Abstract
In nerve cells, a select group of RNAs has been localized to dendritic domains. Here we have examined dendritic RNA transport in sympathetic neurons in primary culture, using a microinjection protocol with neuronal BC1 RNA and with BC1-derived sequence segments. After cytoplasmic microinjection, full-length BC1 RNA was selectively transported to dendrites; in contrast, control RNAs such as nuclear RNAs and random-sequence irrelevant RNAs remained restricted to cytoplasmic areas proximal to the injection sites. Chimeric RNAs were constructed that contained the full-length BC1 sequence inserted upstream or downstream of the coding regions of nondendritic mRNAs. After microinjection, such chimeric RNAs were specifically targeted to dendrites; microinjected corresponding nonchimeric mRNAs were not. Dendritic transport of BC1 RNA was rapid: the average dendritic delivery rate within the first hour after microinjection was 242 ± 25 μm/hr. Whereas a 5′-BC1 segment of 62 nucleotides was transported to dendrites to extents and at levels similar to full-length BC1 RNA, a 3′-BC1 segment of 60 nucleotides did not exit injected somata to any significant degree. A cis-acting dendritic targeting element is thus contained in the 5′ part of neuronal BC1 RNA. These results demonstrate that mechanisms exist in neurons for fast and specific transport of selected RNAs to dendrites.
Keywords: neuronal BC1 RNA, RNA transport, dendrites, targeting element, sympathetic neurons, primary cultures of nerve cells, microinjection
The individual protein repertoire of each dendritic microdomain is likely to be the result not only of directed protein transport, but also of local synthesis of selected proteins. This notion was prompted by earlier ultrastructural studies that revealed preferential localization of polyribosomes underneath synaptic sites in dendrites (Steward and Levy, 1982; Steward and Reeves, 1988). The hypothesis of dendritic translation has been strengthened in recent years by the discovery in dendrites of various types of mRNAs and nonmessenger RNAs. Dendritic mRNAs include, among others, those encoding: (1) the high molecular mass forms of microtubule-associated protein 2 (MAP2a/b; Garner et al., 1988; Bruckenstein et al., 1990;Kleiman et al., 1990), (2) the α-subunit of Ca2+/calmodulin-dependent protein kinase type II (CaMKIIα; Burgin et al., 1990); (3) the inositol 1,4,5-trisphosphate receptor type 1 (InsP3R1; Furuichi et al., 1993); (4) neurogranin (Ng/RC3; Landry et al., 1994); (5) a number of amino acid receptor subunits (Miyashiro et al., 1994; Racca et al., 1997); and (6) the cytoskeleton-associated protein Arc (Link et al., 1995; Lyford et al., 1995). Dendritic nonmessenger RNAs include BC1 RNA (Tiedge et al., 1991), ribosomal RNAs (rRNAs) (Kleiman et al., 1993; 1994), and tRNAs (Tiedge and Brosius, 1996). BC1 RNA, a short RNA polymerase III transcript, is associated with proteins to form a ribonucleoprotein particle (RNP; Kobayashi et al., 1991; Cheng et al., 1996) that may be involved functionally in the transport and/or translation of dendritic mRNAs (for review, see Brosius and Tiedge, 1995).
BC1 RNA and dendritic mRNAs were found colocalized in preparations of dendritic spines and of synaptosomes (Chicurel et al., 1993; Rao and Steward, 1993). Active protein synthesis has been demonstrated in dendrites in vitro (Torre and Steward, 1992; Crino and Eberwine, 1996) and in vivo (Mayford et al., 1996). Neurotrophin-induced synaptic plasticity in the hippocampus has been reported to depend on local protein synthesis in CA1 pyramidal cell dendrites (Kang and Schuman, 1996). Local translation may thus play an important role in the development of synaptic connections and in their long-term structural and functional modulation (for review, see Steward and Banker, 1992; Steward, 1994, 1995, 1997).
A prerequisite for the local synthesis of distinct proteins in dendritic microdomains is the targeted delivery of RNAs, including cognate mRNAs and other RNAs, to such domains. However, the mechanisms that allow a select population of RNA molecules to be transported to dendrites are poorly understood. The objective of this work was therefore to examine the directed transport of a specific RNA to dendrites. For this purpose, we adopted a microinjection protocol with sympathetic neurons in primary culture. Transport competence of dendritic and nondendritic RNAs, generated by in vitrotranscription, was analyzed in eels that had been injected in the perinuclear cytoplasmic region. As a dendritic RNA, we chose neuronal BC1 RNA. We report here that BC1 RNA is selectively transported to dendrites, and that it is competent to direct dendritic targeting of normally nondendritic mRNAs. Contained within the 5′ region of BC1 RNA is a cis-acting element that is responsible for dendritic targeting.
