Abstract
Translocation t(15;19)(q13;p13.1) defines a lethal midline carcinoma arising adjacent to respiratory tract in young people. To characterize molecular alterations responsible for the distinctly aggressive biological behavior of this cancer, we mapped the chromosome 15 and 19 translocation breakpoints by fluorescence in situ hybridization (FISH) and Southern blotting. To evaluate preliminarily the frequency, anatomical distribution, and histological features of t(15;19) cancer, we developed a FISH assay for paraffin sections. Our findings reveal a novel oncogenic mechanism in which the chromosome 19 translocation breakpoint interrupts the coding sequence of a bromodomain gene, BRD4. These studies implicate BRD4 as a potential partner in a t(15;19)-associated fusion oncogene. In addition, we localized the chromosome 15 breakpoint to a 9-kb region in each of two cases, thereby identifying several candidate oncogenes which might represent the BRD4 fusion partner. FISH evaluation of 13 pediatric carcinomas revealed t(15;19) in one of four sinonasal carcinomas, whereas this translocation was not detected in thymic (n = 3), mucoepidermoid (n = 3), laryngeal (n = 2), or nasopharyngeal (n = 1) carcinomas. Our studies shed light on the oncogenic mechanism underlying t(15;19) and provide further evidence that this highly lethal cancer arises from respiratory mucosa.
Characteristic recurrent translocations are common in sarcomas and lymphomas/leukemias but are uncommon in epithelial neoplasms. Highly effective and biologically rational therapies have been developed for some leukemias and sarcomas, often enabled by the cell biology insights obtained from the cloning and functional characterization of translocation-associated oncogenes. For example, chronic myeloid leukemia is characterized by a t(9;22) fusion gene encoding the BCR-ABL oncoprotein, resulting in constitutive ABL tyrosine kinase activity. Dramatic therapeutic responses have been achieved in this disease using the ABL ATP-binding pocket inhibitor, Gleevec. 1
Most clinically aggressive, widely metastatic, treatment-refractory carcinomas have complex karyotypes, without identifiable recurrent chromosomal translocations. However, it is unclear whether pathognomonic translocations are truly absent in these cancers, or whether they are simply masked by the overall cytogenetic complexity. It is generally assumed that carcinoma cytogenetic complexity is a manifestation of underlying genetic instability, permitting rapid acquisition of the sequential mutations that bestow the collective “aggressive” phenotype on an epithelial progenitor cell. On the other hand, there are several types of carcinoma, typically low-to-medium grade, which have relatively simple karyotypes, and which often feature balanced translocations. For example, recurrent translocations are found in follicular and papillary carcinoma of the thyroid, 2-5 and in papillary renal cell carcinoma, 6 none of which are particularly aggressive clinically.
Balanced translocation t(15;19) has been reported in a small number of carcinomas in children and young adults, and the clinical correlations with this translocation are striking. 7-11 All t(15;19) carcinomas were poorly differentiated histologically and extremely aggressive (average survival 18 weeks) with rapid clinical progression despite multi-modality therapies. Hence, this is the first recurring translocation identified in a particularly aggressive form of carcinoma. Characterization of the translocation breakpoints might therefore enable development of models which evaluate “aggressive” clinicopathological features, including invasiveness, high proliferative rate, and metastatic potential in epithelial tumors. In the present report we identify the putative chromosome 19 translocation target, and we also localize chromosome 15 breakpoints to a 9-kb genomic region. Further, we develop a paraffin-based fluorescence in situ hybridization (FISH) assay, and show application of this method in evaluating potential t(15;19) events in pediatric carcinomas.
Materials and Methods
Cytogenetic Analysis
Cytogenetic analysis was performed on Giemsa-banded metaphase spreads as described previously. 12
Cell Lines
We established a rapidly growing cell line from a lymph node metastasis in t(15;19) case 1. The primary tumor cells were grown in RPMI 1640 medium (Life Technologies, Inc. (Gibco BRL), Rockville, MD) and 15% fetal bovine serum and were held at confluence in T25 flasks, being fed thrice weekly between passaging. Cells were passed by releasing the confluent monolayers with trypsin and splitting the cells 1:2 to new T25 flasks. The cells grew rapidly and after passage 5 could be passed at 1:4. A cell line was also established from t(15;19) case 2, as reported previously. 13 Both cell lines were cytogenetically stable, with persistence of the t(15;19) translocation in all cells after more than 20 passages.
