Abstract
Inflammatory myofibroblastic tumor (IMT) is a rare, but distinctive mesenchymal neoplasm composed of fascicles of bland myofibroblasts admixed with a prominent inflammatory component. Genetic studies of IMTs have demonstrated chromosomal abnormalities of 2p23 and rearrangement of the anaplastic lymphoma kinase (ALK) gene locus. In a subset of IMTs, the ALK C-terminal kinase domain is fused with a tropomyosin N-terminal coiled-coil domain. In the current study, fusion of ALK with the clathrin heavy chain (CTLC) gene localized to 17q23 was detected in two cases of IMT. One of these cases exhibited a 2;17 translocation in addition to other karyotypic anomalies [46,XX,t(2;17)(p23;q23),add(16)(q24)].
Inflammatory myofibroblastic tumors (IMTs) occur in the soft tissues as well as the viscera and have a proclivity for children and young adults. 1 Histopathologically, IMTs are composed of a variable admixture of bland-appearing spindle-shaped cells (myofibroblasts), inflammatory cells (lymphocytes, eosinophils, and plasma cells), and collagen fibers. The nosology of IMTs has been controversial. IMTs have also been referred to as atypical fibromyxoid tumors, pseudosarcomatous fibromyxoid tumors, plasma cell granulomas, pseudosarcomatous myofibrotic proliferations, postoperative spindle cell nodules, and inflammatory pseudotumors. 1 Many of these terms suggest a nonneoplastic origin. In contrast, recent genetic studies have demonstrated clonal chromosomal aberrations involving 2p23 2 and associated TPM3-ALK or TPM4-ALK fusion gene transcripts in a subset of these lesions, 3 supporting IMT as a neoplastic process. In this study, a novel chimeric transcript involving the ALK and CLTC genes was detected in two cases of IMT. These findings further expand the variety of ALK fusions in IMT and their parallels with the variant ALK fusions described in anaplastic large cell lymphoma (ALCL).
Materials and Methods
Representative fresh and paraffin-embedded tissues were obtained from an IMT of the neck of a 3-year-old female at the University of Nebraska Medical Center (UNMC) (case 1) and an IMT of the pelvis of a 37-year-old male at Memorial Sloan-Kettering Cancer Center (MSKCC) (case 2). UNMC and MSKCC tumor procurement was performed under Institutional Review Board no. 281-96 and IRB no. 90-024, NCI-P30 CA36727, and NIH PO1 CA47179, respectively. These two cases were identified as follows: 68 cases from UNMC, MSKCC, and St. Jude Children’s Research Hospital with a histological diagnosis of IMT were examined by ALK immunohistochemistry (described below), of which 41 were ALK immunostain-positive. Of the 41 positive cases, 6 exhibited a granular ALK-staining pattern that we demonstrate herein to be associated with the expression of CLTC-ALK (see Results). Unfortunately, other than for the two cases reported here, suitable RNA for reverse transcriptase-polymerase chain reaction (RT-PCR) analysis could not be obtained from the formalin-fixed samples available on the remaining cases to demonstrate expression of CLTC-ALK transcripts (manuscript in preparation).
ALK Immunohistochemistry
ALK immunostaining was performed using monoclonal mouse anti-human antibody ALK-1 (DAKO, Carpinteria, CA) at a dilution of 1:25 and polyclonal rabbit anti-human ALK-11 4 at a dilution of 1:200 after heat-induced epitope retrieval. Antibody detection was achieved using a modified avidin-biotin peroxidase complex method.
