Abstract
The diagnosis of mantle cell lymphoma (MCL) is particularly important for clinical management because of a remarkable prognostic difference between MCL and other types of B-cell lymphoma. In addition to immunohistochemical analysis, we have established a 5′ exonuclease-based real-time reverse transcriptase-mediated quantitative polymerase chain reaction (RQ-PCR) method to detect cyclin D1 overexpression for the diagnosis of MCL. The RQ-PCR could detect cyclin D1 overexpression in all nine examined MCL cases, in contrast genomic PCR detected t(11;14) in only two of nine cases. By RQ-PCR the expression of G6PDH was significantly higher in myeloid leukemias than those in B-cell lymphomas (P = 0.018). As a result, cyclin D1/G6PDH ratio ranged from 0.78 to 12.4 (mean, 1.83) in MCL, exclusively higher than those in other B-cell lymphoma (0.00009 ∼ 0.16) and myeloid leukemia (0.00011 ∼ 0.085). The high expression of cyclin D1 in certain myeloid leukemias was identified to reflect their proliferative activity and not to represent the oncogenic overexpression. The 95% confidence interval of the cyclin D1/G6PDH ratio was 0.29 ∼ 11.1 for MCL, 0.014 ∼ 0.25 for other B-cell lymphomas and 0.000014 ∼ 0.083 for myeloid leukemia, suggesting that a cutoff value can be set at 0.25. The RQ-PCR of cyclin D1 is convenient and especially useful for the diagnosis of MCL.
Mantle cell lymphoma (MCL) is a distinct entity of non-Hodgkin’s lymphoma with characteristic clinicopathological and molecular-genetic features and poor prognosis. 1 Cyclin D1 overexpression as a result of t(11;14)(q13;q32) translocation plays an important role in the pathogenesis of MCL. 1 We recently clarified that the overexpression of cyclin D1 plays a key role in the diagnosis of MCL, especially in the differential diagnosis from MCL-like low-grade B-cell lymphoma. 2 However, the overexpression of cyclin D1 has not yet been included in the diagnostic criteria of MCL in the World Health Organization classification, 3 which might be caused by some technical problems for immunohistochemistry. Cyclin D1 overexpression at the mRNA level can be detected by Northern blotting 4 or by reverse transcriptase-mediated polymerase chain reaction (RT-PCR), but Northern blotting is sometimes hampered by RNA degradation in the specimens and by complicated procedures. The RT-PCR assay is likely to amplify faint physiological cyclin D1 derived from nonoverexpressing lymphomas or contaminating normal cells, 5 thus necessitating special techniques such as competitive 6 or quantitative PCR. 7 However, these techniques require modifications after PCR and can be complicated and time consuming, so that these methods are currently not considered convenient for routine diagnostic use. 8 This prompted us to investigate a simple, clear, reliable, and reproducible procedure. In this report, we describe a real-time reverse transcriptase-mediated quantitative polymerase chain reaction (RQ-PCR) detection method of cyclin D1 overexpression for the diagnosis of MCL.
Materials and Methods
Patient Samples
A total of 37 biopsy lymph node samples that were snap-frozen and stored were used in this study. They consisted of 9 MCLs, 3 MCL-like low-grade B-cell lymphomas, 10 diffuse large B-cell lymphomas, 10 follicular lymphomas, and 5 reactive lymphadenitis. Because of occasional cyclin D1 expression in myeloid leukemia, 4,6 frozen bone marrow cells from acute myeloid leukemia (AML) patients were also included. Diagnostic immunohistochemistry for cyclin D1 overexpression in B-cell lymphomas was performed as previously described. 2 The patient materials were used with the informed consent and approval by the institutional review board of the Aichi Cancer Center.