FISH Mapping on Metaphase Preparations
YACs, BACs, and cosmids were biotin or digoxigenin-labeled by random octamer priming (Life Technologies, Inc. (Gibco BRL), Rockville, MD). 14 Metaphase preparations from cell line cells or control normal lymphocytes were spread on slides and denatured according to standard protocols. Hybridization and washing steps were performed as described. 14 Detection was with FITC-anti-digoxigenin (Roche, Indianapolis, IN) and 594 rhodamine streptavidin (Molecular Probes, Eugene, OR), and nuclei were counterstained with 1 mg/ml 4,6-diamidino-2-phenylindole-dihydrochloride (DAPI). Images were captured using a charge-coupled device camera (Photometrics, Tucson, AZ). FISH mapping of the chromosome 15 and 19 breakpoints was performed by “walking” with YAC, BAC, then cosmid (cosmids not used for chromosome 15) probes inward to the breakpoint region using dual-color, split-apart FISH.
Southern Analysis
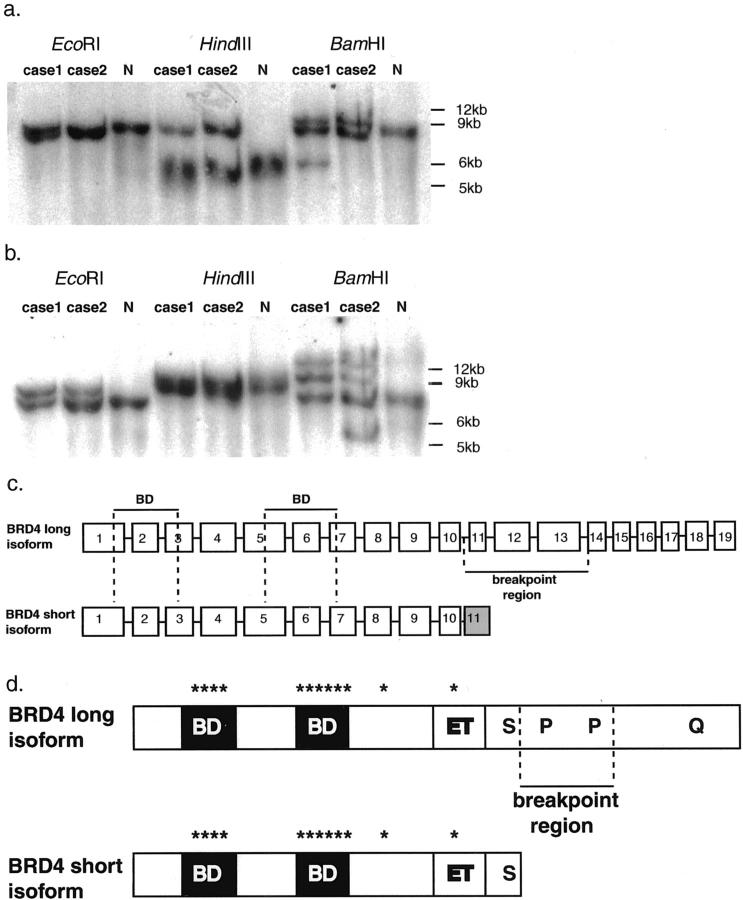
Cell line DNA isolation and blotting techniques were performed as described. 15 Polymerase chain reaction-generated, gel-isolated DNA probe templates (intron 12 and exon 13 of BRD4 (GenBank Accession No. AF386649) (see Figure 3b ▶ ); intron 1 and exon 2 of Nop10p) were labeled by random nanonucleotide priming (Prime-It II, Stratagene, Cedar Creek, TX) and [α-32P]dCTP.
Figure 3.