MATERIALS AND METHODS
Cell culture. Primary cultures of sympathetic neurons were generated as described (Higgins et al., 1991). Briefly, superior cervical ganglia from embryonic day 19–21 Sprague Dawley rat embryos were dissociated by mechanical and enzymatic treatment. Neurons were then maintained on glass coverslips (Carolina Biological Supply, Burlington, NC) that had been coated with filter-sterilized poly-d-lysine (Sigma, St. Louis, MO) at a concentration of 100 μg/ml. The basic nutrient solution consisted of a 50% (v/v) mixture of Ham’s F12 medium (Life Technologies, Gaithersburg, MD) and DMEM (Life Technologies). Basic media were supplemented with bovine serum albumin (BSA, 500 μg/ml; Calbiochem, La Jolla, CA), rat transferrin (20 μg/ml; Jackson ImmunoResearch, West Grove, PA),l-glutamine (20 μg/ml; Life Technologies), sodium selenite (5 ng/ml; Sigma), bovine insulin (10 μg/ml; Sigma), and nerve growth factor (β-NGF, 100 ng/ml; Harlan Bioproducts, Indianapolis, IN). Dendritic growth was induced by supplementing the medium with basement membrane extract (final concentration, 100 μg/ml; Matrigel, Collaborative Biomedical Products, Bedford, MA) on the third day in vitro. Non-neuronal proliferation was discouraged by adding cytosine arabinofuranoside (2 μm; Sigma) on the second and fifth days after plating.
RNA Preparation. For microinjection experiments, RNA was transcribed in vitro in the presence of35S-uridine triphosphate (UTP). The following transcripts were generated: (1) full-length BC1 RNA [152 nucleotides (nt)] from plasmid pBCX607 (Cheng et al., 1996), (2) a 5′ segment of BC1 RNA (nt 1–62) from plasmid pBCX607, (3) a 3′ segment of BC1 RNA (nt 93–152) from plasmid pMK1 (Tiedge et al., 1991), (4) nuclear U4 RNA (145 nt) from plasmid pSP6-U4 (Hausner et al., 1990), (5) nuclear U6 RNA (107 nt) from plasmid pSP6-U6 (Hausner et al., 1990), and (6) 64- and 144-nt random irrelevant RNAs from the polylinker region of plasmid pSL300 (Brosius, 1989). Radiolabeled U4 and U6 RNAs and polylinker irrelevant RNAs were used as controls.
Two constructs were used in this work for in vitrotranscription of chimeric RNAs. (1) A plasmid (pGFP-BC1) was generated in which the full-length BC1 sequence was cloned 3′ to the coding region of the green fluorescent protein (GFP) mRNA sequence. The original GFP plasmid (a BamHI fragment cloned into theBamHI site of pRSETB, 714 bp coding region) was obtained from Drs. R. Heim and R. Tsien (University of California, San Diego, CA; see also Heim et al., 1995). The BC1 sequence was cloned into the KpnI site just downstream from the duplicate stop codon. The plasmid was linearized withHindIII before in vitro transcription. (2) In a second chimeric construct (pCMV-BC1-bcd), the full-length BC1 sequence was cloned 5′ to the coding region (1467 bp) of thebicoid (bcd; Berleth et al., 1988) mRNA sequence (courtesy of Drs. C. Garner, University of Alabama, Birmingham, AL, and R. Müller, University of Bremen, Bremen, Germany; the originalbcd clone pTN3bcd was obtained from Dr. W. Driever, Massachusetts General Hospital, Charlestown, MA). The BC1 sequence was cloned into a HindIII site 55 nt upstream of the bcd start codon; 3′ to the bcd stop codon, plasmid pCMV-BC1-bcd contains 88 nt of the bcd 3′ untranslated region. This plasmid was linearized with Asp718 I beforein vitro transcription. The corresponding nonchimeric clones pGFP and pCMV-bcd were identical to the chimeric clones pGFP-BC1 and pCMV-BC1-bcd, respectively, except for the fact that they did not contain the BC1 sequence.
RNAs were prepared from linearized plasmids, using SP6, T3, or T7 RNA polymerase, according to protocols of the manufacturer (Promega, Madison, WI). After transcription in the presence of35S-UTP, excess unlabeled UTP was added to the reaction mixture to ensure that labeled transcripts were full length. Allin vitro transcripts were checked for size and integrity by PAGE. In Figure 1, we analyzed preinjection and postinjection aliquots of the 3′-BC1 segment, compared with preinjection full-length BC1 RNA. The results show that each sample produced a single band at the expected relative position, indicating that the injected transcripts were full length and that no degradation had occurred during handling of the samples.
Fig. 1.