Cytogenetic Analysis
A 0.25-cm 3 sterile, representative tissue sample from case 1 was submitted for cytogenetic analysis. Standard culture and harvesting procedures were performed, as described previously. 5
Fluorescence in Situ Hybridization (FISH)
FISH studies using 2p23 (ALK) breakpoint spanning and flanking probes (Vysis, Inc., Downers Grove, IL) were executed on metaphase preparations of case 1 and on cytological touch preparations of both cases according to the manufacturer’s instructions. Examination for the presence of split signals and image preparation were performed as previously described. 6
RT-PCR Analysis and Rapid Amplification of cDNA Ends (RACE) Analysis
Total RNA was extracted from fresh or frozen tissue using Trizol reagent (Life Technologies, Inc., Gaithersburg, MD) or RNA Stat-60 (Tel-Test, Inc., Friendswood, TX). To exclude the presence of a TPM-ALK fusion transcript, RT-PCR studies were performed on both cases using a consensus TPM3/TPM4-FWD primer and an ALK-REV primer (5′-GGAAAAGACAATTGATGAC and 5′-GCAGTAGTTGGGGTTGTAGTC, respectively). Methodological details of the RT-PCR assays were essentially as previously described. 6 The PCR products were electrophoresed in a 2% NuSieve agarose gel (FMC Bioproducts, Rockland, ME) and visualized by ethidium bromide staining.
RACE studies were performed using the 5′RACE system from Life Technologies, Inc. (Rockville, MD) according to the manufacturer’s instructions. Briefly, after annealing of an ALK-specific reverse primer (ALK/RT, 5′-TTCAGGCAGCGTCTTCACAG), 5 μg of RNA from each case was used for cDNA synthesis by SuperScript II reverse transcriptase (Life Technologies, Inc., Rockville, MD). The RNA was then degraded with RNase and the cDNA purified using a Glass MAX Spin Cartridge. Subsequently, the purified cDNA was tailed with dCTP using TdT and amplified by PCR using the manufacturer’s Abridged anchor primer as forward primer and a nested ALK-specific reverse primer (ALK/TK2, 5′-GGCTTGGGTC GTTGGGCATTC). Reamplification of the primary PCR product was completed using the manufacturer’s Abridged universal anchor primer and a nested ALK-specific reverse primer (ALK-3′, 5′-CGAGGT GCGGAGCTTGCTCAGC). The products were analyzed on agarose gels. Sharp product bands were subjected to direct sequencing, whereas nondiscrete products were cloned and multiple independent clones were sequenced.
After completion of the RACE studies, a hemi-nested RT-PCR method was used to confirm the presence of a CLTC-ALK fusion gene in both cases. Thirty-five thermal cycles were used for the first-round PCR with CLTC-FWD primer (5′-TTAGATGCTTCAGAATC ACTG) and ALK-REV primer at the following temperatures: 95°C for 1 minute, 60°C for 1 minute, 72°C for 1 minute, with a final extension of 72°C for 10 minutes. Second-round PCR was performed with CLTC-FWD primer and ALK-3′ primer using the same PCR conditions described for the first round. The amplified fragments were identified by gel electrophoresis and ethidium bromide staining, followed by direct sequencing of the product band. The integrity of the tumor RNAs was assessed by an independent amplification using primers for the ubiquitously expressed hypoxanthine phosphoribosyl-transferase (HPRT) or phosphoglycerate kinase (PGK) genes. A negative control devoid of a template and a negative control lacking reverse transcriptase were included in all RT-PCR studies.
Results
Histology and Immunohistochemistry
Cases 1 and 2 were composed of spindle-shaped myofibroblasts arranged focally in fascicular or storiform-like patterns, admixed with lymphocytes and plasma cells. The presence of vimentin, smooth muscle actin, muscle-specific actin, and desmin immunoreactivity and/or prominent, dilated rough endoplasmic reticulum and abundant thin microfilaments forming dense bodies beneath cell membranes ultrastructurally, confirmed the myofibroblastic nature of the lesions (not shown). Case 1 exhibited strong granular cytoplasmic immunoreactivity for both ALK-1 and ALK-11 in 70 to 80% of the myofibroblasts (Figure 1A) ▶ . Case 2 was not immunoreactive for ALK-1, but did show faint-to-moderate immunoreactivity for ALK-11 localized to the cytoplasm of many myofibroblasts (not shown). In our anecdotal experiences, as well as those of a number of other investigators (S. W. Morris, unpublished observations), the ALK-1 monoclonal antibody is substantially less sensitive for detection of low-level ALK fusion protein expression than the ALK-11 polyclonal antibody; the discordant staining of case 2 observed with these two antibodies likely reflects these sensitivity differences, given that our FISH and RT-PCR studies clearly revealed this case to contain ALK rearrangement and fusion (below).
