Abstract
FUBI (failure of ureteric bud invasion) is a highly inbred strain of mouse with a high spontaneous incidence of uni- or bilateral renal agenesis (60%). Bilateral renal agenesis is lethal within 2 days after birth. The primary defect of FUBI is failure of the ureteric bud to penetrate into the metanephric mesenchyme at around embryonic day 11, resulting in apoptosis of metanephric cells and leading to renal agenesis on the affected side. The metanephros seemed to be normal because co-culturing of the FUBI metanephros with homologous spinal cord induced differentiation of the rudiment, but co-culturing with the homologous ureteric bud frequently did not. Genetic analysis revealed that more than two genes were involved in this malformation and we mapped one of the modifier loci, fubi1, on chromosome 2, at ∼65 cM from the centromere. In this region, there are two possible candidate genes, Wilms’ tumor 1 and formin, that play important roles in kidney development. Some of formin mutants shared a similar phenotype with FUBI; however, there was no difference in the expression of formin in embryonic kidneys between FUBI and control NFS/N mice. Studies of fubi1 congenic mice indicated that interaction of two or more loci is essential for the FUBI phenotype.
Renal agenesis is a relatively frequent congenital anomaly in humans, although its incidence varies widely. 1-3 Renal agenesis can be bilateral, as represented by Potter’s syndrome, or unilateral. Unilateral agenesis does not cause any detectable health problem and thus most affected individuals remain asymptomatic throughout life. 4 Bilateral agenesis is fatal within a few days after birth, and thus not subject to medical intervention. For these reasons, little attention has been paid to renal agenesis despite its relatively high incidence. There is strong evidence that renal agenesis is a complex of different developmental anomalies. Detailed study of these anomalies may provide key information on the sequence of complicated steps in the process of kidney organogenesis. 5,6
In higher vertebrates, renal organogenesis is initiated by condensation of metanephric mesenchyme in the intermediate mesoderm. Subsequently, the ureteric bud extends from the Wolffian duct and invades the metanephric mesenchyme. Further differentiation occurs depending on a series of reciprocal inductive interactions between the epithelial ureteric bud and the metanephric mesenchyme. Any defects in these steps may cause arrest of renal rudiment formation. There are several experimental animal models of renal agenesis, either spontaneous 7-9 or induced by gene manipulation. 10-16 Anomalies in the urogenital or skeletal system are not infrequently associated with the renal agenesis, 8,9 and the penetrance of the renal agenesis phenotype is usually not 100%. 9,14,15,17 These observations suggest that kidney development is a multistep and rather complicated process and that many genes are involved in the process. Wilms’ tumor 1 (WT1), 10 c-ret oncogene, 11,17 formin, 9,15 and Wnt4 13 are some of the genes involved in renal organogenesis.
In this report, we describe a novel renal agenesis model, the highly inbred FUBI (failure of ureteric bud invasion) strain of mice. The basic observation is that the penetration of the ureteric bud into the metanephric blastema is partially impaired at day 11 of gestation, resulting in unilateral renal agenesis (50%) or lethal bilateral renal agenesis (10%). The kidney of the unaffected side is normal in both morphology and function. In vitro culturing of the kidney rudiment of embryonic day 11.5 revealed a specific defect in the ureteric bud penetration into the metanephric blastema. The metanephric mesenchyme, into which the ureteric bud failed to invade, eventually died out by apoptosis. Genetic analysis suggested that more than two genes are essential for manifesting the mutant phenotype in FUBI mouse. We have mapped one of the relevant loci on mouse chromosome 2.
Materials and Methods
Mice
DDY is an inbred strain of mice established from closed-colony ddY mice by brother-sister matings throughout 20 generations at Saitama Cancer Center Research Institute (Saitama, Japan). A mutant mouse with a unilateral kidney was first found among DDY mice, and several littermates were found to have a similar anomaly. Thereafter this anomaly was maintained by selectively mating a male with unilateral renal agenesis with a female littermate for >50 generations. Unilateral renal agenesis could easily be identified by palpation and confirmed by postmortem examination. We named the mouse strain bearing this anomaly FUBI according to its phenotypic defect. The NFS/N strain maintained in our laboratory had normal kidneys and was used as a normal control strain.