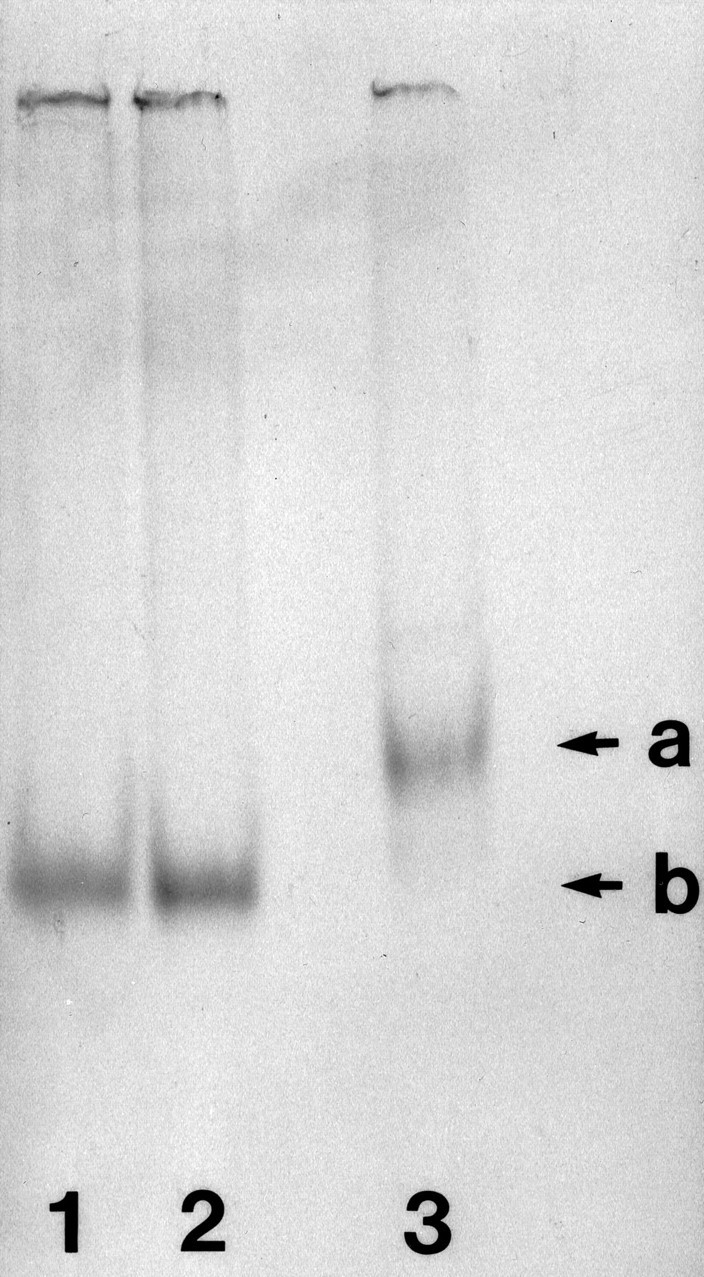
Integrity of 35S-labeled in vitro transcripts, as ascertained by PAGE. Lane 1, 3′-BC1 segment, preinjection aliquot; lane 2, 3′-BC1 segment, postinjection aliquot; lane 3, full-length BC1 RNA, preinjection aliquot; 8% acrylamide, Tris-borate/EDTA buffer. Arrow a indicates the position of full-length BC1 RNA (152 nt); arrow b indicates the position of the 3′-BC1 segment (60 nt).
Microinjection. Fine-tipped (<0.5 μm) microinjection needles were used to pressure inject neurons with RNA. Injected RNAs were 35S-radiolabeled at 3 × 106cpm/μl. RNA was microinjected at volumes of several femtoliters per pulse; total injected volume per cell was <5% of cell body volume for the standard injection routine. Lucifer yellow (0.4%) was coinjected for calibration purposes. In low-amount injections, the estimated injected amounts of RNA were five times lower than in standard injections. Neurons were incubated for 4 hr at 35°C (unless stated otherwise) before fixation and emulsion autoradiography. Although cultured sympathetic neurons were viable for extended periods (up to several weeks) after microinjections, we found an incubation period of 4 hr sufficient to ensure that BC1 RNA or derivatives would reach distal dendritic segments. Fixation and emulsion autoradiography were performed as described (Tiedge, 1991). Coverslips were mounted, cell side up, on microscope slides with DPX (Fluka, Ronkonkoma, NY), dipped in Kodak (Rochester, NY) NTB-2 emulsion (diluted 1:1 with HPLC-grade H2O), and exposed for 3–4 weeks. After developing (Kodak D-19 developer, 50% strength; Kodak Rapid-Fix), cells were coverslipped with Kaiser’s glycerol jelly (Banker and Goslin, 1991). Cells were analyzed and photographed (Ektachrome-160T film, Kodak) on a Nikon Microphot-FXA microscope, using dark-field, phase-contrast, and Nomarski (DIC) optics.