Figure 1.
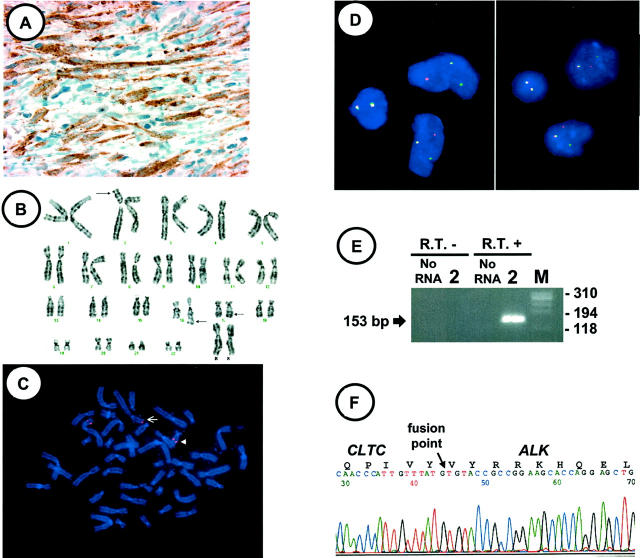
A: ALK-11 immunostaining performed on case 1 shows granular cytoplasmic staining confined to the myofibroblastic cells. B: Representative karyotype of case 1 (arrows indicate breakpoints). C: FISH performed on a destained preparation of the metaphase cell in B with a 2p23 breakpoint spanning probe shows a signal split [der(2), arrow, der(17), arrowhead]. Bicolor FISH studies performed on cytological touch preparations of cases 1 (D, left) and 2 (D, right) with probes flanking the 2p23 breakpoint show a split of the Spectrum Orange and Spectrum Green signals in two cells each, indicating disruption of ALK. Normal FISH signals are present in the remaining morphologically smaller and rounder cells (inflammatory cells). E: Portion of agarose gel showing products of second step hemi-nested RT-PCR for CLTC-ALK for case 2. The first step used the CLTC-FWD primer with ALK-REV; the second step used the CLTC-FWD primer with the ALK-3′ primer (see Materials and Methods for primer sequences). No RNA and no R.T. controls refer to lack of RNA or lack of reverse transcriptase, respectively, in the first step. M: portion of size marker PhiX174/HaeIII. F: CLTC-ALK fusion junction sequence from case 1. The identity of all transcripts was confirmed by sequencing.
Cytogenetic Analysis
Case 1 exhibited the following complements: 46,XX,t(2;17)(p23;q23),add(16)(q24)[5]/92,idemx2[1]/46,XX[4] (Figure 1B) ▶ . In case 2, material suitable for cytogenetic analysis was not available.
FISH
Case 1 metaphase cell FISH confirmed the presence of a 2;17 translocation involving the ALK locus (Figure 1C) ▶ . Case 1 and 2 interphase cell FISH revealed a split of one set of the two-color probe signals, indicating a disruption of the 2p23 breakpoint (ALK) on one chromosome 2 homologue in 52 and 46% of the cells, respectively (Figure 1D) ▶ .