Cell Lines
Cell lines used in this study were SP-49, 9 SUDHL-4, 10 SUDHL-6 10 (B-cell lymphoma), HL-60, 11 MEG-01, 12 Kasumi-1, 13 NKM-1, 14 NOMO-1, 15 ME-1R, 16 IMS-M1, 17 HEL, 18 CMK, 19 K562, 20 U937 21 (myeloid leukemia), AST-1, 22 and Hut102 23 (T-cell lymphoma). SP-49, HL-60, MEG-01, Kasumi-1, NKM-1, NOMO-1, ME-1R, IMS-M1, and AST-1 overexpressed cyclin D1 by Northern blotting. 4
Northern Blotting and Real-Time RT-PCR
Total RNA was extracted from the patient samples, as well as from 16 leukemia/lymphoma cell lines, and Northern blotting was performed as described previously. 4
The real-time quantitative cyclin D1 assay was performed in a PRISM 7700 Sequence Detector (Applied Biosystems Japan, Tokyo, Japan). cDNA transcribed from 100 ng of total RNA was mixed with 0.5 μmol/L cyclin D1 primers and 0.2 μmol/L TaqMan probe labeled with 5′-FAM (6-carboxy fluorescein) and 3′-TAMRA (6-carboxy-tetramethyl rhodamine), and was amplified in a 25 μl volume using the TaqMan PCR core reagents kit (Applied Biosystems Japan). Samples were amplified with a precycling hold at 95°C for 10 minutes, followed by 45 cycles of denaturation at 95°C for 15 seconds, annealing at 60°C for 30 seconds, and extension at 72°C for 30 seconds. The primers used were 5′-ACAAACAGATCATCCGCAAACAC-3′ (sense) and 3′-TGTTGGGGCTCCTCAGGTTC-5′ (anti-sense), and the TaqMan probe was 5′-FAM-ACATCTGTGGCACAGAGGGCAACG-TAMRA-3′. The copy number of cyclin D1 in each sample was calculated with a standard curve generated from serially diluted (100 to 10 7 copies) plasmids containing cyclin D1 cDNA.
For external control, the glucose-6-phospate dehydrogenase (G6PDH) gene was amplified using oligonucleotides 5′-CATGGTGCTGAGATTTGCCAAC-3′ (sense) and 5′-TCAACACCTTGACCTTCTCATCAC-3′ (anti-sense), and was analyzed with 5′-FAM-ATCCGGGACGTGATGCAGAACCACCTAC-TAMRA-3′ TaqMan probe under the same conditions as that for cyclin D1. The amplification was duplicated for each sample and the mean values were used to calculate cyclin D1/G6PDH ratio.
Genomic PCR for the Detection of t(11;14)
Genomic PCR was performed as previously described 24 with some modifications. DNA amplification was performed with a hold at 94°C for 3 minutes, followed by 35 cycles of denaturation at 94°C for 1 minute, followed by annealing at 66°C for 2 minutes, and extension at 72°C for 1 minute. PCR products were electrophoresed on 2% agarose gels, transferred to a nylon membrane (Hybond-N; Amersham-Japan, Tokyo, Japan) and hybridized with a [γ-32P]ATP-labeled MCL-2 oligonucleotide.
For the control of genomic PCR, a variable region of the immunoglobulin heavy chain gene was amplified with FR2A, LJH, and VLJH primers as previously described. 25
Statistical Analysis
The Mann-Whitney test and the Kruskal-Wallis test were performed to examine the difference of results by disease subtypes. The Smirnov’s test was used to identify an extreme data. Data were analyzed with the Microsoft Excel 97 (Microsoft-Japan, Tokyo, Japan) and Statcel softwares (OMS, Tokorozawa, Japan).