Observation of Embryos
Embryonic day 0.5 (E0.5) was defined as the day on which a vaginal plug was found after overnight mating. Embryos at various embryonic stages were recovered and dissected under a surgical microscope. After removal of the abdominal viscera, the retroperitoneal structures were carefully examined to confirm the presence or absence of the kidney and ureter on either side. For histopathological sections, the embryos were fixed with 10% neutral buffered formalin, embedded in paraffin, serially sectioned, and stained with hematoxylin and eosin.
In Situ End-Labeling
Apoptotic cells were detected by direct immunoperoxidase detection of digoxigenin-labeled genomic DNA in thin sections using an ApopTaq Plus kit (Intergene, Purchase, NY). 18
Immunohistochemistry
Paraffin sections of embryos were stained with peroxidase-labeled anti-laminin antibody (rabbit anti-laminin, no. Z0097; DAKO Inc., Glostrup, Denmark).
Organ Culture
Kidney rudiments and dorsal spinal cords were microdissected from E11.5 embryos in Improved Minimum Essential Medium (Life Technologies, Rockville, MD). Each kidney rudiment was co-cultured with dorsal spinal cord of the same embryo. The spinal cord was placed on the opposite side of the ureteric bud, and the organs were cultured for 1 to 7 days in 5% CO2/95% air at 37°C in a Trowell-type organ culture in Improved Minimum Essential Medium supplemented with 10% fetal calf serum on 0.1-μm pore size Nucleopore polycarbonate membranes (Corning Inc.-Life Sciences, Acton, MA) placed on top of a metal grid. 19 The appearance of clear condensates was considered to indicate the induction of differentiation of the metanephric mesenchyme. As a control, kidney rudiments from E11.5 NFS/N embryos were cultured in the same manner.
Genetic Analysis
Genomic DNA was isolated from brains according to standard protocols. For linkage analysis, the simple sequence repeat (microsatellite) length polymorphism assay was performed. All primers for microsatellite analysis were purchased from Research Genetics, Inc. (Huntsville, AL). Methods for polymerase chain reaction (PCR) and agarose electrophoresis of PCR products have been described previously. 20 Of 363 microsatellite loci examined, 107 were polymorphic between FUBI and NFS/N. For genome-wide screening, we selected 52 loci, that were distributed so as to cover ∼75% of the entire genome.
Statistical Analysis
The association of renal agenesis with alleles of the markers was evaluated by a chi-square test of the independence (degree of freedom = 1) of the frequencies in heterozygous (FUBI/NFS) and homozygous (FUBI/FUBI) mice with renal agenesis. Associations were considered significant when the chi-square value exceeded 11.7 (a value equivalent to 95% probability of linkage in mouse backcrosses). 21,22
Construction of Congenic Strain
Reciprocal congenic strains were constructed between FUBI and NFS/N with regard to the locus fubi1, which is responsible for renal agenesis. 23,24 In brief, the congenic strain FUBI-fubi1NFS was constructed by repeated backcrossing of the (FUBI × NFS/N) F1 to FUBI and selecting males that were heterozygous for fubi1 by PCR of microsatellite markers D2Mit91, D2Mit58, D2Mit63, and D2Mit223. Males heterozygous at fubi1 were again backcrossed to FUBI females, and their male progeny, heterozygous for fubi1, were used for the next generation. This process was repeated until generation N10, at which point heterozygous males and females were intercrossed and the progeny homozygous for fubi1NFS were selected and mated. Subsequently, the congenic strain FUBI-fubi1NFS was maintained by brother-sister mating, and is now at generation N10F10. The reciprocal congenic strain NFS-fubi1FUBI was constructed by the reciprocal backcrossings and selections. The line is now at generation N10F10.
Sequencing of Formin cDNA
Fifteen cDNA fragments, which covered the entire coding sequence of isoform Ia and IV of murine formin gene 9 with several overlapping sequences, were amplified by reverse transcriptase (RT)-PCR from FUBI and NFS kidney RNA. Each PCR product was cloned into the pCR-TOPO II vector using TOPO TA cloning kit (Invitrogen, Carlsbad, CA). The plasmid DNAs were sequenced with the AmpliTaq Cycle Sequence kit (Perkin Elmer, Foster City, CA) using an ABI PRISM 377 DNA sequencer. The homology search was performed with NCBI BLAST (http://www.ncbi.nlm.nih.gov/BLAST/).