Data evaluation. To calculate the extent of dendritic labeling, photomicrographs of cells to be analyzed were printed (at 625× magnification), and the distance of labeling was measured for each dendrite. For the determination of the distal-most point of dendritic labeling, a signal was considered significant if it exceeded background levels by a factor of 3 or more. The average rate of BC1 delivery to dendrites during the first hour was calculated as the mean of the measured distances that the labeling signal had reached after this time. The SD was calculated assuming normal distribution. To calculate the hypothetical “initial velocity” of BC1 RNA (a limit as time approaches zero, when any retrograde translocation and the finite length of dendrites become irrelevant), we fitted several mathematical functions to the experimental data, using nonlinear regression analysis. The software used was SPSS for Windows, version 6.1.2. The best fits (R2 = 0.916 in both cases) were obtained with an exponential function of the formd = A[1 − exp(−t/B)] and with a hyperbolic function of the formd = At/(B + t). For both equations, d is the labeling distance, t the time after injection, A a distance constant (dmax), and B a time constant. On the basis of the exponential function, we calculated an initial velocity of 388 μm/hr (lower and upper limits for the 95% confidence interval were 373 and 403 μm/hr, respectively). On the basis of the hyperbolic function, we obtained an initial velocity of 469 μm/hr (95% confidence interval, 441–497 μm/hr). For the exponential function, the assumed average length of dendrites that resulted in the best fit was 354 μm; for the hyperbolic function, this value was 447 μm. Because we rarely observed dendrites that exceeded 400 μm in length in the cultures, the hyperbolic function was rejected.
Primary and secondary structure analyses were performed according to the methods of Devereux et al. (1984) and Zuker (1989), respectively, using GenBank/EMBL sequence data with the Genetics Computer Group (Madison, WI) sequence analysis software package. Because the BC1 RNA gene has been suggested to be the master gene for ID repeats (for review, see Deininger et al., 1996), we also performed GenBank/EMBL database searches for ID-containing neuronal mRNAs. Although several such mRNAs were identified, the subcellular location of none of these has yet been established.
RESULTS
Dendritic transport of BC1 RNA in sympathetic neurons in culture
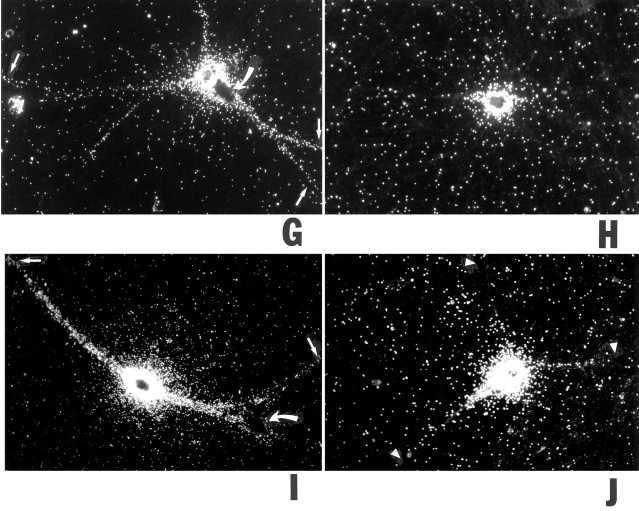
Radiolabeled RNA, generated by in vitro transcription, was microinjected into the perinuclear somatic region of sympathetic neurons in primary culture. Full-length BC1 RNA was delivered specifically to dendrites (Fig.2A,B). Analysis of >600 dendrites confirmed that distal regions of virtually all dendritic branches were reached within a 4 hr postinjection incubation period. Dendritic labeling appeared clustered at times, although this was not generally the case in all dendritic segments analyzed. As a control, we injected U4 RNA, a short nuclear RNA (Bringmann et al., 1984; Hashimoto and Steitz, 1984; Lührmann et al., 1990) that is similar in size to BC1 RNA. No significant dendritic transport was observed: after 4 hr, labeling was restricted to somata and proximal-most dendritic segments (Fig. 2C,D). Analogous results were obtained with a random irrelevant RNA of equivalent size, generated from the polylinker sequence of plasmid pSL300; again, the labeling signal was mainly restricted to cell bodies and proximal dendritic regions (Fig.2E,F). In contrast, somatic restriction of the labeling signal was never observed with cells that had been injected with BC1 RNA. This was further confirmed by experiments in which the estimated amount of injected RNA was lowered by a factor of 5 to probe for possible effects of injection amounts on the extent of dendritic labeling. As shown in Figure 2G, cells that had been injected with such low levels of BC1 RNA nevertheless showed significant labeling throughout the entire dendritic extent, although at lower overall levels. In contrast, U4 RNA injected at low levels remained restricted to the cell body (Fig.2H). These results indicate that the subcellular distribution of injected RNAs was not a function of the amounts of RNA injected. In summary, the data show that BC1 RNA is transported selectively and specifically to dendrites of sympathetic neurons in culture.
Fig. 2.