Results
Quantitative Cyclin D1 Expression
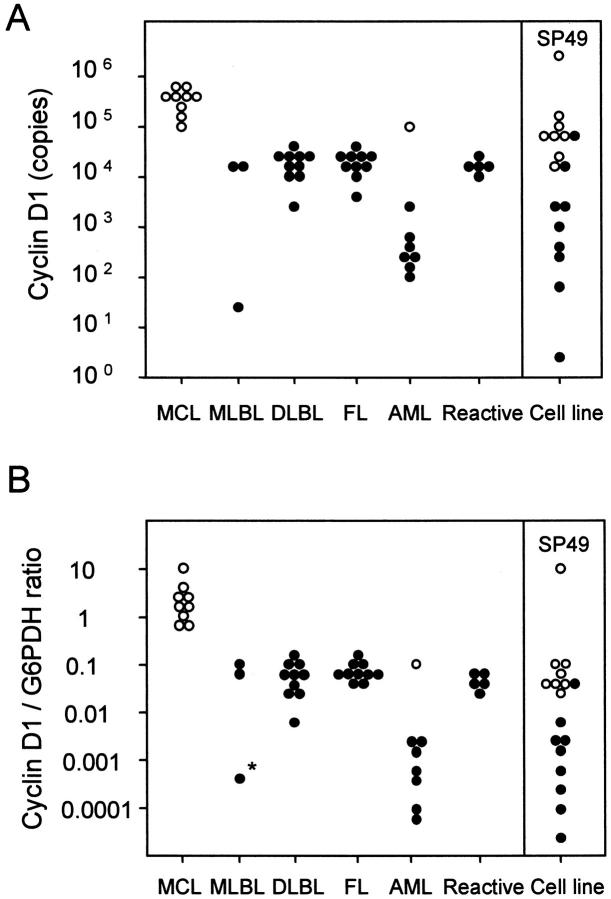
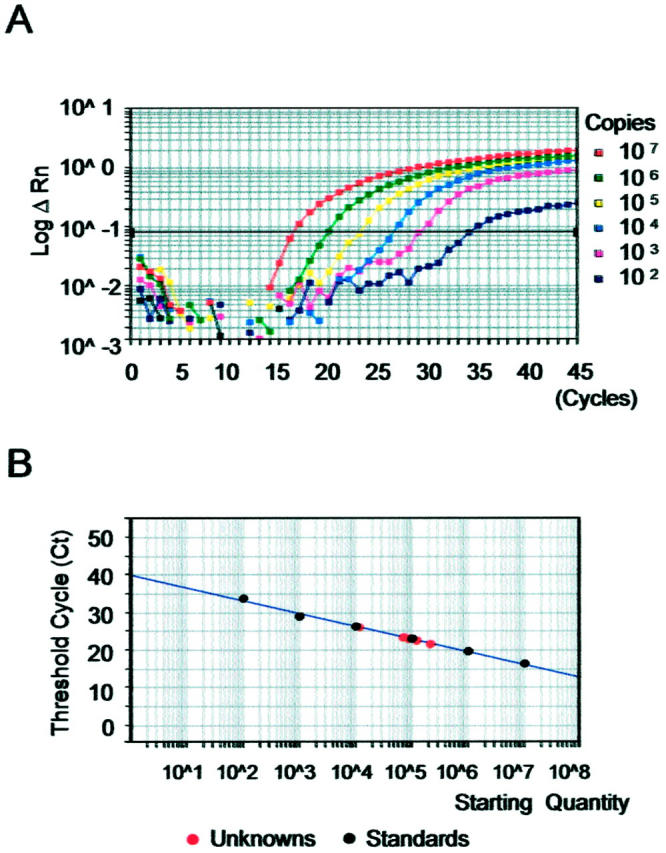
The real-time RT-PCR amplification of the serially diluted cyclin D1 plasmid controls showed a logarithmic signal increase (Figure 1A) ▶ . The standard curve was generated by using the Ct at which the fluorescence signal of the reporter dye rose above the baseline signal (Figure 1B) ▶ . The results from the cell line cDNA and patient samples were plotted on the standard curve, and the estimated copy numbers of the cyclin D1 gene was calculated as well as for the G6PDH gene (Figure 2) ▶ . For example, SP-49, a MCL cell line, 9 was found to contain 2.9 × 10 6 copies of the cyclin D1 gene in 100 ng cDNA, and HL60 1.9 × 10 4 copies. Patient samples of MCL contained an average of 3.6 × 10 5 copies of cyclin D1, and those of MCL-like low-grade B-cell lymphomas 1.1 × 103, of DLB 2.0 × 104, of follicular lymphomas 2.0 × 104, and of reactive lymphadenitis 1.7 × 10 4 (Table 1) ▶ . One AML patient shown to be positive for cyclin D1 by Northern blotting had 8.0 × 10 4 copies of cyclin D1 in the sample, whereas those from other AML patients contained an average of 7.0 × 10 2 copies (Figure 3A) ▶ . These results correlated well with those obtained with Northern blotting and immunohistochemistry (data not shown).