RT-PCR
Expression of candidate genes, Wilms’ tumor 1 (WT1) 10 and formin (Fmn), 9,25 was analyzed using a semiquantitative RT-PCR method. Beta-actin was used as an internal control. Total RNA was isolated from a kidney of an E11.5 FUBI embryo and an E11.5 NFS/N embryo using an RNeasy mini kit (Qiagen, Hilden, Germany) and treated with RNase-free DNase I (Qiagen). A 50-ng aliquot of total RNA was reverse-transcribed with 0.2 μg of random hexadeoxynucleotides (pd(N)6) primer using a first-strand cDNA synthesis kit (Amersham Pharmacia Biotech, Uppsala, Sweden) and diluted to a final volume of 50 μl with distilled water. For each PCR reaction, a 100-μl mixture containing 2 μl of cDNA, PCR buffer (10 mmol/L Tris-HCl) (pH 8.3), 50 mmol/L KCl, 1.5 mmol/L MgCl2), 0.2 mmol/L dNTP, 0.5 μmol/L forward and reverse primers, and 1 U of Taq polymerase (Perkin Elmer, Foster City, CA) was divided into four aliquots. These aliquots were then subjected to different numbers of PCR cycles. The PCR conditions were 30, 35, 40, or 45 cycles of 94°C for 1 minute, 58°C (for WT1) or 59°C (for Fmn and β-actin) for 1 minute, and 72°C for 1 minute. A final extension reaction was performed at 72°C for 10 minutes. Eight μl of each PCR product was analyzed by electrophoresis in 3% agarose. DNA bands were visualized by staining with ethidium bromide. The sequences of the primers were as follows: WT1: forward, 5′-CGGGATCCTTCACCTTGCACTTCTCGG-3′, reverse 5′-CGAATTCGTGCTGTATCCTTGGTTGCG-3′; Fmn isoform I-III: forward 5′-CCTAGCCATCATCACATGCC-3′, reverse 5′-TCCGTAGCGAAGAGGTAAGC-3′; Fmn isoform IV:forward 5′-GCTGCTCAACATTGACATGC-3′, reverse 5′- CTTGCTGTTACTAATGCGTGC-3′; β-actin: forward 5′-CCTCATGAAGATCCTGACCG-3′, reverse 5′-TCCACACAGAGTACTTGCGC-3′.
Results
Renal Agenesis in FUBI Mice
Among 256 FUBI mice, three kinds of renal phenotypes were observed: 133 mice with unilateral agenesis (Figure 1A) ▶ , 20 with bilateral agenesis (Figure 1B) ▶ , and 103 with apparently normal kidneys. The neonates with bilateral renal agenesis died within 2 days after birth. Both sexes were equally affected. As seen in Table 1 ▶ , from any combination of parental phenotypes, progeny with all three phenotypes were born. Hydronephrosis was occasionally observed in older mice (48 of 368, 13%). In all mice, the adrenal glands were present bilaterally. No abnormality was found in the genital or skeletal system in either sex. Out of 253 NFS/N mice used as a control strain in this study, none had kidney abnormalities.
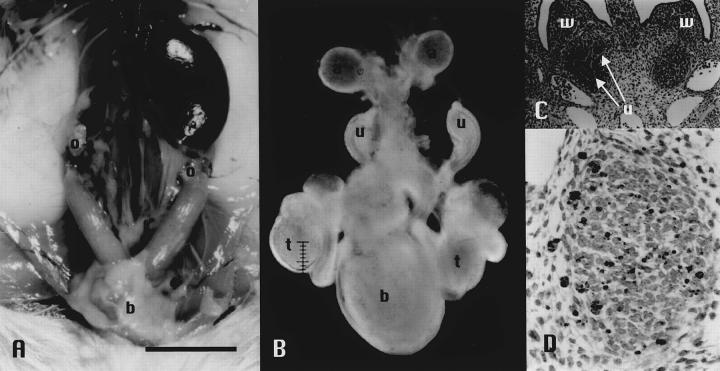
Figure 1.
Macroscopic and microscopic findings of FUBI mice. A: Unilateral renal agenesis in an adult FUBI mouse. Scale bar, 1 cm. B: Bilateral renal agenesis in an E16.5 FUBI embryo. Scale bar, 0.1 mm. C: Serial sections from E11.5 FUBI embryos showing failure of left ureteric bud to invade into condensed metanephric blastema. Normal development in the right kidney. White arrows show the right ureteric bud that invades and bifurcates in the metanephric blastema (H&E stain, original magnification, ×120). D: Apoptotic signals detected in the metanephric blastema without ureteric bud invasion. Direct immunoperoxidase detection of digoxigenin-labeled genomic DNA in sections using an ApopTaq Plus kit (Intergen, Purchase, NY). 18 Original magnification, ×330. a, adrenal gland; b, bladder; o, ovary; t, testis; u, ureter/ureteric bud; w, Wolffian duct.