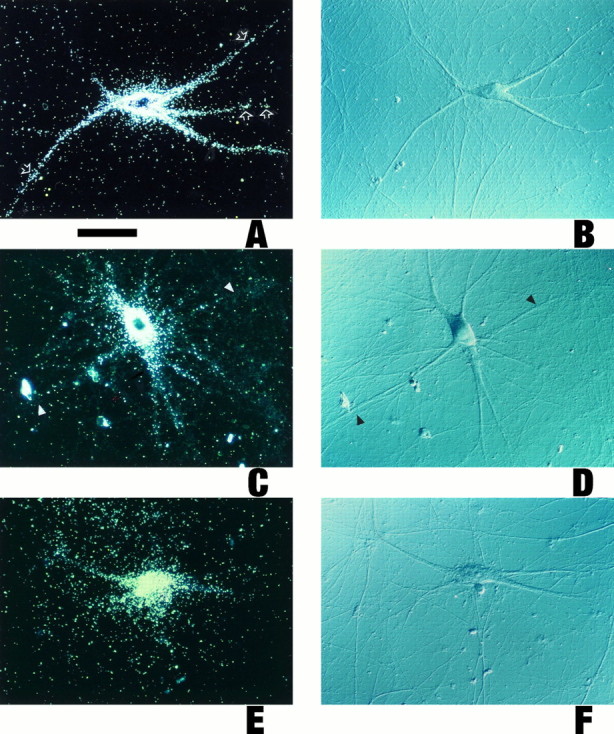
Dendritic transport of microinjected BC1 RNA. Sympathetic neurons in culture were injected with the radiolabeledin vitro transcripts as follows (with the respective numbers of analyzed cells/dendrites given inparentheses). A, B, Full-length BC1 RNA (128/612). Cell bodies and dendrites (including medial and distal segments) of neurons injected with full-length BC1 RNA were labeled strongly.
Open arrows point to clusters of autoradiographic silver grains over some dendritic segments. C, D, Nuclear U4 RNA (33/154). No specific labeling was observed in medial or distal dendritic regions (arrowheads), whereas a low labeling signal was apparent in proximal regions of some dendrites. In such cases, scattering from the soma may also have contributed to the signal in proximal dendritic regions. E, F, A random irrelevant RNA (144 nt), generated from the polylinker sequence of plasmid pSL300 (27/122). As with U4 RNA, no specific labeling was evident in medial and distal dendritic regions. G, Full-length BC1 RNA, low-amount injection routine. Arrows indicate labeling in distal dendritic segments. Although the overall labeling signal was significantly lower than in A, the somatodendritic distribution of the injected RNA was the same. Curved arrow indicates a noninjected cell. H, U4 RNA, low-amount injection routine. No dendritic labeling was observed.I, Chimeric GFP-BC1 RNA (21/99); J, Nonchimeric GFP mRNA (16/71). The chimeric RNA was targeted to dendrites (arrows in I indicate labeling in distal dendritic regions), whereas the nonchimeric RNA was not (arrowheads in J indicate lack of labeling in dendrites). Curved arrow in Iindicates a noninjected cell. Equivalent results were obtained with BC1-bcd RNA and with bcd mRNA, respectively, and in experiments in the low-amount injection routine (data not shown). Microinjection of 35S-UTP (12/44 cells/dendrites analyzed) did not result in any significant dendritic labeling after 4 hr of incubation (data not shown). A, C, E, G-J, Dark-field optics; B, D, F, Nomarski (DIC) optics. Scale bar, 100 μm.
We next asked whether BC1 RNA would be able to impart dendritic targeting competence on RNAs that are normally not delivered to dendrites. For this purpose, we generated chimeric constructs in which the full-length BC1 sequence was inserted either 5′ or 3′ to the coding region of a nondendritic mRNA. In one such construct, BC1 RNA was inserted directly downstream from the coding region of Aequorea victoria GFP mRNA (Heim et al., 1995). Chimeric RNA transcribed from this construct, when injected into the cytoplasm of sympathetic neurons, was transported to dendrites at significant levels (Fig.2I). Corresponding nonchimeric GFP mRNA, in contrast, remained restricted to neuronal somata (Fig.2J). In a second chimeric construct, the BC1 sequence was inserted 55 nt upstream from the coding region ofbcd mRNA (Berleth et al., 1988). Chimeric RNA generated from this construct was again specifically targeted to dendrites, whereas nonchimeric bcd mRNA was not (data not shown). As with BC1 RNA, the differential distribution of the labeling signal to dendrites was independent of the amounts of chimeric RNAs injected. The combined data obtained with chimeric RNAs thus demonstrate that BC1 RNA contains a cis-acting element that is sufficient to direct the dendritic targeting of normally nondendritic mRNAs.
