Abstract
Gain of the long arm of chromosome 8 (8q) is one of the most common gains found in the advanced prostate cancer by comparative genomic hybridization. We have previously identified a putative target gene for the 8q gain, EIF3S3, that encodes a p40 subunit of eukaryotic translation initiation factor 3 (eIF3). Here, we studied the frequency of the EIF3S3 amplification in different stages of prostate cancer and co-amplification of EIF3S3 and oncogene MYC. In addition, prognostic utility of the EIF3S3 copy number alteration was evaluated. The analyses were done with fluorescence in situ hybridization and tissue microarray technology. High-level amplification of EIF3S3 was found in 11 of 125 (9%) of pT1/pT2 tumors, 12 of 44 (27%) of pT3/pT4 tumors, and 8 of 37 (22%) of lymph node metastases as well as in 26 of 78 (33%) and 15 of 30 (50%) of hormone refractory locally recurrent tumors and metastases, respectively. The amplification was associated with high Gleason score (P < 0.001). One of the 79 tumors with EIF3S3 amplification had only two copies of MYC, whereas all tumors with amplification of MYC had also amplification of EIF3S3 indicating common co-amplification of the genes. Gain of EIF3S3 was associated with poor cancer-specific survival in incidentally found prostate carcinomas (P = 0.023). In the analyses of prostatectomy-treated patients, the amplification was not statistically significantly associated with progression-free time. In conclusion, amplification of EIF3S3 gene is common in late-stage prostate cancer suggesting that it may be functionally involved in the progression of the disease.
During the past decades prostate cancer has become the most commonly diagnosed cancer of men in many Western countries. 1 Despite the substantial clinical importance of the disease, the molecular mechanisms underlying the development and progression of the disease are incompletely understood. 2 Chromosomal aberrations in prostate cancer have been studied with several techniques, such as classical cytogenetics, loss of heterozygosity analysis, fluorescence in situ hybridization (FISH), and especially, by comparative genomic hybridization (CGH). 2 These studies have implicated several chromosomal regions, such as 6q, 8p, 10q, 13q, 16q, and Xq, that may harbor genes involved in the tumorigenesis of prostate cancer. 3-9
Using CGH, we and others have previously shown that one of the most common genetic aberrations in advanced prostate cancer is the gain of the long arm (q-arm) of chromosome 8. 3-7 It is found in up to 80% of hormone-refractory tumors and distant metastases but only in ∼5% of untreated primary prostate carcinomas. 3,4 In the prostatectomy-treated patients, the gain of 8q seems to be associated with advanced stage and poor prognosis. 7,10 In addition to prostate cancer, gain of 8q is commonly found in several other malignancies, such as breast, bladder, and ovarian cancers. 11 For example, almost half of the breast carcinomas contain gain of 8q. And, the gain seems to be associated also with poor survival. 12
In most of the prostate tumors gain of 8q comprises the whole q-arm. However, CGH studies have indicated that there are, at least two independently amplified subchromosomal regions, 8q21 and 8q23-q24, suggesting the presence of several target genes. 4,5 The well-known oncogene, MYC, located at 8q24.1, is considered to be a putative target gene for the gain. 4 To identify other possible target genes, we recently used the subtraction hybridization technique to clone overexpressed genes in breast and prostate tumors. 13 We found that EIF3S3, located at 8q23, was amplified and overexpressed in approximately one-third of the hormone-refractory prostate carcinomas. 13,14 The EIF3S3 gene encodes for the p40 subunit of the eukaryotic translation initiation factor 3 (eIF3). eIF3 is the largest (∼600 kd) translation initiation factor protein complex, which has a central role in the initiation of translation. It binds to 40S ribosomal subunits in the absence of other initiation factors and preserves the dissociated state of 40S and 60S ribosomal subunits. It also stabilizes eIF2 · GTP · Met-tRNA binding to 40S subunits and mRNA binding to ribosomes. 15 However, very little is known about the p40 subunit itself and how it could be functionally involved in tumorigenesis. 13,16 There are, however, suggestions that aberrant regulation of translation could be important in the development of cancer. For example, overexpression of initiation factors EIF4E and EIF4G1 has been shown to transform normal cells. 17,18 In addition, amplification of EIF4G1, and overexpression of EIF4E have been found in squamous cell lung and breast carcinomas, respectively. 19,20 Also, allelic imbalance of INT6, which encodes for p48 subunit of eIF3, has been detected in breast cancer. 21 Recently, a new gene EIF5A2, encoding a putative translation initiation factor was cloned and shown to be amplified in the subset of ovarian cancer. 22,23
The aim of this study was to investigate the frequency of the EIF3S3 amplification in different stages of prostate cancer and to study the co-amplification of EIF3S3 and MYC. In addition, prognostic utility of the EIF3S3 amplification was evaluated. The analyses were done using FISH and new tissue microarray technology allowing large number of tumors to be rapidly analyzed.
