that missense mutations in the N-terminal MH1 region of SMAD4 cause rapid degradation of the protein in vitro. 16,17
Previous studies have found germline SMAD4 mutations in ∼25 to 60% of JPS cases, but one common mutation (4-bp deletion, codons 414 to 416, stop at codon 434) accounts for many patients in some studies. Howe and colleagues 1 used SSCP analysis and sequencing to find mutations in five of nine patients studied. All of these were frameshift changes, including three examples of the 4-bp deletion and two other mutations producing stop codons at 235 and 350. Friedl and colleagues 18 used direct sequencing in 11 cases to detect the common 4-bp deletion in two patients and a codon 277 frameshift in one another. Roth and colleagues 19 used direct sequencing in seven JPS cases to find one missense change (codon 353), one nonsense mutation (codon 177), and one patient with the common 4-bp deletion. Kim and colleagues 20 found three SMAD4 mutations in five patients using SSCP analysis, comprising a nonsense change at codon 388 and two missense changes at codons 390 and 361. We ourselves have found 7 mutations in 44 cases (summarized in Table 2 ▶ ). Thus, germline SMAD4 mutations seem to occur most commonly, but not exclusively, after codon 200, affecting the C-terminal of the gene that is involved in trimerization of the SMAD4 protein. Nonsense and frameshift changes predominate, but pathogenic missense mutations and splice variants can occur. These data from JPS are consistent with the spectrum of somatic mutations found in colorectal and pancreatic cancers, with the exception of the higher frequency of homozygous deletions found in the sporadic tumors. 7
Apart from the distinct phenotype of Cowden syndrome, in which JPS polyps occur, genotype-phenotype associations are difficult to analyze in JPS, because the number of families is relatively small. Juvenile polyps in Cowden syndrome are said to be indistinguishable from those in classical JPS, although no formal assessment has been made. We wondered whether the morphology of JPS polyps might be different in patients with and without germline SMAD4 mutations and found that this was indeed the case. All families had polyps with some dysplasia or hyperplasia, consistent with the juvenile polyp in JPS being a premalignant lesion. However, SMAD4 mutation carriers’ polyps had less prominent stroma and a richer epithelial component than the classical juvenile polyps of those patients without SMAD4 mutations. These results are consistent with suggestions that some carriers of SMAD4 mutations may have higher cancer risk than patients without SMAD4 mutations. 1 Polyp morphology is, however, variable within the same individual and between patients from the same family, so that it cannot be used reliably for any one polyp as an indicator of the likelihood of a germline SMAD4 mutation.
Finally, our data show that with the confident exclusion of SMAD4 as the causative gene, with the aid of mutation screening, linkage analysis, and immunohistochemistry, the SMAD4 mutation-negative cohort is as homogenous as possible. Without these false-negatives the identification of new JPS genes will be facilitated. A combination of mutation screening, immunohistochemistry, and morphological assessment is reliable for eliminating families with SMAD4 mutations from the analysis. Moreover, families and individuals may be selected for SMAD4 mutation detection in the diagnostic molecular pathology laboratory using immunohistochemistry as an initial screen. With the recent discovery of BMPRIA/ALK3 mutations in some JPS patients, 21 this becomes even more important.
Acknowledgments
We thank the patients and their clinicians for supply samples and data; George Elia (ICRF) for performing the immunohistochemistry; Ian Frayling for providing valuable data regarding the mutation in family 20; Caroline Hill and colleagues for providing date regarding the anti-SMAD4 B8 antibody epitope; and the Equipment Park, Imperial Cancer Research Fund for the important role of sequencing.
Footnotes
Address reprint requests to Kelly L. Woodford-Richens, Molecular and Population Genetics Laboratory, Imperial Cancer Research Fund, London WC2A 3PX, UK. E-mail: woodford@icrf.icnet.uk.
Supported by an EU Biomed2 grant (K.W.-R).