Dendritic targeting of BC1 segments
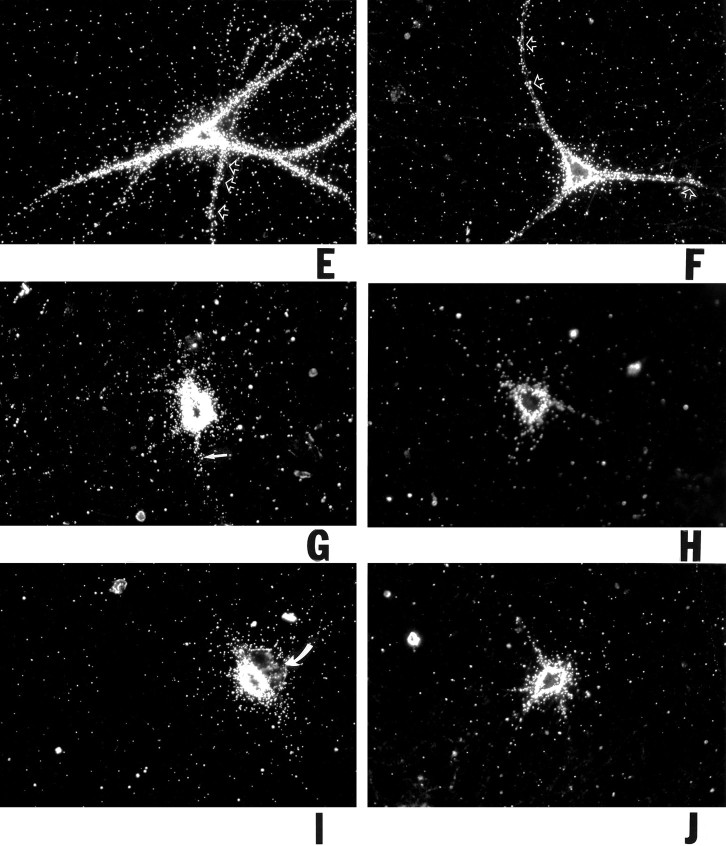
BC1 RNA can be subdivided into three distinct subdomains (DeChiara and Brosius, 1987) of which the 5′ domain (and possibly, although to a much smaller degree, also the 3′ domain) has the potential to form stable secondary structures (Deininger et al., 1996). We therefore decided to examine the dendritic transport competence of individual 5′ and 3′-BC1 segments. (The central region of 22 consecutive A-residues was assumed transport-irrelevant and was therefore not analyzed here.) As shown in Figure 3we found that a 5′-BC1 segment (62 5′-most nt) was transported to dendrites to an extent, at levels and at rates that were comparable to full-length BC1 RNA. After microinjection with the 5′-BC1 segment, cell body and dendrites of a neuron shown in Figure3A,B were strongly labeled. In clear contrast, a 3′-BC1 segment (60 3′-most nt) remained restricted to the soma and, at significantly lower levels, to proximal-most dendritic segments (Fig. 3C,D). Medial and distal parts of dendrites were devoid of specific labeling. Even in proximal dendrites, the labeling produced by the 3′-BC1 segment was significantly lower compared with the 5′-BC1 segment. Dendritic labeling produced by the 3′-BC1 segment was also insubstantial in comparison with full-length BC1 RNA.
Fig. 3.
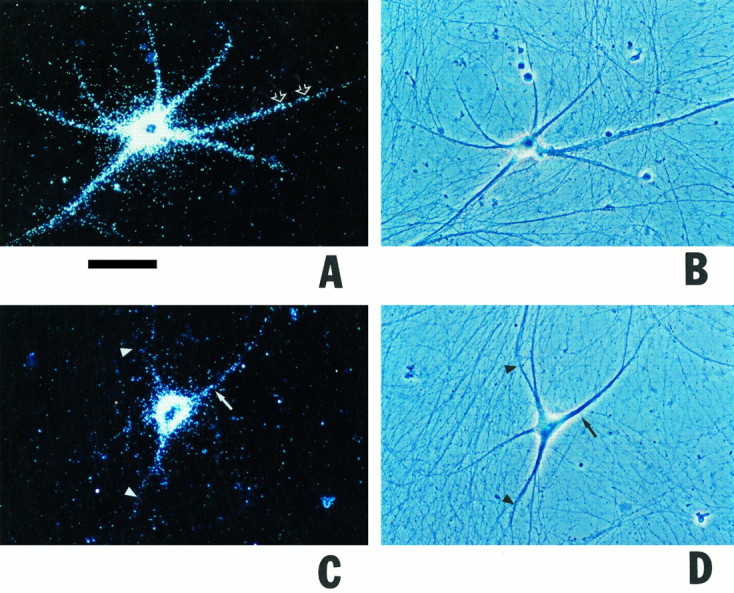
Differential dendritic transport competence of 5′ and 3′ segments of BC1 RNA. Sympathetic neurons in culture were microinjected with radiolabeled in vitro transcripts as follows (with the respective numbers of analyzed cells/dendrites given in parentheses). A, B, 5′ Segment of BC1 RNA (98/377). Cell body and dendrites of a neuron injected with the 5′-BC1 segment showed significant labeling; a signal was detected extending to distances of >250 μm from the soma. C, D, 3′ Segment of BC1 RNA (48/199). The injected cell was labeled over cell body and proximal-most (<100 μm from somata) dendritic regions (arrows). Medial and distal parts of dendrites were devoid of specific labeling (arrowheads).E, F, 5′ Segment of BC1 RNA. The cell inE was injected with standard amounts, cell inF with low amounts. In both cases, labeling was clearly dendritic. Note that in the cell in E, labeling was equally strong in two secondary dendritic branches. G,H, 3′ Segment of BC1 RNA. The cell in Gwas injected with standard amounts, cell in H with low amounts. In both cases, labeling was restricted to the soma (arrow in G indicates low level labeling in a proximal-most dendritic segment). I, U6 RNA (31/161); J, a random irrelevant RNA (64 nt), generated from the polylinker sequence of plasmid pSL300 (39/167). Injection amounts were intermediate (I) and low (J), respectively. Curved arrow in I indicates a noninjected cell.Open arrows in A, E,F point to clusters of autoradiographic silver grains in dendrites. A, C, E–J, Dark-field optics; B, D, phase-contrast optics. Scale bar, 100 μm.