Materials and Methods
Tissue Samples
The material consisted of three sets of prostate tumors. Group I included 21 benign prostatic hyperplasias, 42 prostatic intraepithelial neoplasias, 183 radical prostatectomy specimens, 20 Tru-Cut needle biopsy specimens of stage T3/T4 prostate tumors, 95 hormone refractory prostate tumors, and 39 distant metastases (obtained from University of Basel and Tampere University Hospitals). Fifty-four untreated local lymph node metastases were obtained from Lund University Hospital. Clinical stage and Gleason score of the tumors were available.
Group II included 112 incidentally found T1a/b tumors from transuretral resections for benign prostatic hyperplasia obtained from the University of Basel. The age of the patients at the time of diagnosis varied between 58 and 94 years with a mean of 76 years. FISH analysis was successful in 105 of 112 specimens. Of those 105, there were 20 Gleason 2-4, 60 Gleason 5-7, and 21 Gleason 8-10 tumors. Gleason score of four tumors was not available. The patients had been treated with standard therapies. Overall and prostate cancer-specific survival data were available.
Group III included 145 radical prostatectomy specimens from the University of Basel. The age of the patients at time of diagnosis was between 45 and 82 years with a mean of 65.4 years. The TNM stage distribution of the successfully hybridized cases was: 1 T1N0M0, 26 T2N0M0, 46 T3N0M0, 4 T2N1M0, 11 T3N1M0, and 6 T3N2M0. The TNM distribution was not available for 41 tumors. The Gleason score distribution was 100 Gleason 5-7, 33 Gleason 8-10 tumors, and 2 unknown. The progression-free time of the patients was available. The progression was defined either by increase in prostate-specific antigen levels (86% of cases), a positive finding in bone scan (11% of cases), or by biopsy proven local recurrence (3% of cases). The average recurrence-free time was 4.5 years (range, 0.6 to 15.1 years).
FISH
Multitissue blocks were made from the original formalin-fixed paraffin-embedded tumor blocks according to published guidelines. 24 Routine hematoxylin and eosin-stained slides were used to evaluate the representativeness of the samples. For the FISH analyses 5-μm sections from the multitissue blocks were either cut onto SuperFrost Plus slides (Menzel-Gläser, Braunschweig, Germany) and baked overnight, or an adhesive-coated tape sectioning system (Instrumedics, Hackensack, NJ) was used. A locus-specific PAC probe for EIF3S3 or MYC 13 and pericentromeric probe for chromosome 8 (pJM128) were labeled by nick translation with digoxigenin (locus-specific probes) and fluorescein-isothiocyanate (centromere-specific probe). The deparaffinized slides were treated with 1 mol/L NaSCN for 10 minutes at 80°C, followed by incubation in 4 mg/ml pepsin (P-7012, in 0.9% NaCl, pH 1.5; Sigma Chemical Co., St. Louis, MO) for 15 minutes at 37°C. The slides were then washed in H2O and 2× standard saline citrate, followed by dehydration in an ethanol series, and air-dried. The probes were applied on the slides in a hybridization mix (50% formamide, 10% dextran sulfate in 1× standard saline citrate, pH 7) and then co-denatured with the samples at 80°C for 8 minutes. After hybridization for 2 to 3 days in a humid chamber, the slides were washed and the locus-specific probes were detected immunohistochemically by anti-digoxigenin rhodamine. The slides were counterstained with 0.1 mol/L 4,6-diamidino-2-phenylindole in Vectashield anti-fade solution (Vector Laboratories Inc., Burlingame, CA).
The FISH signals were scored from nonoverlapping epithelial cells using an Olympus BX50 epifluorescence microscope (Tokyo, Japan). A Photometrics charge-coupled device camera (Photometrics, Tucson, AZ) and IPLab software program (Scananalytics Inc., Fairfax, VA) were used to capture images. The previously published criteria for amplification 13 were slightly modified because tissue sections, instead of isolated nuclei, were analyzed here. Briefly, the tumors were classified into three groups: nonamplified (no increase in EIF3S3 or c-myc copy number), low-level amplification (3 to 5 copies per cell), and high-level amplification (≥5 copies of the genes per cell). Tumors that showed >10% of malignant cells with increased copy number of either EIF3S3 or c-myc were considered to have copy number alterations.