References
- 1.Howe JR, Roth S, Ringold JC, Summers RW, Jarvinen H, Sistonen P, Tomlinson IPM, Houlston RS, Bevan S, Mitros FA, Stone EM, Aaltonen LA: Mutations in the SMAD4/DPC4 gene in juvenile polyposis. Science 1998, 280:1086-1088 [DOI] [PubMed] [Google Scholar]
- 2.Houlston R, Bevan S, Williams A, Young J, Dunlop M, Rozen P, Eng C, Markie D, Woodford-Richens K, Rodriguez-Bigas M, Leggett B, Neale K, Phillips R, Sheridan E, Hodgson S, Iwama T, Eccles D, Bodmer W, Tomlinson I: Mutations in DPC4 (SMAD4) cause juvenile polyposis syndrome, but only account for a minority of cases. Hum Mol Genet 1998, 7:1907-1912 [DOI] [PubMed] [Google Scholar]
- 3.Veale AM, McColl I, Bussey HJ, Morson BC: Juvenile polyposis coli. J Med Genet 1966, 3:5-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marsh DJ, Roth S, Lunetta KL, Sistonen P, Dahia PLM, Hemminki A, Zheng Z, Caron S, van Orsouw NJ, Bodmer WF, Cottrell SE, Dunlop MG, Eccles D, Hodgson SV, Jarvinen H, Kellokumpu I, Markie D, Neale K, Phillips R, Rosen P, Syngal S, Vijg J, Tomlinson IPM, Aaltonen LA, Eng C: Exclusion of PTEN/MMAC1/TEP1 and 10q22–24 as the susceptibility locus for juvenile polyposis syndrome (JPS). Cancer Res 1997, 57:5017-5020 [PubMed] [Google Scholar]
- 5.Woodford-Richens K, Bevan S, Churchman M, Dowling B, Norbury G, Hodgson S, Desai D, Neale K, Phillips KS, Young J, Leggett B, Dunlop M, Rozen P, Eng C, Markie D, Rodriguez-Bigas MA, Sheridan E, Iwama T, Eccles D, Kim JC, Kim KM, Bodmer WF, Tomlinson IPM, Houlston RS: Analysis of genetic and phenotypic heterogeneity in juvenile polyposis. Gut 1999, 9:9-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woodford-Richens K, Williamson J, Bevan S, Young J, Leggett B, Frayling I, Thway Y, Hodgson S, Kim JC, Iwama T, Novelli M, Sheer D, Poulsom R, Wright N, Houlston R, Tomlinson I: Allelic loss at SMAD4 in polyps from juvenile polyposis patients and use of fluorescence in situ hybridization to demonstrate clonal origin of the epithelium. Cancer Res 2000, 60:2477-2482 [PubMed] [Google Scholar]
- 7.Hahn SA, Hoque AT, Moskaluk CA, da Costa LT, Schutte M, Rozenblum E, Seymour AB, Weinstein CL, Yeo CJ, Hruban RH, Kern SE: Homozygous deletion map at 18q21.1 in pancreatic cancer. Cancer Res 1996, 56:490-494 [PubMed] [Google Scholar]
- 8.Thiagalingam S, Lengauer C, Leach FS, Schutte M, Hahn SA, Overhauser J, Willson JK, Markowitz S, Hamilton SR, Kern SE, Kinzler KW, Vogelstein B: Evaluation of candidate tumour suppressor genes on chromosome 18 in colorectal cancers. Nat Genet 1996, 13:343-346 [DOI] [PubMed] [Google Scholar]
- 9.de Caestecker MP, Piek E, Roberts AB: Role of transforming growth factor-beta signaling in cancer. J Natl Cancer Inst 2000, 92:1388-1402 [DOI] [PubMed] [Google Scholar]