As with full-length BC1 RNA and with BC1-chimeric RNAs, the specific distribution pattern of microinjected BC1 segments was not dependent on the amounts injected. Figure 3E,F compares the dendritic delivery of the 5′-BC1 segment observed with the standard-amount (E) and low-amount (F) injection routines. In both cases the labeling clearly extended into distal dendritic segments, although the overall level of labeling was considerably lower in the low-amount experiment. As with full-length BC1 RNA, dendritic labeling appeared clustered in some dendritic segments. Such clustering seems to suggest that the targeted delivery to dendrites is of a discontinuous nature, with supramolecular particles operating as transport vehicles; alternatively, discontinuity in the dendritic labeling signal may indicate preferential docking to different target sites along the dendritic extent. In contrast to the experiments with the 5′-BC1 segment, no significant dendritic labeling was observed after microinjection of the 3′-BC1 segment, either at standard- or at low-amount injection routines (Fig.3G,H). As a control, we injected U6 RNA, a short nuclear RNA (Hausner et al., 1990); the resulting labeling was restricted to neuronal somata, as shown in Figure 3I. Similarly, injection of a random irrelevant RNA of 64 nt, generated from the polylinker sequence of plasmid pSL300, did not result in any significant dendritic labeling (Fig. 3J).
Taken together, the experiments with individual BC1 segments indicate that a dendritic targeting element is contained within the 62 5′-most nucleotides of BC1 RNA.
Extent of dendritic transport: quantitative analysis
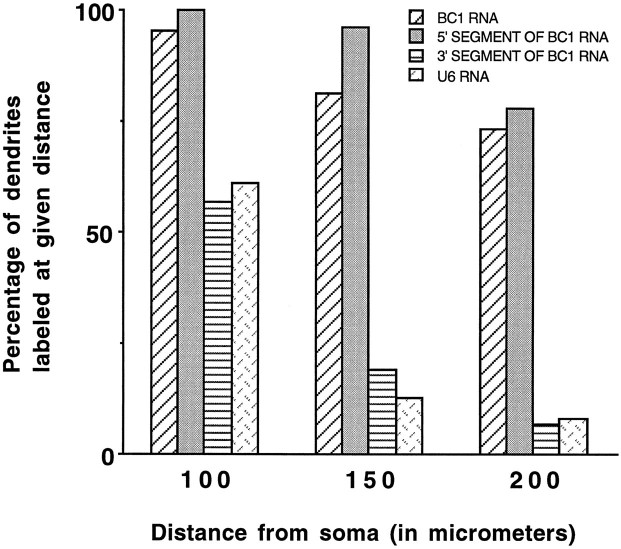
We performed a quantitative analysis of the distribution of microinjected RNAs along dendrites of sympathetic neurons in culture. For this, the extent of postinjection dendritic labeling was calculated as the percentage of dendrites that exhibited significant labeling at given distances from the soma, as measured from the center of the nucleus (Fig. 4). Thus, in cells injected with full-length BC1 RNA or with a 5′-BC1 segment, significant labeling could be detected in most dendrites (74 and 78%, respectively) at a distance of 200 μm from the soma after 4 hr of postinjection incubation. In contrast, <10% of all dendrites were labeled at 200 μm from the cell body after injection of a 3′-BC1 segment, of U4 or U6 RNA, or of random irrelevant RNAs. At distances from the soma of 250 μm or more (in dendrites that extended to such distances), full-length BC1 and 5′-BC1 labeling was still significant, whereas 3′-BC1 and control labeling (U4, U6, and irrelevant RNAs) was negligible. This analysis confirms that microinjected BC1 RNA was selectively transported to dendrites, and that injected nuclear and random irrelevant RNAs were retained in the cell body or proximal-most dendritic segments. It further confirms that dendritic targeting competence of BC1 RNA resides within a 5′ segment of ≤62 nt.
Fig. 4.
Distribution of microinjected RNAs along dendrites of sympathetic neurons in culture. Most dendrites of neurons injected with BC1 RNA (74%) or with a 5′-BC1 segment (78%) exhibited a signal that could be detected at a distance of 200 μm from the soma after 4 hr of postinjection incubation. A 3′-BC1 segment and nuclear U6 RNA produced a labeling signal at a distance of 200 μm from the soma in <10% of all dendrites. Data for distances of >200 μm were not plotted, because not all dendrites extended to such lengths. The labeling signal over dendrites was considered significant if it was at least three times higher than the background. The numbers of cells/dendrites analyzed for this figure were as follows: BC1 RNA, 81/350; 5′-BC1 segment, 46/192; 3′-BC1 segment, 48/199; U6 RNA, 10/30. Data for U4 RNA and for 144 and 64 nt irrelevant RNAs (data not shown) were very similar to the data obtained with the 3′-BC1 segment or with U6 RNA.