Southern blot of genomic DNAs from t(15;19) cases 1 and 2, and normal lymphocytes (N). Novel restriction fragments are seen in the EcoRI, HindIII, and BamHI t(15;19) cell digests after hybridization to BRD4 (a) and Nop10p (b) probes. c: Exon-intron maps for long and short transcripts of BRD4. The t(15;19) translocation breakpoints interrupt the coding sequence of the long transcript (intron 10), and are approximately 4-kb 3′ to the end of the short transcript. d: Schematic of the BRD4 isoforms: BD, bromodomain; ET, ET domain; S, serine-rich region; P, proline-rich regions; Q, glutamine-rich region; *, kinase-like motifs. Vertical dotted lines define the translocation breakpoint region.
Paraffin FISH Screening
Paraffin sections from 13 carcinomas (1975 to present; age range, 3 to 53 years; mean age, 16 years; M/F = 3/10), including 3 thymic, 3 mucoepidermoid (2 parotid gland, 1 lung), 2 laryngeal, 1 nasopharyngeal, and 4 sinonasal from Children’s Hospital, Boston, Massachusetts, were examined for the t(15;19) by FISH. Chromosome 15 and 19 breakpoints were evaluated using a dual color, split-apart FISH assay. For chromosome 15, a telomeric YAC, 908c5, and a centromeric YAC, 733c7, were used; for chromosome 19, a telomeric BAC, 87 m17, and a centromeric YAC, 766e7, were used.
FISH on paraffin sections was performed using a modification of the technique described by Bull and Harnden. 16 Briefly, formalin-fixed, 4-μm thick paraffin-embedded sections were baked overnight (60°C) and deparaffinized in xylene. Slides were immersed in 100 mmol/L Tris-base, 50 mmol/L EDTA, and incubated at 100°C in a temperature-controlled microwave for 30 to 60 minutes. Tissue digestion was performed with pepsin solution (Digest-All III, Zymed, San Francisco, CA), applied directly to the slides at 37°C for 10 minutes, and postfixed in 10% formaldehyde for 1 minute. The biotin and digoxigenin-labeled YAC DNA and Cot1 mixture was diluted in a 50% formamide solution containing 10% dextran sulfate, applied and coverslipped onto the slide. The slide was denatured on a flatbed polymerase chain reaction machine at 94°C for 3 minutes. Hybridization and washing steps were performed as described 14 and signal detection was as described above.
Results
Clinicopathological features for all known t(15;19) carcinomas are summarized in Table 1 ▶ , and are remarkable for poor survival and a treatment-refractory course. The histologies of the t(15;19) cancers evaluated in this study are shown in Figure 1 ▶ . Cases 1 and 2 provided the cell lines used for mapping studies. The tumor in case 1 arose adjacent to thymus and respiratory epithelium in a 22-year-old female. The tumor in case 2 arose from the epiglottis in a 13-year-old female.
Table 1.
Clinicopathological Features of t(15:19)+ Carcinomas
| Case no. | Age/sex | Undifferentiated cells | Differentiation | Keratin | Survival (weeks) | Therapy | Reference no. |
|---|---|---|---|---|---|---|---|
| 1* | 22 /F | + | Sq | + | 14 | CR | 7 |
| 2* | 13 /F | + | + | 38 | CR | 11 | |
| 3 | 12 /F | + | Sq | + | 13 | CR | |
| 4 | 11 /F | + | Sq | + | 18 | CR | 8 |
| 5 | 5 /M | + | Sq/G | + | 8 | C | 9 |
| 6 | 34 /F | + | NS | + | NS | NS | 10 |
Average Survival Was 18 Weeks. C, chemotherapy; G, glandular; NS, not specified; R, radiation; Sq, squamous.
*A cell line derived from this case was used for mapping studies described herein.
Figure 1.