Statistical Analysis
Statistical analysis of the data were done using BMDP Statistical Software Package. 25 Pearson chi-square test was used to evaluate the associations of the gene copy number and tumor type, clinical stage, and Gleason score. The survival differences of patients were evaluated by Kaplan-Meier method, and the statistical significance of survival differences between the patient groups was determined with Mantel-Cox and Breslow tests.
Results
Figure 1 ▶ demonstrates a FISH analysis of a hormone-refractory prostate tumor with high-level amplification of EIF3S3.
Figure 1.
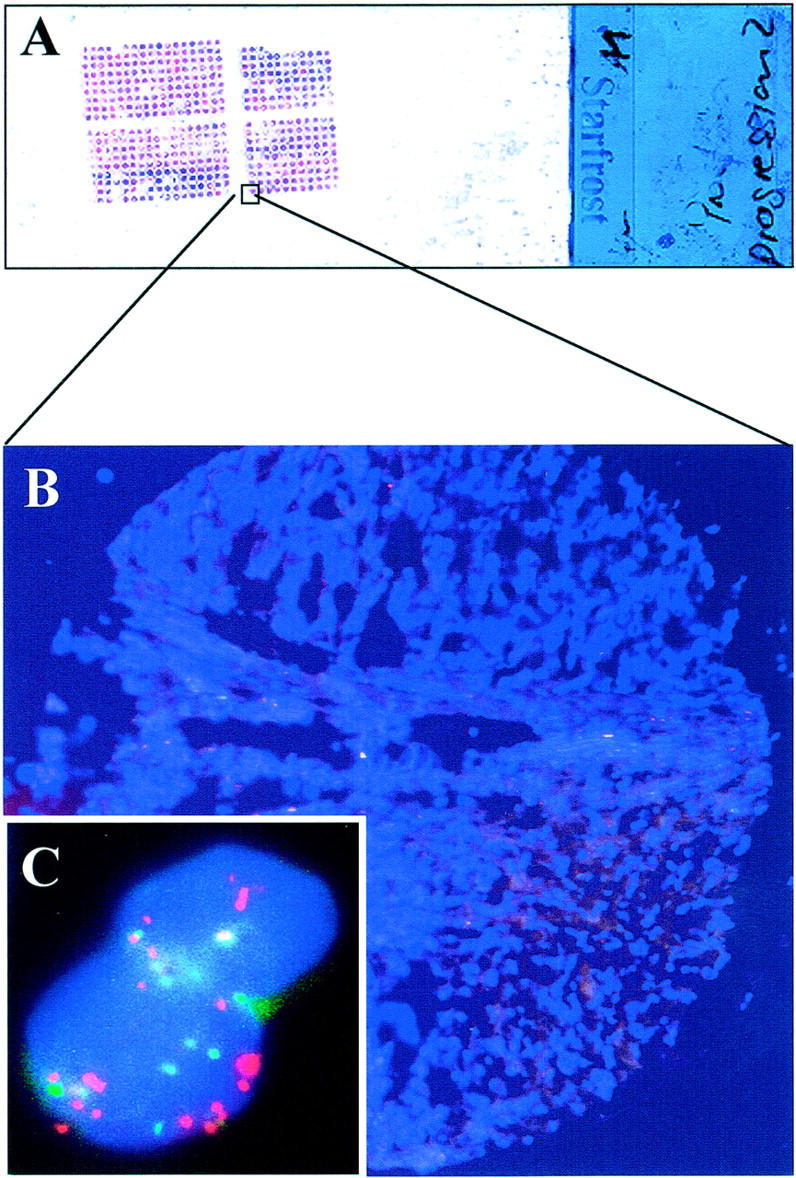
FISH analysis of the copy number of EIF3S3 gene in prostate cancer. The analysis was performed on multitissue array format in which one slide (A) contained up to ∼500 small (diameter, 0.6 mm) samples (B) counterstained with 4,6-diamidino-2-phenylindole. C: Red signals indicated >10 copies of the EIF3S3 gene and green color approximately six copies of chromosome 8 centromere in a hormone-refractory prostate tumor.
Group I: this set of samples was used to study the frequency of EIF3S3 amplification across different grades and stages of prostate tumors. FISH analysis of the EIF3S3 was successful in 369 of 454 (81.3%) of cases. Table 1 ▶ summarizes the frequency of the EIF3S3 amplification in prostate tumors. No copy number changes were found in nonmalignant (benign prostatic hyperplasia) or premalignant (prostatic intraepithelial neoplasia) prostate. The high-level amplification of EIF3S3 was found only in <10% of local (pT1 and pT2) prostate cancers, whereas hormone-naïve lymph node metastases as well as hormone-refractory tumors showed the amplification in ∼20 to 50% of the cases. Gain (or low-level amplification) of EIF3S3 was found in ∼30 to 50% of the prostate cancers. The amplification of the gene was statistically significantly associated with advanced stage of disease (P < 0.001) and high Gleason score (P < 0.001).