Table 1.
Kidney Phenotypes of Progeny of FUBI Parents with Various Phenotypes
| Phenotype of parents | No. of progeny | Phenotype of progeny | |||
|---|---|---|---|---|---|
| Normal | Unilateral | Bilateral | |||
| Right | Left | ||||
| N× N | 66 | 29 | 15 | 17 | 5 |
| N× U | 67 | 22 | 24 | 14 | 7 |
| U× N | 70 | 33 | 15 | 17 | 5 |
| U× U | 53 | 19 | 11 | 20 | 3 |
N, Normal kidney; U, unilateral agenesis (right or left).
Renal Agenesis in FUBI Embryos
To identify the early changes in renal organogenesis, mouse embryos were examined at various embryonic stages. In normal embryos of NFS/N mice, the ureteric buds contacted the metanephric rudiments at E11.5 and subsequently invaded the latter. These buds actively sprouted branches and showed well-developed arborization in the renal rudiments on both sides. In some FUBI embryos at E11.5, the ureteric bud invaded the metanephric mesenchyme on one side but failed to do so on the other side (Figure 1C) ▶ . Such abnormalities were observed in many embryos, and failure to invade was sometimes observed on both sides. At E12.5, remarkable apoptosis was observed in the metanephros without ureteric bud invasion (Figure 1D) ▶ . A serial section from the same embryo stained with anti-laminin antibody revealed that the basement membrane of the ureteric bud remained intact, indicating that the ureteric bud failed to contact and invade the metanephric mesenchyme (data not shown). Figure 1B ▶ shows a male with bilateral renal agenesis, in which the ureters on both sides remained blind ends and the renal rudiments had been completely absorbed. Other urogenital organs and the skeletal system were normally developed. Taken together, these results indicate that the essential process in this mutation is failure of the ureteric bud to invade the metanephric rudiment at around E11. Once the ureteric bud invades the metanephros, subsequent development seems to proceed normally, but when the invasion fails, the metanephric rudiment is absorbed entirely by E13 to E14 (data not shown).
Renal Development in Organ Culture
The defect of the ureteric bud in FUBI embryos was confirmed in organ cultures of the kidney anlage. Kidney rudiment and spinal cord were microdissected from E11.5 FUBI and NFS/N embryos and cultured in a Trowell-type organ culture for up to 7 days according to the method described by Saxen and Lehtonen. 19 In this culture system, bifurcation of the ureteric bud and the subsequent induction of metanephric mesenchyme differentiation were observed in all of the NFS/N (control strain) kidney rudiments cultured (14 of 14, 100%). On the other hand, bifurcation of the ureteric bud was observed in only 43% (6 of 14) of cultured FUBI kidney rudiments. The induction of metanephric mesenchyme differentiation progressed in FUBI kidney rudiments with ureteric bud bifurcation (6 of 6, 100%), but did not in those without ureteric bud bifurcation (0 of 8, 0%). In this experiment, induction of metanephric mesenchyme differentiation by spinal cord progressed normally in every case, indicating that the specific defect is not in the metanephric mesenchyme but in the ureteric bud.
Inheritance of Renal Agenesis in FUBI Mice
To determine the mode of inheritance and to map the mutant locus, reciprocal F1 and F2 between FUBI and NFS/N and backcrosses to FUBI were prepared. Renal agenesis was observed in 153 of 256 mice (60%) in the parental FUBI strain, but in none of 253 NFS/N mice and none of 108 F1 hybrids. Therefore, the mutation is inherited recessively at relatively low penetrance. Out of 228 mice of the backcross generation, FUBI × (NFS/N × FUBI)F1 and (NFS/N × FUBI)F1 × FUBI, 39 mice (17.1%) showed either bilateral or unilateral renal agenesis. Out of 275 F2 mice, 7 (2.5%) developed agenesis. The low penetrance in the parental FUBI mice combined with these observations suggests that two or more genes may be involved in the manifestation of the mutant phenotype.