Histological features of t(15;19) cases 1–3. a: Case 1 is remarkable for sheets of very discohesive undifferentiated cells with scant cytoplasm and prominent, central nucleoli. The tumor was found adjacent to thymus, but as seen here also invaded bronchial seromucinous glands, and was found nearby to undermine and replace the ciliated columnar epithelium (H&E, ×400). b: Case 2 has syncitial sheets of undifferentiated cells with frequent mitoses, single cell necrosis, and percolating lymphocytes giving a lymphoepithelial-like appearance (H&E, ×600). c: Case 3 has prominent nests of undifferentiated cells with focal keratinization (H&E, ×600). Inset: dual-color FISH reveals rearrangement, as evidenced by splitting apart of red-green probe doublet, in paraffin section of this sinonasal carcinoma. d: Immunoperoxidase studies (case 2) show reactivity for pan-keratin (MNF116-alkaline phosphatase, red), indicating epithelial differentiation (hematoxylin counterstain, ×400).
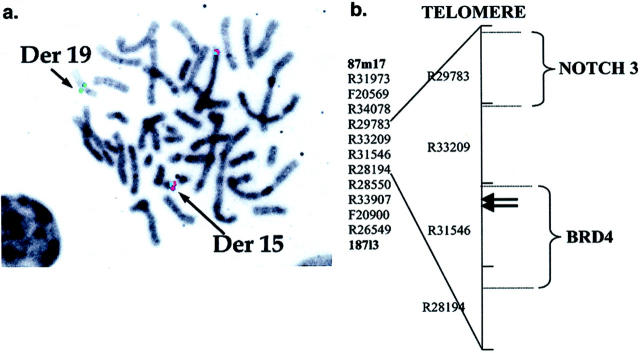
FISH mapping analyses are summarized in Figure 2 ▶ . The chromosome 19p13.1 cosmid clone R31546 was split by the t(15;19) breakpoints in cases 1 and 2. The split cosmid FISH signal was approximately 80:20, with the smaller signal being on the telomeric side of the breakpoint. This localization is consistent with involvement of the 3′ end of the BRD4 coding sequence. The chromosome 15q13 BAC clone 122p18 was also split approximately 50:50 by the translocation breakpoints in t(15;19) cases 1 and 2. BAC 122p18 contains three genes, Nop10p, Golgin-67, and KCC3 which have not yet been localized relative to the breakpoints.
Figure 2.
a: FISH localization of t(15;19) translocation breakpoint within the BRD4-containing cosmid clone R31546. Green and red FISH signals correspond to cosmid clone R31546 and BAC 87 m17, respectively. DAPI counterstain is shown as Giemsa emulation. Normal chromosome 19 has paired red and green signals (top, center), whereas the der(19) has only a green signal (centromeric end of R31546), and the der(15) has a red signal paired with a very weak green signal (telomeric end of R31546). b: Cosmid and BAC map of the 19p13.1 translocation breakpoint region (map framework from Lawrence Livermore National Laboratory). BACs 87m17 and 187l3 delimit the telomeric and centromeric ends of the region, respectively. All clones between the BACs are cosmids. Candidate genes in the region are BRD4 and NOTCH3. Translocation breakpoints in each of two t(15;19) carcinomas were FISH mapped to a 5-kb region, interrupting the BRD4 coding sequence, at the telomeric end of cosmid R31546 (arrows).
Southern hybridization with a probe to BRD4 intron 12 and exon 13 revealed rearranged fragments in t(15;19) cases 1 and 2 (Figure 3a) ▶ . A restriction map of the translocation breakpoint region was created by evaluating the cosmid R31546 sequence using Webcutter, Baylor College of Medicine Sequence Utilities. These correlations narrowed the translocation breakpoints to a 3.5-kb region delimited by BamHI and HindIII restriction sites at cosmid R31546 nucleotides 6433 and 9945, respectively. Hence, two independent genomic mapping methods, FISH and Southern blotting, demonstrated localization of the chromosome 19 translocation breakpoints to the 3′ end of the BRD4 coding sequence in each of two t(15;19) cancers.
There are two major isoforms of BRD4 encoded by alternate splicing products of 3179 nucleotides (GenBank Accession No. XM009302), and 5198 nucleotides (GenBank Accession No. AF386649). The t(15;19) breakpoints map between intron 10 and intron 13 of the longer BRD4 transcript (Figure 3b) ▶ . This is consistent with a breakpoint splitting the coding sequence of the longer BRD4 transcript approximately in half, while leaving the short transcript unaltered. At the protein level, the translocation is predicted to result in separation of the BRD4 N-terminal end, containing bromodomains, kinase-like motifs, and a serine-rich domain, from the C-terminal glutamine-rich and proline-rich domains (Figure 3c) ▶ .