Table 1.
Association of EIF3S3 Amplification with Clinicopathological Variables
| Variable | EIF3S3 copy number (%) | P value | ||
|---|---|---|---|---|
| Normal | Low-level amplification | High-level amplification | ||
| Specimen type | ||||
| Androgen-dependent | ||||
| BPH | 19/19 (100) | 0/19 (0) | 0/19 (0) | <0.001* |
| High-grade PIN | 36/36 (100) | 0/36 (0) | 0/36 (0) | |
| Prostatectomy specimen (T1/T2) | 74/125 (59) | 40/125 (32) | 11/125 (9) | |
| Locally advanced (T3/4) | 18/44 (41) | 14/44 (32) | 12/44 (27) | |
| Lymph-node metastases | 10/37 (27) | 19/37 (51) | 8/37 (22) | |
| Hormone-refractory | ||||
| Local | 20/78 (26) | 32/78 (41) | 26/78 (33) | |
| Metastases | 4/30 (13) | 11/30 (37) | 15/30 (50) | |
| Gleason score | ||||
| ≤7 | 79/115 (69) | 31/115 (27) | 5/115 (4) | <0.001 |
| ≥8 | 22/78 (28) | 33/78 (42) | 23/78 (30) | |
*Comparison made between all groups under the title “specimen type.”
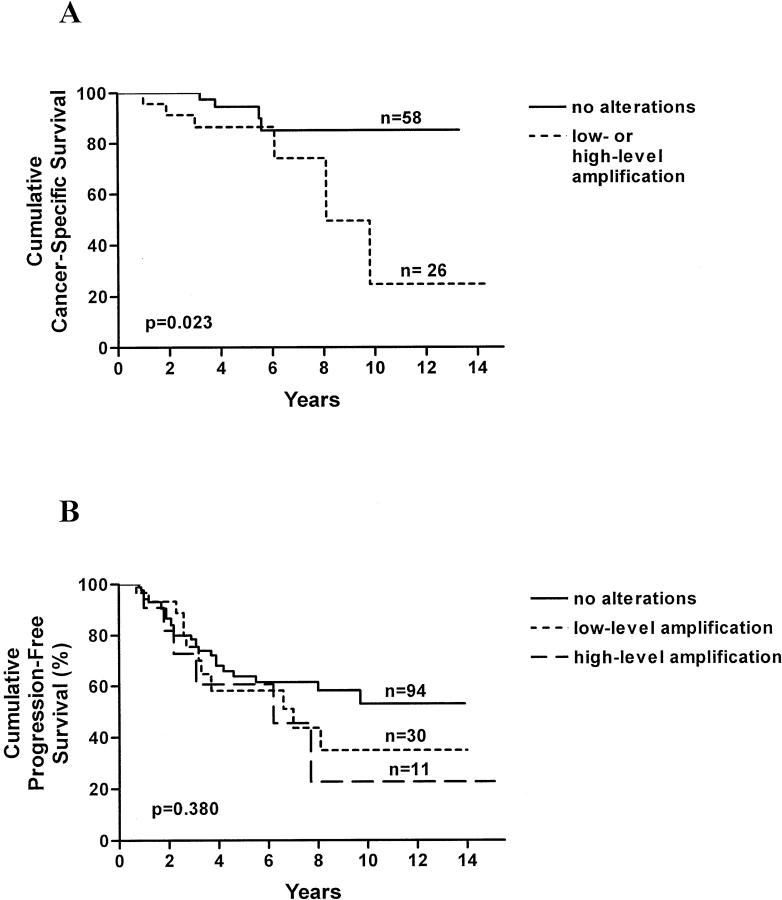
Group II: this set of samples was used to evaluate the prognostic significance of EIF3S3 amplification in prostate carcinomas that were incidentally found in transurethral resection specimens. FISH analysis was successful in 105 of 112 (93%) of cases. High-level amplification of EIF3S3 was found only in 6 of 105 cases (5.7%), whereas low-level amplification was found in 27 of the 105 (26%). Of the six cases with high-level amplification, five (83.3%) had died during the follow-up period. In contrast, 18 of the 27 (55.7%) cases with low-level amplification and 37 of the 72 (51.4%) cases without copy number alterations had died. However, there was no association between overall survival and EIF3S3 copy number alterations. The prostate cancer-specific follow-up data were available from 85 of the 105 cases. One tumor from the 10 (10%) patients who had died of prostate cancer showed the gene amplification, whereas only 2 tumors out of the 75 (3%) patients who had not died of prostate cancer showed the amplification (Table 2) ▶ . The difference was not quite statistically significant (P = 0.084). In the disease-specific survival analysis (Figure 2A) ▶ , the increased copy number (either low- or high-level amplification) of EIF3S3 was associated with poor survival (P = 0.023).