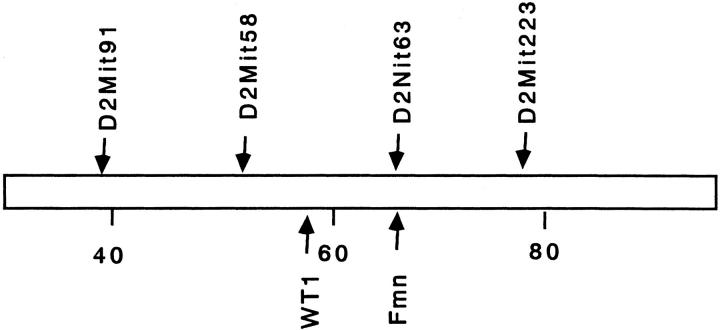
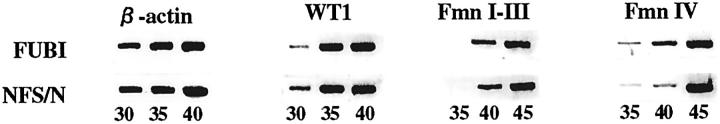
Subsequently we attempted to map the responsible genes on the mouse chromosomes using 39 backcross mice with renal agenesis. A total of 52 microsatellite markers polymorphic between FUBI and NFS/N were selected to cover ∼75% of the entire genome and a genome-wide screening for linkage disequilibrium was done. As shown in Table 2 ▶ , significant (chi-square > 13.8; P < 0.001) linkage was found on chromosome 2 at ∼65 cM from the centromere. We named this locus fubi1 (failure of ureteric bud invasion 1). In this region (Figure 2) ▶ , two important candidate genes that play pivotal roles in kidney development, WT1 10 and Fmn, 9,25 are mapped. However, no difference was found in the sequence of the Fmn-coding region between FUBI and NFS/N (data not shown). The levels of expression of these two genes in the kidney at E11.5 were examined by semiquantitative RT-PCR (Figure 3) ▶ . There was no significant difference in their expression between FUBI and NFS/N. We could not exclude the possibility, however, that the level of expression on the affected side falls below a critical level at E11.5.
Table 2.
Genome-Wide Screening of the Loci Affecting FUBI Phenotype Using 39 Backcross Mice with Renal Agenesis
| Chromosome | Marker locus | Position (cM) | Homo | Hetero | Chi-square value |
|---|---|---|---|---|---|
| 2 | D2Mit32 | 11.0 | 19 | 20 | |
| D2Mit91 | 37.0 | 23 | 16 | ||
| D2Mit15 | 50.0 | 29 | 10 | 9.3 | |
| D2Mit58 | 51.4 | 31 | 8 | 13.6 | |
| D2Mit63 | 65.2 | 32 | 7 | 16.0 | |
| D2Mit223 | 76.7 | 29 | 10 | 9.3 | |
| D2Mit174 | 105.0 | 22 | 17 | ||
| 4 | D4Mit103 | 1.3 | 19 | 20 | |
| D4Mit27 | 42.5 | 25 | 14 | ||
| D4Mit59 | 78.9 | 30 | 9 | 11.3 | |
| 12 | D12Mit12 | 6.0 | 22 | 17 | |
| D12Mit3 | 32.0 | 28 | 11 | 7.4 | |
| D12Mit20 | 58.0 | 21 | 18 |
Chi-square value was calculated using the following formula; chi-square = (a-19.5)2/19.5 + (b-19.5)2/19.5. Data for chromosomes on which chi-square values were <5.0 are not shown.
Figure 2.
Fubi1 region on chromosome 2. Scale bar shows distance from the centromere. Map positions of all loci are from Chromosome Committee Reports for the Mouse Genome (http://www.informatics.jax.org/). In congenic lines for fubi1, the genotypes of all four microsatellite loci were confirmed.
Figure 3.
Semiquantitative analysis of expression of WT1 and Fmn by RT-PCR using β-actin as an internal control. The numbers below each gel image indicate the numbers of cycles.