Figure 1.

Amplification plot of RQ-PCR for cyclin D1. A: Serially diluted plasmids containing cyclin D1 cDNA (100 to 10 7 copies per tube) were analyzed for controls. B: Standard curve generated from the mean value of duplicated examinations. The slope and the Y-intercept of the curve were −3.261 and 36.999 (correlation coefficient, 0.997). Black circles represent controls and red circles tested materials of cell lines and patient samples. The tested materials plotted are (from left to right) SUDHL-6, SUDHL-4, HL-60, Meg-01, patient 390 (MCL), patient 400 (MCL), and SP-49.
Figure 2.
Amplification plot of RQ-PCR for G6PDH. A: Serially diluted plasmids containing cyclin D1 cDNA (102 to 10 7 copies per tube) were analyzed for controls. B: Standard curve generated from the mean value of duplicated examinations. The slope and the Y-intercept of the curve were −3.406 and 40.084 (correlation coefficient, 0.994). The tested materials plotted are those examined in Figure 1 ▶ .
Table 1.
Results of the Quantitative PCR
| Cyclin D1 (copies) | G6PDH (copies) | Cyclin D1/G6PDH ratio | ||||
|---|---|---|---|---|---|---|
| Average | (Range) | Average | (Range) | Average* | (Range) | |
| MCL | 3.6 × 105 | (1.0 × 105 ∼ 7.1 × 105) | 2.9 × 105 | (1.3 × 104 ∼ 4.2 × 105) | 1.8 | 0.78 ∼ 12.4 |
| MLBL | 1.1 × 103 | (3.0 × 101 ∼ 1.9 × 104) | 2.6 × 105 | (2.0 × 105 ∼ 3.6 × 105) | 0.0079 | 0.000085 ∼ 0.097 |
| DLB | 2.0 × 104 | (2.5 × 103 ∼ 4.4 × 104) | 4.0 × 105 | (1.0 × 105 ∼ 1.1 × 106) | 0.048 | 0.0052 ∼ 0.16 |
| FL | 2.0 × 104 | (4.9 × 103 ∼ 4.9 × 104) | 2.6 × 105 | (1.1 × 105 ∼ 4.4 × 105) | 0.071 | 0.044 ∼ 0.13 |
| AML | 1.1 × 104 | (1.2 × 102 ∼ 8.0 × 104) | 9.5 × 105 | (1.4 × 105 ∼ 2.1 × 106) | 0.0011 | 0.000076 ∼ 0.085 |
| Reactive | 1.7 × 104 | (8.5 × 103 ∼ 3.2 × 104) | 3.6 × 105 | (2.0 × 105 ∼ 5.4 × 105) | 0.044 | 0.032 ∼ 0.071 |
*, Geometric average.
MCL, mantle cell lymphoma; MLBL, MCL-like B-cell lymphoma; DLB, diffuse large B-cell lymphoma; FL, follicular lymphoma; AML, acute myeloid leukemia.
Figure 3.
Cyclin D1 expression level and cyclin D1/G6PDH ratio by real-time RT-PCR. Patient samples and cell lines that are positive (open circles) and negative (filled circles) for cyclin D1 as detected by Northern blotting are plotted for cyclin D1 expression level (A) and cyclin D1/G6PDH (B). The cyclin D1/G6PDH ratio from all patients with MCL exceeded 0.7 (range, 0.78 ∼ 12.4) and the geometric mean value was 1.8 [95% confidence interval (CI), 0.29 ∼ 11.1]. The ratios from other patients were exclusively <0.2 and the geometric mean value was 0.044 (95% CI: 0.014 ∼ 0.25) for other B-cell lymphomas excluding one extreme value (*, judged by Smirnov’s test). The cyclin D1/G6PDH ratio of SP-49 was 8.2, whereas that of other cell lines ranged from 0.11 to 3.1 ×10 −5.