RT-PCR and RACE Analyses
RT-PCR analysis was negative for a TPM3-ALK or TPM4-ALK fusion gene product in both cases (data not shown). Rather, 5′ RACE analysis of case 1 revealed an ALK fusion with the clathrin heavy chain gene (CLTC) localized to 17q23. 7 This CLTC-ALK fusion incorporates 1634 residues of the 1675-amino acid clathrin heavy chain. Hemi-nested RT-PCR analysis confirmed the presence of a CLTC-ALK fusion gene product in case 1 and demonstrated the identical fusion product in case 2 (Figure 1, E and F) ▶ . The ALK and CLTC breakpoints in these IMTs were identical to those recently reported in CLTC-ALK fusions in ALCL. 8
Discussion
Inflammatory pseudotumors have been reported under a wide variety of descriptive terms, with IMT recently gaining wide acceptance. The histopathogenesis of IMT has historically been a subject of dispute. In part, this is because of frequently overlapping clinical and pathological features with reactive processes in some cases, and sarcomatous processes in others. 1 Recent studies, however, demonstrating clonal karyotypic rearrangements and tumor-specific fusion gene products, provide evidence for a monoclonal, neoplastic origin for IMT. 2,3,9-13
Here, we describe a case of IMT cytogenetically exhibiting a 2;17 translocation similar to that reported recently by Griffin and colleagues. 2 Additional studies including FISH and immunohistochemistry confirmed that this tumor, as well as a separate IMT lesion included in the current study, featured ALK rearrangements and ALK expression. Initially, RT-PCR studies were performed to determine whether previously defined fusion transcripts involving ALK and TPM3 or TPM4 characterized either of the IMTs but these were negative. These findings prompted us to perform 5′ RACE studies in an effort to identify the fusion partner of ALK in these two cases. These studies revealed a fusion transcript involving ALK and the clathrin heavy chain gene (CLTC) in both tumors, not previously described in IMT.
Clathrin is the major protein constituent of the coat that surrounds the cytoplasmic face of the organelles (coated vesicles) mediating selective protein transport. 14 Clathrin coats are involved in receptor-mediated endocytosis, localization of resident membrane proteins to the trans-Golgi network, and transport of proteins to the lysosome/vacuole. 15-17 Recently, clathrin has also been immunodetected in the mitotic spindle, suggesting a novel role for clathrin in mitosis or a novel regulatory mechanism for localization of clathrin in mitotic cells. 18
Clathrin is a three-legged molecule, termed a triskelion, composed of heavy and light chains. Two clathrin heavy chain genes exist in the genome, clathrin heavy chain gene (CLTC), which has been localized to 17q23, 7 and clathrin heavy chain polypeptide-like gene (CLTCL) that has been localized to 22q11.2. 19 In the present IMT cases, the fusion point in the CLTC transcript was close to its 3′ end, thus conserving nearly all of the clathrin heavy chain, including the motifs responsible for triskelion assembly. Presumably, the clathrin moiety in this CLTC-ALK fusion promotes constitutive activation and relocalization of the ALK kinase domain from its normal position at the inner surface of the cell membrane in neural cells to the cytoplasm of myofibroblastic cells.
The sequence of the CLTC-ALK fusion transcript identified in the IMTs in this study is identical to that recently described in ALCL. The partner gene in this ALK fusion was originally inadvertently misidentified as CLTCL, 8 possibly because of changes in the designation of the corresponding GenBank accession NM[lowhy]004859. The CLTC nucleotides adjacent to the CLTC-ALK fusion junction are unlike CLTCL allowing distinction between the two genes. Although cytogenetic studies were not available in the report by Touriol and colleagues, 8 it is likely that the lymphomas they studied contain a t(2;17)(p23;q23). Analogous to the CLTC-ALK fusion-positive ALCL, the ALK immunostaining in our IMT cases was confined to the cytoplasm of the tumor cells and was characterized by a granular appearance.
CLTC-ALK is the second known fusion oncogene that transforms, in vivo, both mesenchymal and lymphoid human cell lineages. A TPM3-ALK fusion oncogene identical to that observed in some ALCL was identified by Lawrence and colleagues 3 in two IMT cases. The most common ALK fusion gene partner in ALCL, detected in ∼80% of cases, is nucleophosmin (NPM). The NPM-ALK fusion gene results from a t(2;5)(p23;q35). 20 Interestingly, an NPM-ALK fusion oncogene has not yet been identified in IMT.