To further examine the role of fubi1 and other genes in renal agenesis, we generated fubi1 congenic strains by extensive marker loci-assisted selective backcrossing (Table 3) ▶ . In this attempt, the fubi1 region examined was somewhat broader consisting of ∼40 cM of chromosome 2 between D2Mit91 and D2Mit223. To our surprise, NFS-fubi1FUBI did not develop renal agenesis at all, indicating that fubi1 is not a causative gene but rather one of multiple modifier loci. The effect of fubi1 was somehow dose-dependent, because the incidence of renal agenesis in FUBI, (FUBI-fubi1NFS × FUBI)F1, and FUBI-fubi1NFS was 60%, 15%, and 3%, respectively. The effect of non-fubi1 loci was also dose-dependent, because the incidence in FUBI, (NFS-fubi1FUBI × FUBI)F1 and NFS-fubi1FUBI was 60%, 3%, and 0%. Herein, for the sake of discussion, we assumed that the non-fubi1 loci were loci on chromosomes other than 2. The observations with these congenic stains indicated that collaboration of at least two loci is essential for renal agenesis in this mutant.
Table 3.
Contribution of fubi1 Allele and Other Genetic Background to Renal Agenesis
| Strains | Genotype | Renal agenesis/no. of mice | % Mutant | |
|---|---|---|---|---|
| fubi1 | Loci on other chromosomes | |||
| FUBI | FUBI /FUBI | FUBI /FUBI | 153 /256 | 60% |
| (FUBI× NFS)F1 | FUBI /NFS | FUBI /NFS | 0 /108 | 0% |
| (FUBI-fubi1 NFS× FUBI)F1 | FUBI /NFS | FUBI /FUBI | 9 /59 | 15% |
| FUBI-fubi1 NFS | NFS /NFS | FUBI /FUBI | 8 /266 | 3% |
| (NFS-fubi1 FUBI× FUBI)F1 | FUBI /FUBI | FUBI /NFS | 2 /62 | 3% |
| NFS-fubi1 FUBI | FUBI /FUBI | NFS /NFS | 0 /275 | 0% |
| NFS | NFS /NFS | NFS /NFS | 0 /253 | 0% |
FUBI-fubi1 NFS; FUBI congenic strain for NFS/N allele of fubi1 (fubi1 NFS); NFS-fubi1 FUBI; NFS/N congenic strain for fubi1 FUBI allele; FUBI/FUBI: FUBI allele homozygous; FUBI/NFS; heterozygous NFS/NFS: NFS allele homozygous.
Discussion
Mutation-induced congenital anomalies are promising tools for investigating critical genetic events in organogenesis. So far several spontaneous murine models of defects in renal organogenesis have been reported, including Danforth Short Tail (Sd), 7,26 urogenital syndrome (us) 8 and limb deformity (ld). 9 Furthermore, genetic manipulations have identified a number of genes that affect the organogenesis of the urogenital system. 10-15 Unlike the anomalies in most other models, the anomaly in FUBI mice is pure renal agenesis without accompanying urogenital or skeletal system malformations.
Organogenesis of the kidney consists of at least four steps. 5,6 Step 1 is formation of the metanephric mesenchyme in the intermediate mesoderm and subsequent outgrowth of the ureteric bud from the Wolffian duct. WT1 seems to play an important role in this step. WT1 expression is detected first in the intermediate mesoderm and then in the mesenchyme before initiation of the ureteric bud formation. 27 In WT1 knockout mice, the ureteric bud fails to grow out of the Wolffian duct and the metanephric blastema degenerates completely. 10 Step 2 is invasion of the ureteric bud into the metanephric mesenchyme. Several genes, including the c-ret oncogene, are considered to be involved in this step. Knockout of c-ret 11,17 or its ligand, GDNF, 12 in mice perturbs the invasion of the ureteric bud and induces renal agenesis of variable manifestation. Step 3 involves reciprocal inductive interactions between the ureteric bud and the metanephric mesenchyme. Many genes are thought to participate in this step. In Wnt-4 mutant embryos, for example, the ureteric bud invades the mesenchyme but no epithelial tubules develop. 13 Step 4 is further differentiation including vascularization and innervation, but the molecular mechanism of these processes remains to be clarified. Organ culture studies revealed that FUBI metanephric mesenchyme is fully susceptible to induction by the homologous spinal cord but is frequently not susceptible to the ureteric bud that fails to bifurcate. There seems to be some stochastic process in which the FUBI ureteric bud fails or does not fail to induce metanephros differentiation. Subtle timing of the secretion of inductive factors or the spacial relationship between the mesenchyme and the ureteric bud may generate the ultimate three phenotypes. Similar stochastic determination of phenotypes is observed in other renal agenesis models. 9,15