Genomic PCR for t(11;14)
Genomic PCR detected t(11;14) only in two of the nine MCL patients with the primer sets examined. No other patient samples of B-cell lymphomas or B-cell lines including SP49 showed positive PCR results for the genomic PCR for t(11;14), although clonal immunoglobulin heavy-chain gene rearrangement was identified in each sample.
Correction of Cyclin D1 Expression by G6PDH
The average copy numbers of control G6PDH was 2.9 × 10 5 for MCL, 3.2 × 10 5 for other B-cell lymphomas, and 9.5 × 10 5 for AML, thus with a significantly higher copy numbers for AML (P = 0.018, Mann-Whitney’s U test). The resultant cyclin D1/G6PDH ratio showed a sharp division into two categories. The cyclin D1/G6PDH ratio of all MCL cases exceeded 0.7 (range, 0.78 ∼ 12.4) and that of the SP-49 cell line was 8.2, whereas all of the other cases showed the ratio below 0.2 (Table 1) ▶ . One case of MCL-like low-grade B-cell lymphomas (* in Figure 3B ▶ ) showed exceptionally low levels of both the cyclin D1 expression and the cyclin D1/G6PDH ratio (P < 0.00001 by Smirnov’s extreme test). The 95% confidence interval of the cyclin D1/G6PDH ratio was 0.29 ∼ 11.1 for MCL, 0.014 ∼ 0.25 for other B-cell lymphoma excluding the one case with extreme value, 0.023 ∼ 0.085 for reactive lymphadenitis, and 0.00014 ∼ 0.083 for AML. These data showed that a cutoff value of cyclin D1/G6PDH for the diagnosis MCL can be set at 0.25 in our system.
Discussion
We have identified that the RQ-PCR detection of cyclin D1 supplemented by the external control G6PDH is particularly useful for the diagnosis of MCL. Although the overexpression of cyclin D1 readily detectable by Northern blotting, 4 this methodology is not widely used for clinical diagnosis, which might be because of complicated procedures or some technical problems. 8 The RQ-PCR is a rapid and reproducible method, and does not require detection steps after PCR, such as gel electrophoresis, densitometry, Southern blotting, and hybridization. Although our present system requires two separated reactions, the PQ-PCR is convenient enough for the routine diagnostic use as well as immunohistochemistry 2 or fluorescence in situ hybridization. 26 A multiplex RQ-PCR using probes labeled with different fluorescent dyes may enable the examination with one reaction.
The PQ-PCR of cyclin D1 is more sensitive than genomic PCR of t(11;14) for the molecular diagnosis of MCL. The genomic PCR could detect t(11;14) only in two of the nine MCL patients and the detection rate was consistent with the results in the literature. Because of the widely scattered locations of the breakpoint of t(11;14) on chromosome 11, its detection rate by the genomic PCR has been reported as 30 to 50%. 7,27-30 Also, the genomic detection by Southern blotting is not sufficient for the initial diagnosis of MCL because of the wide-range breakpoint distribution 31 or the presence of variant translocations. 32 Although the real-time PCR for genomic t(11;14) is sensitive enough for the detection of minimal residual disease, the application is restricted to the patients with a detectable breakpoint. 33,34
Increased expression of cyclin D1 has been described in a part of myeloid leukemias by Northern blotting 4 as well as competitive quantitative PCR, 6 although its pathological significance was unclear. Our present result with RQ-PCR of cyclin D1 alone also showed high expression level in AML samples. However, the expression level of G6PDH was also significantly high in AML, which might be dismissed in Northern blot or nonquantitative PCR analyses. The high copy number of G6PDH in AML patients and cell lines indicates their proliferative activity. Evaluating cyclin D1 expression with the cyclin D1/G6PDH ratio allowed for correction of the cyclin D1 expression accompanying cell proliferation, and makes it possible to identify aberrant cyclin D1 expression in MCL specifically. The high expression of cyclin D1 in a part of myeloid leukemias reflects their proliferative activity and does not represent the oncogenic overexpression. Weak expression of cyclin D1 without any 11q13 translocations has also been described in hairy cell leukemia, 35 but we could not examine hairy cell leukemia cases because of its uncommonness in Japan. The expression mechanism of cyclin D1 in hairy cell leukemia should be clarified by future investigations.