Table 2.
Association of EIF3S3 Amplification with Prostate Cancer-Specific Death in Incidentally Found Prostate Cancer
| No. of cases (%) | ||||
|---|---|---|---|---|
| EIF3S3 copy number | Total | |||
| Normal | Low-level amplification | High-level amplification | ||
| Alive or died of some other reason than prostate cancer | 55 (73) | 18 (24) | 2 (3) | 75 (100) |
| Dead | 4 (40) | 5 (50) | 1 (10) | 10 (100) |
P = 0.084.
Figure 2.
A: Prostate cancer-specific survival of incidentally found (n = 84) prostate cancer patients according to the EIF3S3 copy number. The patients had been treated with various forms of therapies according to routine clinical practice. B: Progression-free survival of the prostatectomy treated men (n = 135) according to the EIF3S3 copy number.
Group III: this set of samples was used to study prognostic significance of the EIF3S3 amplification in patients treated by radical prostatectomy. The FISH analysis was successful in 135 of 145 (93.1%) cases. Eleven of 135 (8.1%) and 30 of 135 (22%) of the samples showed high-level and low-level amplification of EIF3S3, respectively. In a single case only one copy of chromosome 8 centromere and EIF3S3 were found. According to Kaplan-Meir analysis, the progression-free survival was slightly poorer for patients with EIF3S3 amplification than in patients without the amplification (Figure 2B) ▶ . However, the difference was not statistically significant (P = 0.380).
By combining the data from all above-mentioned samples (n = 461), the co-amplification of EIF3S3 and c-myc in almost all cases became evident (Table 3) ▶ . There was only one case with high-level EIF3S3 amplification with two copies of MYC.
Table 3.
Co-Amplification of EIF3S3 and MYC
| MYC copy number | No. of tumors | ||
|---|---|---|---|
| EIF3S3 copy number | |||
| Normal | Low-level amplification | High-level amplification | |
| Normal | 218 | 2 | 1 |
| Low-level amplification | 2 | 157 | 10 |
| High-level amplification | 0 | 3 | 68 |
Discussion
Gain of 8q is one of the most common chromosomal alterations in late-stage prostate cancer detected by CGH. 3-5 We have earlier identified a putative target gene for the 8q gain, EIF3S3. 13,14 Here, we analyzed the frequency of the amplification of EIF3S3 in prostate carcinomas of different stages. None of the benign prostatic hyperplasia or prostatic intraepithelial neoplasia lesions showed amplification of the gene. In addition, the high-level amplification was found to be rare in untreated stage pT1/pT2 tumors. However, in locally advanced and metastatic tumors, the amplification was found approximately in one-fourth of the cases. In hormone-refractory tumors the amplification was detected in 30 to 50% of the cases. The findings are in good agreement with our earlier FISH analyses showing high-level amplification of the gene in 13 of 44 (30%) of locally recurrent hormone-refractory prostate carcinomas. 13 The frequency of low-level amplification in the locally recurrent hormone-refractory tumors was somewhat lower (41% versus 70%) than we have previously reported. 13 This may be because of the fact that more tumors were analyzed here. In addition, the FISH analysis was performed on tissue sections instead of isolated nuclei as in our previous study. 13 Lost signals because of nuclear slicing may possibly lead to underdetection of low-level amplifications. The amplification was found almost eight times more often in poorly differentiated tumors (Gleason score ≥ 8) than moderately or well-differentiated tumors (Gleason score ≤ 7). Altogether, the results suggest that the amplification of EIF3S3 is involved in the late progression of prostate cancer.
The FISH analyses were done using tissue sections making the evaluation of actual copy number of the gene difficult. However, in the case of amplification, typically 5 to 10 signals per nucleus were seen indicating that the level of amplification was quite moderate. The finding is consistent with the earlier CGH studies, which most often have shown gain of the whole q-arm of the chromosome, and only rarely high-level regional amplification. 3-5,9 This feature makes the gain of 8q clearly different from, for example gain of Xq, the second most common gain in hormone-refractory prostate cancers. According to FISH analysis, the amplification of the target gene of the Xq gain, androgen receptor gene, typically consists of 10 or more copies of the gene. 26 However, we have previously shown that even the moderately increased copy number of EIF3S3 may lead to overexpression of the gene in prostate cancer. 13 Therefore, it is possible that amplification of lower level but larger chromosomal region harboring many genes in 8q is selected for during the progression of prostate cancer.