Southern hybridization with a probe to intron 1 and exon 2 of Nop10p revealed rearranged fragments in each of two t(15;19) cancers (Figure 3b) ▶ . This observation localizes the chromosome 15q13 breakpoint to a ∼9-kb region containing the Nop10p candidate gene. The proximity of two other known genes, Golgin-67 and KCC3, to this region, remains to be determined.
FISH screening of 13 paraffin-embedded, supradiaphragmatic pediatric carcinomas revealed a sinonasal carcinoma with chromosome 19p13.1 and 15q13 rearrangements (Figure 1c ▶ and Table 1 ▶ , case 3), consistent with t(15;19). The remaining 12 carcinomas, including 3 thymic, 3 mucoepidermoid, 2 laryngeal, 1 nasopharyngeal, and 3 sinonasal, lacked chromosome 15 or 19 rearrangements.
Discussion
We describe herein the molecular characterization of t(15;19)(q13;p13.1), which is the cytogenetic hallmark of a particularly aggressive form of carcinoma in children and young adults. Our studies implicate the bromodomain-encoding gene, BRD4, as the chromosome 19 target of the t(15;19)(q13;p13.1) translocation, and they also localize the chromosome 15 breakpoints to a 9-kb genomic region. Notably, the BRD4 breakpoints in each of two t(15;19) cancers interrupt the BRD4 coding sequence, and these findings are strong evidence for a BRD4 fusion oncogene mechanism. This is in distinction to recurrent cancer translocations associated with non-fusion oncogenes, which generally involve the untranslated or flanking regions of those gene(s). Examples of non-fusion oncogenic mechanisms include those targeting c-myc, Bcl-2, and cyclin D1 in non-Hodgkin’s lymphomas, where the translocation breakpoints are scattered and vary by as much as 400 kb among tumors whose cytogenetically determined translocations are indistinguishable. 17-19 Of note, there are no reported examples of recurrent balanced translocation breakpoints which localize within the coding sequence of a gene, but which fail to alter the function of that gene. Our evidence, albeit based on analysis of only two t(15;19) carcinomas, is that the chromosome 19 genomic breakpoints cluster within a 3.5-kb region in the BRD4 coding sequence. Notably, there is precedent for involvement of bromodomain genes, eg, CBP and p300, in fusion oncogenes. 20 By contrast, bromodomain genes have not been implicated in non-fusion, translocation-associated, cancer cytogenetic mechanisms.
The translocation breakpoints identified thus far in t(15;19) cancers divide the BRD4 long isoform into two pieces wherein the N-terminal component contains chromatin-binding bromodomains, 21 predicted kinase activity, 22 and serine-rich potential transactivation or co-repressor domains (Figure 3c) ▶ . 23 The C-terminal component has a glutamine-rich domain with potential transactivation function, 24 and a proline-rich domain with potential roles in protein-protein interaction. 25 Further studies are needed to determine whether functional t(15;19) BRD4 oncoprotein(s) use one or both of these BRD4 regions.
An understanding of BRD4 oncogenic mechanisms will likely require characterization of the functional differences between the native short and long BRD4 isoforms. For example, relative levels of isoform expression might determine tissue-specific and developmentally relevant programs of cell proliferation or differentiation. Characterization of translocation-associated perturbation of the long BRD4 isoform, and likely sparing of the short isoform, may provide clues to mechanisms of normal and oncogenic BRD4 function. In particular, it is possible that the short isoform may play an inhibitory role by occupying chromosomal positions that would otherwise be available to the long isoform. In this sense, the t(15;19) translocation could function to inhibit BRD4 long isoform function by a dominant negative mechanism.