Genetic analysis suggests that two or more genes are involved in the FUBI phenotype. Low penetrance of the genes made our genetic analysis more difficult than we expected. Such low penetrance is almost ubiquitous among renal agenesis models, and is not specific to FUBI. 9,11,14,15 A genome-wide screening of the mutant mice in the backcross generation revealed a significant linkage disequilibrium on chromosome 2. The fubi1 locus in this region seemed to be one of multiple loci modifying the FUBI phenotype in a dose-dependent manner. Of the two candidate genes, WT1 is unlikely to encode such a modifier because it has a role in step 1. 10,27 The limb deformity (ld) mutants of Fmn seem to be closer to the FUBI defect. 9,15 Fmn is a big transcription factor gene with at least 24 exons and its mutations in several domains have been known to yield a variety of renal agenesis syndromes. 9,15,25 Especially, knockout mice of the isoform IV of Fmn show a pure form of renal agenesis without skeletal abnormality, thus resembling to FUBI. 15 However, there was no mutation in the Fmn-coding sequence and no difference in its expression in the kidney anlage between FUBI and the control strain, at least 1 day before the critical interaction. We cannot exclude an abnormality in Fmn as a candidate of fubi1, but it is not a likely candidate. The fact that the congenic NFS/N mice with fubi1FUBI did not show renal agenesis but the reciprocal congenic FUBI-fubi1NFS did, at least at a low rate, may support this assumption. Identification of the genes involved in FUBI renal agenesis and understanding their interactions will require further investigation.
Acknowledgments
We thank Dr. H. Sariola, K. Sainio, and L. Saxen, University of Helsinki, Finland, for stimulating discussions and instruction in the organ culture technique; and Mr. Y. Toda, H. Koda, and Ms. S. Uemura for expert technical assistance.
Footnotes
Address reprint requests to Hiroshi Hiai, Department of Pathology and Biology of Diseases, Kyoto University Graduate School of Medicine, Yoshida-Konoe-cho, Sakyo-ku, Kyoto 606-8501, Japan. E-mail: hiai@path1.med.kyoto-u.ac.jp.
Supported by Grants-In-Aid for Basic Research from the Ministry of Education, Culture, Sports, and Science, Japan; and the Eiko Norihara Memorial Fund.
References
- 1.Buchta RM, Viseskul C, Gilbert EF, Sarto GE, Opitz JM: Familial bilateral renal agenesis and hereditary renal adysplasia. Z Kinderheilkd 1973, 115:111-129 [DOI] [PubMed] [Google Scholar]
- 2.Roodhooft AM, Birnholz JC, Holmes LB: Familial nature of congenital absence and severe dysgenesis of both kidneys. N Engl J Med 1984, 310:1341-1345 [DOI] [PubMed] [Google Scholar]
- 3.Wilson RD, Baird PA: Renal agenesis in British Columbia. Am J Med Genet 1985, 21:153-169 [DOI] [PubMed] [Google Scholar]
- 4.Argueso LR, Ritchey ML, Boyle ET, Jr, Milliner DS, Bergstralh EJ, Kramer SA: Prognosis of patients with unilateral renal agenesis. Pediatr Nephrol 1992, 6:412-416 [DOI] [PubMed] [Google Scholar]
- 5.Saxen L, Sariola H: Early organogenesis of the kidney. Pediatr Nephrol 1987, 1:385-392 [DOI] [PubMed] [Google Scholar]
- 6.Kuure S, Vuolteenaho R, Vainio S: Kidney morphogenesis: cellular and molecular regulation. Mech Dev 2000, 92:31-45 [DOI] [PubMed] [Google Scholar]
- 7.Gluecksohn-Schoenheimer S: The morphological manifestations of a dominant mutation in mice affecting tail and urogenital system. Genetics 1943, 28:341-348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lane PW, Birkenmeier CS: Urogenital syndrome (us): a developmental mutation on chromosome 2 of the mouse. Mamm Genome 1993, 4:481-484 [DOI] [PubMed] [Google Scholar]