Another putative target gene for the gain of 8q in prostate cancer is MYC located at 8q24.1. 4 We have earlier shown that in a subset of breast carcinomas EIF3S3 and MYC are not co-amplified. 14 Here, in this large series of prostate tumors, we found only one case in which EIF3S3 was amplified without MYC amplification, whereas all cases with MYC amplification had also EIF3S3 amplification. The finding suggests that both EIF3S3 and MYC may be important in the progression of prostate cancer, and therefore they are equally selected for. In addition to MYC and EIF3S3 there are other putative target genes for 8q gain in prostate cancer as well. These include recently cloned prostate stem-cell antigen, and GC79 encoding a zinc-finger protein, both located in the 8q23-24 region. 27,28 The other minimal commonly amplified region in prostate cancer is 8q21 of which target genes are still not known. 4,5,9 Because there are likely to be numerous putative target genes, it will be important to compare the alterations of all of the different putative target genes in large tumor materials as done here for MYC and EIF3S3.
The prognostic significance of the EIF3S3 amplification was retrospectively studied here in two sets of tumors. In the cases of incidental prostate cancers, the amplification was found in only ∼5% of cases. The number of tumors with the amplification was too small for prognostic analyses. However, by combining the groups of low- and high-level amplification, we found that the increased copy number of EIF3S3 was associated with poor disease-specific survival. Thus, it may be that the copy number alteration of EIF3S3 could be useful in predicting which incidental cancers are clinically significant. Evidently larger studies are needed to confirm the finding.
In the second set of cases, 135 prostatectomy specimens, from patients whose progression-free survival data were available, were analyzed. Approximately 55% of the patients with the amplification but only ∼30% of those without amplification experienced progression. Although the progression-free time curves showed a worse prognosis for the patients with amplification of EIF3S3 than for patients without the amplification, the difference was not statistically significant. This is in some contrast to the findings of Sato and co-workers, 29 who have previously suggested that MYC amplification is associated with poor survival in prostatectomy treated stage C disease. Because MYC and EIF3S3 were almost always co-amplified, it was evident that MYC did not have prognostic value in our material either (data not shown). The major difference between the studies by Sato and colleagues 29 and by us is that Sato and co-workers analyzed high-grade, stage pT3 tumors, whereas our material consisted of both moderately and poorly differentiated pT1-pT3 tumors. Thus, for example, the frequency of 8q gain was higher in the study by Sato and colleagues 29 than in our study (54.2% versus 30.4%). The definition of high-level amplification was also different in the two studies. Sato and co-workers 29 found an “additional increase” of MYC in 19.4% of cases, whereas high-level amplification of EIF3S3 (and MYC) was found in only 8% of our prostatectomy series. It may also be that because the cases in our study were lower stage and grade, the prostatectomy had removed these tumors before the effects of EIF3S3 (and/or MYC) on progression had have enough time to affect.
In this study, the FISH analyses were performed in a multitissue section format. The use of multitissue blocks instead of original single tumor blocks has several advantages. It allowed us to screen a large number of tumors in a relatively short period of time. Altogether 609 specimens were analyzed. Because the hybridizations were done on a few slides (five slides per gene), the slide-to-slide variation could also be expected to be low. An additional advantage of multitissue slides is that, at least in our hands, the FISH analyses seemed to work better in this than in the traditional one tumor section per slide format. This may well be because of the fact that in the multitissue blocks the tissue samples are small and equal in size. Therefore, the pretreatment of the slide, which is the most important variable in the FISH analysis, probably affects equally to each specimen on the slide. The disadvantage of the multitissue technology is that only a small proportion of the tumor is analyzed. Because of the known intratumoral heterogeneity of prostate cancer, the absolute frequency of aberrations may thus be somewhat underestimated. However, this possible underestimation of absolute frequencies should not affect the clinicopathological associations based on a large number of specimens, because all specimens on a tissue microarray are subjected to the same sampling limitations.
In conclusion, we have shown here that the high-level amplification of EIF3S3 gene is associated with advanced stage, androgen independence, and poor differentiation of prostate cancer. The gene is in most of the cases co-amplified with MYC. Both association with advanced stage and preliminary prognostic analyses suggest that amplification of EIF3S3 might be important for the progression of prostate cancer. Further studies are warranted to evaluate the function of the gene as well as possible prognostic utility of the amplification.
Acknowledgments
We thank Mrs. Mariitta Vakkuri and Ms. Heli Lehtonen for technical assistance.