- 10.Bevan S, Woodford-Richens K, Rozen P, Eng C, Young J, Dunlop M, Neale K, Phillips R, Markie D, Rodriguez-Bigas M, Leggett B, Sheridan E, Hodgson S, Iwama T, Eccles D, Bodmer W, Houlston R, Tomlinson I: Screening SMAD1, SMAD2, SMAD3, and SMAD5 for germline mutations in juvenile polyposis syndrome. Gut 1999, 45:406-408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilentz RE, Su GH, Dai JL, Sparks AB, Argani P, Sohn TA, Yeo CJ, Kern SE, Hruban RH: Immunohistochemical labeling for dpc4 mirrors genetic status in pancreatic adenocarcinomas: a new marker of DPC4 inactivation. Am J Pathol 2000, 156:37-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woodford-Richens K, Bevan S, Churchman M, Dowling B, Jones D, Norbury CG, Hodgson SV, Desai D, Neale K, Phillips RK, Young J, Leggett B, Dunlop M, Rozen P, Eng C, Markie D, Rodriguez-Bigas MA, Sheridan E, Iwama T, Eccles D, Smith GT, Kim JC, Kim KM, Sampson JR, Evans G, Tejpar S, Bodmer WF, Tomlinson IP, Houlston RS: Analysis of genetic and phenotypic heterogeneity in juvenile polyposis. Gut 2000, 46:656-660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morson B, Dawson I, Day D, Jass J, Price A, Williams G: Morson and Dawson’s Gastrointestinal Pathology, ed 3. Oxford, Blackwell Scientific Publications, 1990
- 14.Takaku K, Miyoshi H, Matsunaga A, Oshima M, Sasaki N, Taketo MM: Gastric and duodenal polyps in Smad4 (Dpc4) knockout mice. Cancer Res 1999, 59:6113-6117 [PubMed] [Google Scholar]
- 15.Shi Y, Hata A, Lo RS, Massague J, Pavletich NP: A structural basis for mutational inactivation of the tumour suppressor Smad4. Nature 1997, 388:87-93 [DOI] [PubMed] [Google Scholar]
- 16.Xu J, Attisano L: Mutations in the tumor suppressors Smad2 and Smad4 inactivate transforming growth factor beta signaling by targeting Smads to the ubiquitin-proteasome pathway. Proc Natl Acad Sci USA 2000, 97:4820-4825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moren A, Itoh S, Moustakas A, Dijke P, Heldin CH: Functional consequences of tumorigenic missense mutations in the amino-terminal domain of Smad4. Oncogene 2000, 19:4396-4404 [DOI] [PubMed] [Google Scholar]
- 18.Friedl W, Kruse R, Uhlhaas S, Stolte M, Schartmann B, Keller KM, Jungck M, Stern M, Loff S, Back W, Propping P, Jenne DE: Frequent 4-bp deletion in exon 9 of the SMAD4/MADH4 gene in familial juvenile polyposis patients. Genes Chromosom Cancer 1999, 25:403-406 [PubMed] [Google Scholar]
- 19.Roth S, Sistonen P, Salovaara R, Hemminki A, Loukola A, Johansson M, Avizienyte E, Cleary KA, Lynch P, Amos CI, Kristo P, Mecklin JP, Kellokumpu I, Jarvinen H, Aaltonen LA: SMAD genes in juvenile polyposis. Genes Chromosom Cancer 1999, 26:54-61 [DOI] [PubMed] [Google Scholar]
- 20.Kim IJ, Ku JL, Yoon KA, Heo SC, Jeong SY, Choi HS, Hong KH, Yang SK, Park JG: Germline mutations of the dpc4 gene in Korean juvenile polyposis patients. Int J Cancer 2000, 86:529-532 [DOI] [PubMed] [Google Scholar]
- 21.Howe JR, Bair JL, Sayed MG, Anderson ME, Mitros FA, Petersen GM, Velculescu VE, Traverso G, Vogelstein B: Germline mutations of the gene encoding bone morphogenetic protein receptor 1A in juvenile polyposis. Nat Genet 2001, 28:184-187 [DOI] [PubMed] [Google Scholar]