Footnotes
Address reprint requests to Ritsuro Suzuki, M.D., Ph.D., Division of Molecular Medicine, Aichi Cancer Center, 1-1 Kanokoden, Chikusa-ku, Nagoya 464-8681, Japan. E-mail: rsuzuki@aichi-cc.pref.aichi.jp.
Supported in part by a Grant-in-Aid for the Second-Term Comprehensive 10-year Strategy for Cancer Control from the Ministry of Health and Welfare, a Grant-in-Aid for Science on Primary Areas (Cancer Research), and a Grant-in-Aid for Encouragement of Young Scientists from the Ministry of Education, Science and Culture, Japan.
References
- 1.Weisenburger DD, Armitage JO: Mantle cell lymphoma—An entity comes of age. Blood 1996, 87:4483-4494 [PubMed] [Google Scholar]
- 2.Yatabe Y, Suzuki R, Tobinai K, Matsuno Y, Ichinohasama R, Okamoto M, Yamaguchi M, Tamaru J, Uike N, Hashimoto Y, Morishima Y, Suchi T, Seto M, Nakamura S: Significance of cyclin D1 overexpression for the diagnosis of mantle cell lymphoma: a clinicopathologic comparison of cyclin D1-positive MCL and cyclin D1-negative MCL-like B-cell lymphoma. Blood 2000, 95:2253-2261 [PubMed] [Google Scholar]
- 3.Harris NL, Jaffe ES, Diebold J, Flandrin G, Muller-Hermelink HK, Vardiman J, Lister TA, Bloomfield CD: World Health Organization classification of neoplastic diseases of the hematopoietic and lymphoid tissues: report of the clinical advisory committee meeting—Airlie House, Virginia, November 1997. J Clin Oncol 1999, 17:3835-3849 [DOI] [PubMed] [Google Scholar]
- 4.Suzuki R, Kuroda H, Komatsu H, Hosokawa Y, Kagami Y, Ogura M, Nakamura S, Kodera Y, Morishima Y, Ueda R, Seto M: Selective usage of D-type cyclins in lymphoid malignancies. Leukemia 1999, 13:1335-1342 [DOI] [PubMed] [Google Scholar]
- 5.Oka K, Ohno T, Kita K, Yamaguchi M, Takakura N, Nishii K, Miwa H, Shirakawa S: PRAD1 gene over-expression in mantle-cell lymphoma but not in other low-grade B-cell lymphomas, including extranodal lymphoma. Br J Haematol 1994, 86:786-791 [DOI] [PubMed] [Google Scholar]
- 6.Uchimaru K, Taniguchi T, Yoshikawa M, Asano S, Arnold A, Fujita T, Motokura T: Detection of cyclin D1 (bcl-1, PRAD1) overexpression by a simple competitive reverse transcription-polymerase chain reaction assay in t(11;14)(q13;q32)-bearing B-cell malignancies and/or mantle cell lymphoma. Blood 1997, 89:965-974 [PubMed] [Google Scholar]
- 7.Aguilera NSI, Bijwaard KE, Duncan B, Krafft AE, Chu W-S, Abbondanzo SL, Lichy JH, Taubenberger JK: Differential expression of cyclin D1 in mantle cell lymphoma and other non-Hodgkin’s lymphomas. Am J Pathol 1998, 153:1969-1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arber DA: Molecular diagnostic approach to non-Hodgkin’s lymphoma. J Mol Diagn 2000, 2:178-190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daibata M, Takasaki M, Hirose S, Kubonishi I, Taguchi H, Ohtsuki Y, Miyoshi I: Establishment of a new human B-cell line carrying t(11;14) chromosome abnormality. Jpn J Cancer Res 1987, 78:1182-1185 [PubMed] [Google Scholar]
- 10.Hecht BK, Epstein AL, Berger CS, Kaplan HS, Hecht F: Histiocytic lymphoma cell lines: immunologic and cytogenetic studies. Cancer Genet Cytogen 1985, 14:205-218 [DOI] [PubMed] [Google Scholar]