Footnotes
Address reprint requests to Dr. Tapio Visakorpi, M.D., Ph.D., Laboratory of Cancer Genetics, Institute of Medical Technology, FIN-33014 University of Tampere, Finland. E-mail: tapio.visakorpi@uta.fi.
Supported by grants from the Academy of Finland, the Cancer Society of Finland, the Reino Lahtikari Foundation, the Medical Research Fund of Tampere University Hospital, the Sigrid Juselius Foundation, and the CaP CURE.
References
- 1.Hsing AW, Tsao L, Devesa SS: International trends and patterns of prostate cancer incidence and mortality. Int J Cancer 2000, 85:60-67 [DOI] [PubMed] [Google Scholar]
- 2.Elo JP, Visakorpi T: Molecular genetics of prostate cancer. Ann Med 2001, 33:130-141 [DOI] [PubMed] [Google Scholar]
- 3.Visakorpi T, Kallioniemi AH, Syvanen AC, Hyytinen ER, Karhu R, Tammela T, Isola JJ, Kallioniemi OP: Genetic changes in primary and recurrent prostate cancer by comparative genomic hybridization. Cancer Res 1995, 55:342-347 [PubMed] [Google Scholar]
- 4.Nupponen NN, Kakkola L, Koivisto P, Visakorpi T: Genetic alterations in hormone-refractory recurrent prostate carcinomas. Am J Pathol 1998, 153:141-148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cher ML, Bova GS, Moore DH, Small EJ, Carroll PR, Pin SS, Epstein JI, Isaacs WB, Jensen RH: Genetic alterations in untreated metastases and androgen-independent prostate cancer detected by comparative genomic hybridization and allelotyping. Cancer Res 1996, 56:3091-3102 [PubMed] [Google Scholar]
- 6.Cher ML, MacGrogan D, Bookstein R, Brown JA, Jenkins RB, Jensen RH: Comparative genomic hybridization, allelic imbalance, and fluorescence in situ hybridization on chromosome 8 in prostate cancer. Genes Chromosom Cancer 1994, 11:153-162 [DOI] [PubMed] [Google Scholar]
- 7.Alers JC, Rochat J, Krijtenburg PJ, Hop WC, Kranse R, Rosenberg C, Tanke HJ, Schroder FH, van Dekken H: Identification of genetic markers for prostatic cancer progression. Lab Invest 2000, 80:931-942 [DOI] [PubMed] [Google Scholar]
- 8.Cunningham JM, Shan A, Wick MJ, McDonnell SK, Schaid DJ, Tester DJ, Qian J, Takahashi S, Jenkins RB, Bostwick DG, Thibodeau SN: Allelic imbalance and microsatellite instability in prostatic adenocarcinoma. Cancer Res 1996, 56:4475-4482 [PubMed] [Google Scholar]
- 9.El Gedaily A, Bubendorf L, Willi N, Fu W, Richter J, Moch H, Mihatsch MJ, Sauter G, Gasser TC: Discovery of new amplification loci in prostate cancer by comparative genomic hybridization. Prostate 2001, 46:184-190 [DOI] [PubMed] [Google Scholar]
- 10.Van Den Berg C, Guan X-Y, Von Hoff D, Jenkins R, Bittner M, Griffin C, Kallioniemi O, Visakorpi T, McGill J, Herath J, Epstein J, Sarosdy M, Meltzer P, Trent J: DNA sequence amplification in human prostate cancer identified by chromosome microdissection: potential prognostic implications. Clin Cancer Res 1995, 1:11-18 [PubMed] [Google Scholar]
- 11.Knuutila S, Bjorkqvist AM, Autio K, Tarkkanen M, Wolf M, Monni O, Szymanska J, Larramendy ML, Tapper J, Pere H, El-Rifai W, Hemmer S, Wasenius VM, Vidgren V, Zhu Y: DNA copy number amplifications in human neoplasms: review of comparative genomic hybridization studies. Am J Pathol 1998, 152:1107-1123 [PMC free article] [PubMed] [Google Scholar]
- 12.Isola JJ, Kallioniemi OP, Chu LW, Fuqua SA, Hilsenbeck SG, Osborne CK, Waldman FM: Genetic aberrations detected by comparative genomic hybridization predict outcome in node-negative breast cancer. Am J Pathol 1995, 147:905-911 [PMC free article] [PubMed] [Google Scholar]
- 13.Nupponen NN, Porkka K, Kakkola L, Tanner M, Persson K, Borg A, Isola J, Visakorpi T: Amplification and overexpression of p40 subunit of eukaryotic translation initiation factor 3 in breast and prostate cancer. Am J Pathol 1999, 154:1777-1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nupponen NN, Isola J, Visakorpi T: Mapping the amplification of EIF3S3 in breast and prostate cancer. Genes Chromosom Cancer 2000, 28:203-210 [PubMed] [Google Scholar]