In conclusion, the CLTC-ALK fusion oncogene represents a novel mechanism of ALK activation in IMT and demonstrates that, similar to ALCL, the fusion partners of the ALK gene in IMT are diverse. ALK protein expression is an independent predictor of survival and serves as a useful biological marker of a specific disease entity within the spectrum of ALCL. Additional studies are warranted to determine whether ALK protein expression is likewise associated with specific clinicopathological traits in IMT.
Acknowledgments
We thank Drs. James Neff, Thomas Seemayer, Sonny Johansson, and Warren Sanger for their valuable comments; and X. Cui and J. Liu for their expert technical assistance.
Footnotes
Address reprint requests to Julia A. Bridge, M.D., Department of Pathology, 983135 Nebraska Medical Center, Omaha, NE 68198-3135. E-mail: jbridge@unmc.edu.
Supported by National Cancer Institute grants NCI-P30 CA36727, the John A. Wiebe Jr. Children’s Health Care Fund and National Childhood Cancer Foundation (to J. A. B.), National Cancer Institute grant NCI CA69129 and CORE grant CA21765 (to S. W. M.), the American Lebanese Syrian Associated Charities, the St. Jude Children’s Hospital (to S. W. M.), and the Fundaçao de Amparo à Pesquisa do Estado de Sao Paulo (to G. W. B. C.).
G. W. B. C. is presently at the Department of Medicine, Universidade Federal de Sao Paulo, Sao Paulo, Brazil.
References
- 1.Coffin CM, Watterson J, Priest JR, Dehner LP: Extrapulmonary inflammatory myofibroblastic tumor (inflammatory pseudotumor). A clinicopathologic and immunohistochemical study of 84 cases. Am J Surg Pathol 1995, 19:859-872 [DOI] [PubMed] [Google Scholar]
- 2.Griffin CA, Hawkins AL, Dvorak C, Henkle C, Ellingham T, Perlman EJ: Recurrent involvement of 2p23 in inflammatory myofibroblastic tumors. Cancer Res 1999, 59:2776-2780 [PubMed] [Google Scholar]
- 3.Lawrence B, Perez-Atayde A, Hibbard MK, Rubin BP, Dal Cin P, Pinkus JL, Pinkus GS, Xiao S, Yi ES, Fletcher CDM, Fletcher JA: TPM3-ALK and TPM4-ALK oncogenes in inflammatory myofibroblastic tumors. Am J Pathol 2000, 157:377-384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morris SW, Naeve C, Mathew P, James PL, Kirstein MN, Cui X, Witte DP: ALK, the chromosome 2 gene locus altered by the t(2;5) in non-Hodgkin’s lymphoma, encodes a neural receptor tyrosine kinase that is highly related to leukocyte tyrosine kinase (LTK). Oncogene 1997, 14:2175-2188 [DOI] [PubMed] [Google Scholar]
- 5.Bridge JA, Swarts SJ, Buresh C, Nelson M, Degenhardt JM, Spanier S, Maale G, Meloni A, Lynch JC, Neff JR: Trisomies 8 and 20 characterize a subgroup of benign fibrous lesions arising in both soft tissue and bone. Am J Pathol 1999, 154:729-733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colleoni GWB, Bridge JA, Garicochea B, Liu J, Filippa DA, Ladanyi M: ATIC-ALK: a novel variant ALK gene fusion in anaplastic large cell lymphoma resulting from the recurrent cryptic chromosomal inversion, inv(2)(p23q35). Am J Pathol 2000, 156:781-789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dodge GR, Kovalszky I, McBride OW, Yi HF, Chu ML, Saitta B, Stokes DG, Iozzo RV: Human clathrin heavy chain (CLTC): partial molecular cloning, expression and mapping of the gene to human chromosome 17q11-qter. Genomics 1991, 11:174-178 [DOI] [PubMed] [Google Scholar]