- 11.Collins SJ, Gallo RC, Gallagher RE: Continuous growth and differentiation of human myeloid leukaemic cells in suspension culture. Nature 1977, 270:347-349 [DOI] [PubMed] [Google Scholar]
- 12.Ogura M, Morishima Y, Ohno R, Kato Y, Hirabayashi N, Nagura H, Saito H: Establishment of a novel human megakaryoblastic leukemia cell line, MEG-01, with positive Philadelphia chromosome. Blood 1985, 66:1384-1392 [PubMed] [Google Scholar]
- 13.Asou H, Tashiro S, Hamamoto K, Otsuji A, Kita K, Kamada N: Establishment of a human acute myeloid leukemia cell line (Kasumi-1) with 8;21 chromosome translocation. Blood 1991, 77:2031-2036 [PubMed] [Google Scholar]
- 14.Kataoka T, Morishita Y, Ogura M, Morishima Y, Towatari M, Kato Y, Inoue H, Saito H: A novel human myeloid leukemia cell line, NKM-1, coexpressing granulocyte colony-stimulating factor receptors and macrophage colony-stimulating factor receptors. Cancer Res 1990, 50:7703-7709 [PubMed] [Google Scholar]
- 15.Morishita Y, Kataoka T, Towatari M, Ito T, Inoue H, Ogura M, Morishima Y, Saito H: Up-regulation of transferrin receptor gene expression by granulocyte colony-stimulating factor in human myeloid leukemia cells. Cancer Res 1990, 50:7955-7961 [PubMed] [Google Scholar]
- 16.Yanagisawa K, Horiuchi T, Fujita S: Establishment and characterization of a new human leukemia cell line derived from M4Eo. Blood 1991, 78:451-457 [PubMed] [Google Scholar]
- 17.Iida S, Seto M, Yamamoto K, Komatsu H, Tojo A, Asano S, Kamada N, Ariyoshi Y, Takahashi T, Ueda R: MLLT3 gene on 9p22 involved in t(9;11) leukemia encodes a serine/proline rich protein homologous to MLLT1 on 19p13. Oncogene 1993, 8:3085-3092 [PubMed] [Google Scholar]
- 18.Martin P, Papayannopoulou T: HEL cells: a new human erythroleukemia cell line with spontaneous and induced globin expression. Science 1982, 216:1233-1235 [DOI] [PubMed] [Google Scholar]
- 19.Sato T, Fuse A, Eguchi M, Hayashi Y, Ryo R, Adachi M, Kishimoto Y, Teramura M, Mizoguchi H, Shima Y, Komori I, Sunami S, Okimoto Y, Nakajima H: Establishment of a human leukaemic cell line (CMK) with megakaryocytic characteristics from a Down’s syndrome patient with acute megakaryoblastic leukaemia. Br J Haematol 1989, 72:184-190 [DOI] [PubMed] [Google Scholar]
- 20.Lozzio CB, Lozzio BB: Human chronic myelogenous leukemia cell-line with positive Philadelphia chromosome. Blood 1975, 45:321-334 [PubMed] [Google Scholar]
- 21.Sundstrom C, Nilsson K: Establishment and characterization of a human histiocytic lymphoma cell line (U-937). Int J Cancer 1976, 17:565-577 [DOI] [PubMed] [Google Scholar]
- 22.Yamamoto K, Osada H, Seto M, Ogura M, Suzuki H, Utsumi KR, Oyama A, Ariyoshi Y, Nakamura S, Kurita S, Takahashi T, Ueda R: Phenotypic and genotypic lineage switch of a lymphoma with shared chromosome translocation and T-cell receptor gamma gene rearrangement. Jpn J Cancer Res 1992, 83:465-476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gazdar AF, Carney DN, Bunn PA, Russell EK, Jaffe ES, Schechter GP, Guccion JG: Mitogen requirements for the in vitro propagation of cutaneous T-cell lymphomas. Blood 1980, 55:409-417 [PubMed] [Google Scholar]