- 9.Maas R, Elfering S, Glaser T, Jepeal L: Deficient outgrowth of the ureteric bud underlies the renal agenesis phenotype in mice manifesting the limb deformity (ld) mutation. Dev Dyn 1994, 199:214-228 [DOI] [PubMed] [Google Scholar]
- 10.Kreidberg JA, Sariola H, Loring JM, Maeda M, Pelletier J, Housman D, Jaenisch R: WT-1 is required for early kidney development. Cell 1993, 74:679-691 [DOI] [PubMed] [Google Scholar]
- 11.Schuchardt A, D’Agati V, Larsson-Blomberg L, Costantini F, Pachnis V: Defects in the kidney and enteric nervous system of mice lacking the tyrosine kinase receptor Ret. Nature 1994, 367:380-383 [DOI] [PubMed] [Google Scholar]
- 12.Sanchez MP, Silos-Santiago I, Frisen J, He B, Lira SA, Barbacid M: Renal agenesis and the absence of enteric neurons in mice lacking GDNF. Nature 1996, 382:70-73 [DOI] [PubMed] [Google Scholar]
- 13.Stark K, Vainio S, Vassileva G, McMahon AP: Epithelial transformation of metanephric mesenchyme in the developing kidney regulated by Wnt-4. Nature 1994, 372:679-683 [DOI] [PubMed] [Google Scholar]
- 14.Muller U, Wang D, Denda S, Meneses JJ, Pedersen RA, Reichardt LF: Integrin alpha8beta1 is critically important for epithelial-mesenchymal interactions during kidney morphogenesis. Cell 1997, 88:603-613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wynshaw-Boris A, Ryan G, Deng CX, Chan DC, Jackson-Grusby L, Larson D, Dunmore JH, Leder P: The role of a single formin isoform in the limb and renal phenotypes of limb deformity. Mol Med 1997, 3:372-384 [PMC free article] [PubMed] [Google Scholar]
- 16.Kreidberg JA: Gene targeting in kidney development. Med Pediatr Oncol 1996, 27:445-452 [DOI] [PubMed] [Google Scholar]
- 17.Schuchardt A, D’Agati V, Larsson-Blomberg L, Costantini F, Pachnis V: RET-deficient mice: an animal model for Hirschsprung’s disease and renal agenesis. J Intern Med 1995, 238:327-332 [DOI] [PubMed] [Google Scholar]
- 18.Gavrieli Y, Sherman Y, Ben-Sasson SA: Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J Cell Biol 1992, 119:493-501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saxen L, Lehtonen E: Embryonic kidney in organ culture. Differentiation 1987, 36:2-11 [DOI] [PubMed] [Google Scholar]
- 20.Yamada Y, Matsushiro H, Ogawa MS, Okamoto K, Nakakuki Y, Toyokuni S, Fukumoto M, Hiai H: Genetic predisposition to pre-B lymphomas in SL/Kh strain mice. Cancer Res 1994, 54:403-407 [PubMed] [Google Scholar]
- 21.Mock BA, Krall MM, Dosik JK: Genetic mapping of tumor susceptibility genes involved in mouse plasmacytomagenesis. Proc Natl Acad Sci USA 1993, 90:9499-9503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamada Y, Shisa H, Matsushiro H, Kamoto T, Kobayashi Y, Kawarai A, Hiai H: T lymphomagenesis is determined by a dominant host gene thymic lymphoma susceptible mouse-1 (TLSM-1) in mouse models. J Exp Med 1994, 180:2155-2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flaherty L: Congenic strains. Foster HL Small JD Fox JG eds. The Mouse in Biomedical Research. 1981, :pp 215-222 Academic Press, New York [Google Scholar]
- 24.Silver LM: Creation of a congenic strain. Silver LM eds. Mouse Genetics. 1995, :pp 46-49 Oxford University Press, New York [Google Scholar]
- 25.Wang CC, Chan DC, Leder P: The mouse formin (Fmn) gene: genomic structure, novel exons, and genetic mapping. Genomics 1997, 39:303-311 [DOI] [PubMed] [Google Scholar]
- 26.Mesrobian HG, Sulik KK: Characterization of the upper urinary tract anatomy in the Danforth spontaneous murine mutation. J Urol 1992, 148:752-755 [DOI] [PubMed] [Google Scholar]
- 27.Pritchard-Jones K, Fleming S, Davidson D, Bickmore W, Porteous D, Gosden C, Bard J, Buckler A, Pelletier J, Housman D, Van Heyningen V, Hastie N: The candidate Wilms’ tumour gene is involved in genitourinary development. Nature 1990, 346:194-197 [DOI] [PubMed] [Google Scholar]