- 8.Touriol C, Greenland C, Lamant L, Pulford K, Bernard F, Rousset T, Mason DY, Delsol G: Further demonstration of the diversity of chromosomal changes involving 2p23 in ALK-positive lymphoma: 2 cases expressing ALK kinase fused to CLTCL (clathrin chain polypeptide-like). Blood 2000, 95:3204-3207 [PubMed] [Google Scholar]
- 9.Treissman SP, Gillis DA, Lee CL, Giacomantonio M, Resch L: Omental-mesenteric inflammatory pseudotumor. Cytogenetic demonstration of genetic changes and monoclonality in one tumor. Cancer 1994, 73:1433-1437 [DOI] [PubMed] [Google Scholar]
- 10.Snyder CS, Dell’Aquila M, Haghighi P, Baergen RN, Kyung Suh Y, Yi ES: Clonal changes in inflammatory pseudotumor of the lung. Cancer 1995, 76:1545-1549 [DOI] [PubMed] [Google Scholar]
- 11.Hojo H, Newton WA, Hamoudi AB, Qualman SJ, Wakase H, Suzuki S, Jaynes F: Pseudosarcomatous myofibroblastic tumor of the urinary bladder in children: a study of 11 cases with review of the literature. Am J Surg Pathol 1995, 19:1224-1236 [DOI] [PubMed] [Google Scholar]
- 12.Sciot R, Dal Cin P, Fletcher CDM, Hernandez JM, Garcia JL, Samson I, Ramos L, Brys P, van Damme B, van den Berghe H: Inflammatory myofibroblastic tumor of bone: report of two cases with evidence of clonal chromosomal changes. Am J Surg Pathol 1997, 21:1166-1172 [DOI] [PubMed] [Google Scholar]
- 13.Su LD, Perez-Atayde A, Sheldon S, Fletcher JA, Weiss SW: Inflammatory myofibroblastic tumor: cytogenetic evidence supporting clonal origin. Mod Pathol 1998, 11:364-368 [PubMed] [Google Scholar]
- 14.Goldstein JL, Anderson RG, Brown MS: Coated pits, coated vesicles, and receptor-mediated endocytosis. Nature 1979, 279:679-685 [DOI] [PubMed] [Google Scholar]
- 15.Schmid SL: Clathrin-coated vesicle formation and protein sorting: an integrated process. Annu Rev Biochem 1997, 66:511-548 [DOI] [PubMed] [Google Scholar]
- 16.Molloy SS, Anderson ED, Jean F, Thomas G: Bicycling the furin pathway: from TGN localization to pathogen activation and embryogenesis. Trends Cell Biol 1999, 9:28-35 [DOI] [PubMed] [Google Scholar]
- 17.Bensen ES, Costaguta G, Payne GS: Synthetic genetic interactions with temperature-sensitive clathrin in Saccharomyces cerevisiae: roles for synaptojanin-like Inp53p and dynamin-related Vps1p in clathrin-dependent protein sorting at the trans-Golgi network. Genetics 2000, 154:83-97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okamoto CT, McKinney J, Jeng YY: Clathrin in mitotic spindles. Am J Physiol 2000, 279:C369-C374 [DOI] [PubMed] [Google Scholar]
- 19.Kedra D, Peyrard M, Fransson I, Collins JE, Dunham I, Roe BA, Dumanski JP: Characterization of a second human clathrin heavy chain polypeptide gene (CLH-22) from chromosome 22q11. Hum Mol Genet 1996, 5:625-631 [DOI] [PubMed] [Google Scholar]
- 20.Morris SW, Kirstein MN, Valentine MB, Dittmer KG, Shapiro DN, Saltman DL, Look AT: Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin’s lymphoma. Science 1994, 263:1281-1284 [DOI] [PubMed] [Google Scholar]