Within 40 kb telomeric of BRD4 is another candidate oncogene, NOTCH3 (Figure 2b) ▶ , which Dang et al 10 have implicated as a potential transcriptionally up-regulated target in a single case of t(15;19) carcinoma. Although the family member, NOTCH1, is a translocation target in human leukemia, 26 we have evaluated NOTCH3 expression by Northern blotting in t(15;19) versus control carcinomas, and have not observed translocation-associated positional effects on NOTCH3 expression (CA French, JA Fletcher, unpublished data). Therefore, although minor NOTCH3 positional effects cannot be excluded, our evidence highlights BRD4 rearrangement as the major event responsible for t(15;19)-mediated epithelial cell transformation.
We have narrowed the 15q13 breakpoint to a 9-kb region containing Nop10p. This highly conserved gene encodes a core small nucleolar ribonucleoprotein (snoRNP) which is essential for snoRNP function, 27 and which potentially interacts with snoRNAs and the RNA subunit of telomerase. 28 It is not yet known whether Nop10p participates with BRD4 in a t(15;19) fusion oncoprotein, and there are several other genes within 100 kb of Nop10p which are alternate targets of the translocation. These include Golgin-67 and the KCl transporter, KCC3. Golgin-67 encodes a Golgi apparatus-associated protein with a coiled-coil, proline-rich domain and a leucine zipper at its N-terminal end, both potential dimerization or protein-protein interaction domains that might contribute to transforming activity in a fusion oncogene. 29-31
The t(15;19) FISH assay described herein is a straightforward method for identification of t(15;19) cancers in archival materials. Therefore, this assay will be useful in establishing clinicopathological correlations, and it will be particularly important to determine whether t(15;19) carcinomas are uniformly associated with the devastating clinical course of those reported to date. Our initial FISH-based screen demonstrates that the t(15;19) is not vanishingly rare in pediatric cancers. Rather, the t(15;19) was detected in 1 of 13 such carcinomas, a sinonasal primary with squamous cell differentiation. The clinical course in this patient, in whom rapid local disease progression was accompanied by widespread bony metastases, was highly unusual for sinonasal squamous cell carcinoma but entirely in keeping with the t(15;19) syndrome. Notably, the FISH assays failed to show t(15;19) in three thymic carcinomas. This finding, together with our observation of t(15;19) in epiglottic and sinonasal primaries, demonstrates that thymic origin, which has been posited in several previous reports, fails to account for all t(15;19) carcinomas. It remains to be determined whether any t(15;19) carcinomas are convincingly thymic, but we favor the possibility that most such cases arise from the respiratory tract.
In sum, t(15;19) carcinoma is a uniquely devastating disease, typically arising in children or young adults, and associated with a distinctive molecular mechanism involving BRD4 rearrangement. Further clinicopathologic and molecular characterizations of this disease will likely provide important insights in the fields of pathology, molecular oncology, and cancer biology.
Acknowledgments
We thank Dr. Sheng Xiao and Dr. Andrew Weng for their valuable suggestions, Dr. Linda Ashworth, of the Lawrence Livermore National Laboratory (LLNL), for generously providing all cosmids used in this work, Dr. Ann Anderson for providing paraffin material, and Ms. Michelle Taffaro for her technical assistance.
Footnotes
Address reprint requests to Chris A. French, or Jonathan A. Fletcher, Department of Pathology, Brigham and Women’s Hospital, 75 Francis Street, Boston, MA 02115. E-mail: cafrench@bics.bwh.harvard.edu or jfletcher@partners.org.
Supported by NIH Institutional NRSA grant T32-HLO7627 to C.A.F.