- 15.Hershey JW, Asano K, Naranda T, Vornlocher HP, Hanachi P, Merrick WC: Conservation and diversity in the structure of translation initiation factor eIF3 from humans and yeast. Biochimie 1996, 78:903-907 [DOI] [PubMed] [Google Scholar]
- 16.Asano K, Vornlocher HP, Richter-Cook NJ, Merrick WC, Hinnebusch AG, Hershey JW: Structure of cDNAs encoding human eukaryotic initiation factor 3 subunits. Possible roles in RNA binding and macromolecular assembly. J Biol Chem 1997, 272:27042-27052 [DOI] [PubMed] [Google Scholar]
- 17.Fukuchi-Shimogori T, Ishii I, Kashiwagi K, Mashiba H, Ekimoto H, Igarashi K: Malignant transformation by overproduction of translation initiation factor eIF4G. Cancer Res 1997, 57:5041-5044 [PubMed] [Google Scholar]
- 18.Lazaris-Karatzas A, Sonenberg N: The mRNA 5′ cap-binding protein, eIF-4E, cooperates with v-myc or E1A in the transformation of primary rodent fibroblasts. Mol Cell Biol 1992, 12:1234-1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brass N, Heckel D, Sahin U, Pfreundschuh M, Sybrecht GW, Meese E: Translation initiation factor eIF-4gamma is encoded by an amplified gene and induces an immune response in squamous cell lung carcinoma. Hum Mol Genet 1997, 6:33-39 [DOI] [PubMed] [Google Scholar]
- 20.Scott PA, Smith K, Poulsom R, De Benedetti A, Bicknell R, Harris AL: Differential expression of vascular endothelial growth factor mRNA vs protein isoform expression in human breast cancer and relationship to eIF-4E. Br J Cancer 1998, 77:2120-2128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyazaki S, Imatani A, Ballard L, Marchetti A, Buttitta F, Albertsen H, Nevanlinna HA, Gallahan D, Callahan R: The chromosome location of the human homolog of the mouse mammary tumor-associated gene INT6 and its status in human breast carcinomas. Genomics 1997, 46:155-158 [DOI] [PubMed] [Google Scholar]
- 22.Jenkins ZA, Haag PG, Johansson HE: Human eIF5A2 on chromosome 3q25–q27 is a phylogenetically conserved vertebrate variant of eukaryotic translation initiation factor 5A with tissue-specific expression. Genomics 2001, 71:101-109 [DOI] [PubMed] [Google Scholar]
- 23.Guan X-Y, Sham JST, Tang TC-M, Fang Y, Huoi K-K, Yang J-M: Isolation of a novel candidate oncogene within a frequently amplified region at 3q26 in ovarian cancer. Cancer Res 2001, 61:3806-3809 [PubMed] [Google Scholar]
- 24.Kononen J, Bubendorf L, Kallioniemi A, Bärlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G, Kallioniemi O-P: Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med 1998, 4:844-847 [DOI] [PubMed] [Google Scholar]
- 25.Dixon WJ: BMDP Statistical Software. 1981. University of California Press, Los Angeles
- 26.Visakorpi T, Hyytinen E, Koivisto P, Tanner M, Keinänen R, Palmberg C, Palotie A, Tammela T, Isola J, Kallioniemi O-P: In vivo amplification of the androgen receptor gene and progression of human prostate cancer. Nat Genet 1995, 9:401-406 [DOI] [PubMed] [Google Scholar]
- 27.Reiter RE, Gu Z, Watabe T, Thomas G, Szigeti K, Davis E, Wahl M, Nisitani S, Yamashiro J, Le Beau MM, Loda M, Witte ON: Prostate stem cell antigen: a cell surface marker overexpressed in prostate cancer. Proc Natl Acad Sci USA 1998, 95:1735-1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang GT, Steenbeek M, Schippers E, Blok LJ, van Weerden WM, van Alewijk DC, Eussen BH, van Steenbrugge GJ, Brinkmann AO: Characterization of a zinc-finger protein and its association with apoptosis in prostate cancer cells. J Natl Cancer Inst 2000, 92:1414-1421 [DOI] [PubMed] [Google Scholar]
- 29.Sato K, Qian J, Slezak JM, Lieber MM, Bostwick DG, Bergstralh EJ, Jenkins RB: Clinical significance of alterations of chromosome 8 in high-grade, advanced, nonmetastatic prostate carcinoma. J Natl Cancer Inst 1999, 91:1574-1580 [DOI] [PubMed] [Google Scholar]