- 24.Segal GH, Masih AS, Fox AC, Jorgensen T, Scott M, Braylan RC: CD5-expressing B-cell non-Hodgkin’s lymphoma with bcl-1 gene rearrangement have a relatively homogeneous immunophenotype and are associated with an overall poor prognosis. Blood 1995, 85:1570-1579 [PubMed] [Google Scholar]
- 25.Kume M, Suzuki R, Yatabe Y, Kagami Y, Miura I, Miura AB, Morishima Y, Nakamura S, Seto M: Somatic hypermutation in the VH segment of immunoglobulin genes of CD5-positive diffuse large B-cell lymphomas. Jpn J Cancer Res 1997, 88:1087-1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coignet LJA, Schuuring E, Kibbelaar RE, Raap TK, Kleiverda KK, Bertheas M-F, Wiegant J, Beverstock G, Kluin PM: Detection of 11q13 rearrangement in hematologic neoplasias by double-color fluorescence in situ hybridization. Blood 1996, 87:1512-1519 [PubMed] [Google Scholar]
- 27.Molot RJ, Meeker TC, Wittwer CT, Perkins SL, Segal GH, Masih AS, Braylan RC, Kjeldsberg CR: Antigen expression and polymerase chain reaction amplification of mantle cell lymphomas. Blood 1994, 83:1626-1631 [PubMed] [Google Scholar]
- 28.Rimokh R, Berger F, Delsol G, Digonnet I, Rouault JP, Tigaud JD, Gadoux M, Coiffier B, Bryon PA, Magaud JP: Detection of the chromosomal translocation t(11;14) by polymerase chain reaction in mantle cell lymphoma. Blood 1994, 83:1871-1875 [PubMed] [Google Scholar]
- 29.Andersen NS, Donovan JW, Borus JS, Poor CM, Neuberg D, Aster JC, Nadler LM, Freedman AS, Gribben JG: Failure of immunologic purging in mantle cell lymphoma assessed by polymerase chain reaction detection of minimal residual disease. Blood 1997, 90:4212-4221 [PubMed] [Google Scholar]
- 30.Samaha H, Dumontet C, Ketterer N, Moullet I, Thieblemont C, Bouafia F, Callet-Bauchu E, Felman P, Berger F, Salles G, Coiffier B: Mantle cell lymphoma: a retrospective study of 121 cases. Leukemia 1998, 12:1281-1287 [DOI] [PubMed] [Google Scholar]
- 31.Raynaud SD, Bekri S, Leroux D, Grosgeorge J, Klein B, Bastard C, Gaudray P, Simon MP: Expanded range of 11q13 breakpoints with differing patterns of cyclin D1 expression in B-cell malignancies. Genes Chromosom Cancer 1993, 8:80-97 [DOI] [PubMed] [Google Scholar]
- 32.Komatsu H, Iida S, Yamamoto K, Mikuni C, Nitta M, Takahashi T, Ueda R, Seto M: A variant chromosome translocation at 11q13 identifying PRAD1/cyclin D1 as the BCL-1 gene. Blood 1994, 84:1226-1231 [PubMed] [Google Scholar]
- 33.Olsson K, Gerard CJ, Zehnder J, Jones C, Ramanathan R, Reading C, Hanania EG: Real-time t(11;14) and t(14;18) PCR assays provide sensitive and quantitative assessments of minimal residual disease (MRD). Leukemia 1999, 13:1833-1842 [DOI] [PubMed] [Google Scholar]
- 34.Luthra R, Sarris AH, Hai S, Paladugu AV, Romaguera JE, Cabanillas FF, Medeiros LJ: Real-time 5′→3′ exonuclease-bases PCR assay for detection of the t(11;14)(q13;q32). Am J Clin Pathol 1999, 112:524-530 [DOI] [PubMed] [Google Scholar]
- 35.Bosch F, Campo E, Jares P, Pittaluga S, Munoz J, Nayach I, Piris MA, De Wolf-Peters C, Jaffe ES, Rozman C, Montserrat E, Cardesa A: Increased expression of the PRAD-1/CCND1 gene in hairy cell leukemia. Br J Haematol 1995, 91:1025-1030 [DOI] [PubMed] [Google Scholar]