References
- 1.Druker BJ, Lydon NB: Lessons learned from the development of an abl tyrosine kinase inhibitor for chronic myelogenous leukemia. J Clin Invest 2000, 105:3-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kroll TG, Sarraf P, Pecciarini L, Chen CJ, Mueller E, Spiegelman BM, Fletcher JA: PAX8-PPARγ1 fusion oncogene in human thyroid carcinoma. Science 2000, 289:1357-1360 [DOI] [PubMed] [Google Scholar]
- 3.Sozzi G, Bongarzone I, Miozzo M, Borrello MG, Blutti MG, Pilotti S, Della Porta G, Pierotti MA: A t(10;17) translocation creates the RET/PTC2 chimeric transforming sequence in papillary thyroid carcinoma. Genes Chromosomes Cancer 1994, 9:244-250 [DOI] [PubMed] [Google Scholar]
- 4.Nakata T, Kitamura Y, Shimizu K, Tanaka S, Fujimori M, Yokoyama S, Ito K, Emi M: Fusion of a novel gene, ELKS, to RET due to translocation t(10;12)(q11;p13) in a papillary thyroid carcinoma. Genes Chromosomes Cancer 1999, 25:97-103 [DOI] [PubMed] [Google Scholar]
- 5.Corvi R, Berger N, Balczon R, Romeo G: RET/PCM-1: a novel fusion gene in papillary thyroid carcinoma. Oncogene 2000, 19:4236-4242 [DOI] [PubMed] [Google Scholar]
- 6.Kardas I, Denis A, Babinska M, Gronwald J, Podolski J, Zajaczek S, Kram A, Lubinski J, Limon J: Translocation (X;1)(p11.2;q21) in a papillary renal cell carcinoma in a 14-year-old girl. Cancer Genet Cytogenet 1998, 101:159-161 [DOI] [PubMed] [Google Scholar]
- 7.Kubonishi I, Takehara N, Iwata J, Sonobe H, Ohtsuki Y, Abe T, Miyoshi I: Novel t(15;19)(q15;p13) chromosome abnormality in a thymic carcinoma. Cancer Res 1991, 51:3327-3328 [PubMed] [Google Scholar]
- 8.Kees UR, Mulcahy MT, Willoughby ML: Intrathoracic carcinoma in an 11-year-old girl showing a translocation t(15;19). Am J Pediatr Hematol Oncol 1991, 13:459-464 [DOI] [PubMed] [Google Scholar]
- 9.Lee AC, Kwong YI, Fu KH, Chan GC, Ma L, Lau YL: Disseminated mediastinal carcinoma with chromosomal translocation (15;19): a distinctive clinicopathologic syndrome. Cancer 1993, 72:2273-2276 [DOI] [PubMed] [Google Scholar]
- 10.Dang TP, Gazdar AF, Virmani AK, Sepetavec T, Hande KR, Minna JD, Roberts JR, Carbone DP: Chromosome 19 translocation, overexpression of Notch3, and human lung cancer. J Natl Cancer Inst 2000, 92:1355-1357 [DOI] [PubMed] [Google Scholar]
- 11.Vargas SO, French CA, Faul PN, Fletcher JA, Davis IJ, Dal Cin P, Perez-Atayde AR: Upper respiratory tract carcinoma with chromosomal translocation 15;19: evidence for a distinct disease entity of young patients with a rapidly fatal course. Cancer 2001, 92:1195-1203 [DOI] [PubMed] [Google Scholar]
- 12.Fletcher JA, Kozakewich HP, Hoffer FA, Lage JM, Weidner N, Tepper R, Pinkus GS, Morton CC, Corson JM: Diagnostic relevance of clonal cytogenetic aberrations in malignant soft-tissue tumors. N Engl J Med 1991, 324:436-442 [DOI] [PubMed] [Google Scholar]
- 13.Kuzume T, Kubonishi I, Takeuchi S, Takeuchi T, Iwata J, Sonobe H, Ohtsuki Y, Miyoshi I: Establishment and characterization of a thymic carcinoma cell line (Ty-82) carrying t(15;19)(q15;p13) chromosome abnormality. Int J Cancer 1992, 50:259-264 [DOI] [PubMed] [Google Scholar]
- 14.Xiao S, Renshaw A, Cibas ES, Hudson TJ, Fletcher JA: Novel fluorescence in situ hybridization approaches in solid tumors: characterization of frozen specimens, touch preparations, and cytological preparations. Am J Pathol 1995, 147:896-904 [PMC free article] [PubMed] [Google Scholar]
- 15.Cleary ML, Mellentin JD, Spies J, Smith SD: Chromosomal translocation involving the β T-cell receptor gene in acute leukemia. J Exp Med 1988, 167:682-687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bull JH, Harnden P: Efficient nuclear FISH on paraffin-embedded tissue sections using microwave pretreatment. Biotechniques 1999, 26:416-418, 422 [DOI] [PubMed] [Google Scholar]
- 17.Cleary ML, Smith SD, Sklar J: Cloning and structural analysis of cDNAs for Bcl-2 and a hybrid Bcl-2/immunoglobulin transcript resulting from the t(14;18) translocation. Cell 1986, 47:19-28 [DOI] [PubMed] [Google Scholar]
- 18.Johnston JM, Carroll WL: c-myc hypermutation in Burkitt’s lymphoma. Leuk Lymphoma 1992, 8:431-439 [DOI] [PubMed] [Google Scholar]
- 19.Dalla-Favera R, Ye BH, Lo Coco F, Chang CC, Cechova K, Zhang J, Migliazza A, Mellado W, Niu H, Chaganti S, Chen W, Rao PH, Parsa NZ, Louie DC, Offit K, Chaganti RSK: BCL-6 and the molecular pathogenesis of B-cell lymphoma. Cold Spring Harb Symp Quant Biol. 1994, 59:117-123 [DOI] [PubMed] [Google Scholar]
- 20.Giles RH, Dauwerse JG, Higgins C, Petrij F, Wessels JW, Beverstock GC, Dohner H, Jotterand-Bellomo M, Falkenburg JH, Slater RM, van Ommen GJ, Hagemeijer A, van der Reijden BA, Breuning MH: Detection of CBP rearrangements in acute myelogenous leukemia with t(8;16). Leukemia 1997, 11:2087-2096 [DOI] [PubMed] [Google Scholar]
- 21.Winston F, Allis CD: The bromodomain: a chromatin-targeting module? Nat Struct Biol 1999, 6:601-604 [DOI] [PubMed] [Google Scholar]
- 22.Dey A, Ellenberg J, Farina A, Coleman AE, Maruyama T, Sciortino S, Lippincott-Schwartz J, Ozato K: A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G(2)-to-M transition. Mol Cell Biol 2000, 20:6537-6549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen C, Agnes F, Gelinas C: Mapping of a serine-rich domain essential for the transcriptional, antiapoptotic, and transforming activities of the v-Rel oncoprotein. Mol Cell Biol 1999, 19:307-316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Escher D, Bodmer-Glavas M, Barberis A, Schaffner W: Conservation of glutamine-rich transactivation function between yeast and humans. Mol Cell Biol 2000, 20:2774-2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alexandropoulos K, Cheng G, Baltimore D: Proline-rich sequences that bind to Src homology 3 domains with individual specificities. Proc Natl Acad Sci USA 1995, 92:3110-3114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ellisen LW, Bird J, West DC, Soreng AL, Reynolds TC, Smith SD, Sklar J: TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell 1991, 66:649-661 [DOI] [PubMed] [Google Scholar]
- 27.Henras A, Henry Y, Bousquet-Antonelli C, Noaillac-Depeyre J, Gelugne JP, Caizergues-Ferrer M: Nhp2p and Nop10p are essential for the function of H/ACA snoRNPs. EMBO J 1998, 17:7078-7090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pogacic V, Dragon F, Filipowicz W: Human H/ACA small nucleolar RNPs and telomerase share evolutionarily conserved proteins NHP2 and NOP10. Mol Cell Biol 2000, 20:9028-9040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McWhirter JR, Galasso DL, Wang JY: A coiled-coil oligomerization domain of Bcr is essential for the transforming function of Bcr-Abl oncoproteins. Mol Cell Biol 1993, 13:7587-7595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao S, McCarthy JG, Aster JC, Fletcher JA: ZNF198-FGFR1 transforming activity depends on a novel proline-rich ZNF198 oligomerization domain. Blood 2000, 96:699-704 [PubMed] [Google Scholar]
- 31.Chaplin T, Bernard O, Beverloo HB, Saha V, Hagemeijer A, Berger R, Young BD: The t(10;11) translocation in acute myeloid leukemia (M5) consistently fuses the leucine zipper motif of AF10 onto the HRX gene. Blood 1995, 86:2073-2076 [PubMed] [Google Scholar]