Rate of dendritic transport
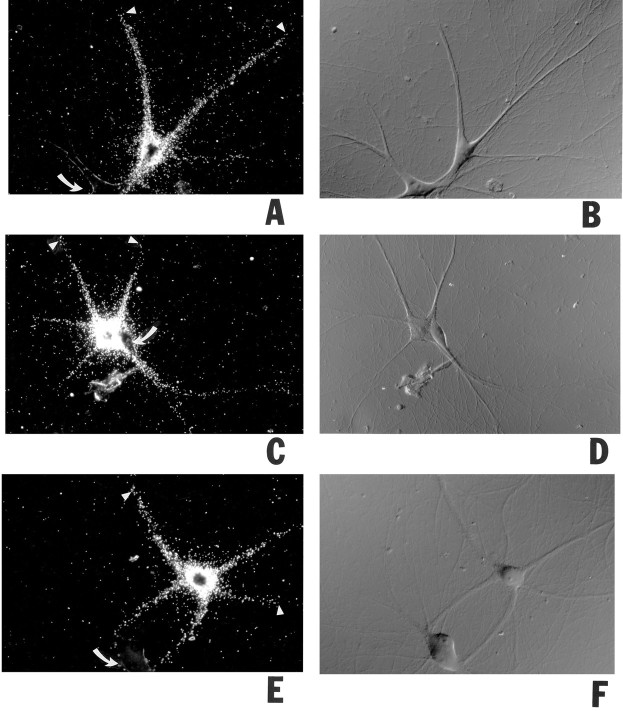
We also asked how fast BC1 RNA was transported into dendrites. Somata of sympathetic neurons in culture were microinjected with full-length BC1 RNA, and cells were incubated at 35°C for various periods before fixation and autoradiography. Figure5 shows that the extent of dendritic labeling is a function of incubation time. The extent of dendritic labeling was defined as the distal-most point along a given dendrite where labeling reached at least three times the background level.
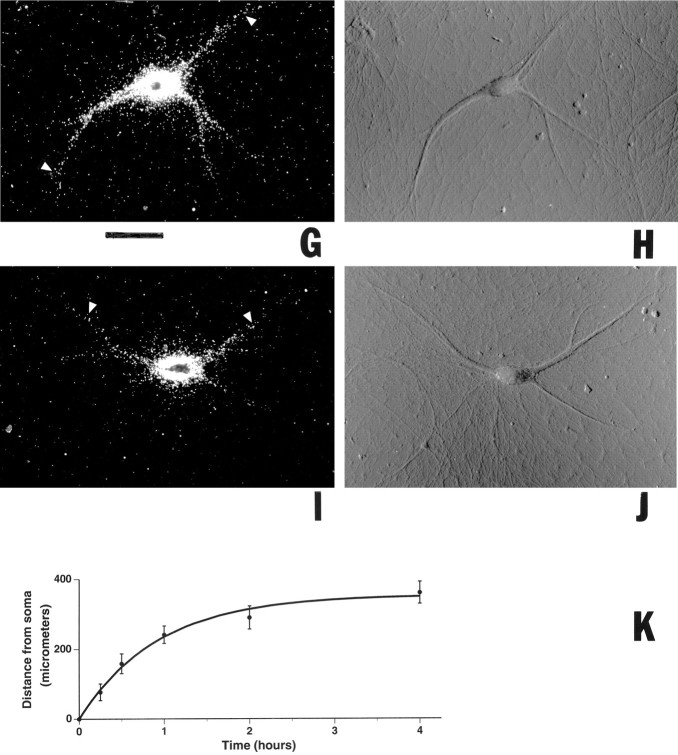
Fig. 5.
Progression of dendritic transport of injected BC1 RNA. Somata of sympathetic neurons in culture were microinjected with35S-labeled full-length BC1 RNA and were incubated at 35°C for 4 hr (A, B), 2 hr (C, D), 1 hr (E,F), 30 min (G,H), and 15 min (I,J), respectively. Arrowheadsindicate the distal-most points in dendrites at which BC1 labeling was detectable for a given time point. Curved arrowsindicate noninjected cells. A, C,E, G, I, Dark-field optics; B, D, F,H, J, Nomarski (DIC) optics. Scale bar, 100 μm. The measured average distances that the labeling signal had reached at each time point were as follows (mean ± SD in μm; sample sizes in parentheses): 15 min, 77 ± 24 (44); 30 min, 159 ± 28 (30); 1 hr, 242 ± 25 (45); 2 hr, 290 ± 33 (21); 4 hr, 360 ± 31 (24). The measured values were not dependent on injection amounts. The data are plotted inK (means, full circles; SD, error bars) with an exponential function (see Materials and Methods) to fit the data.
During the first hour after microinjection, BC1 RNA advanced to an average distance of 242 ± 25 μm. We may thus regard this value as the average rate of dendritic BC1 transport within this period. However, the actual anterograde translocation rate for single BC1 molecules is certainly higher than the average progression rate of the entire BC1 population, this being attributable to factors such as: (1) possible retrograde motility of the RNA and (2) the finite length of dendrites. Such factors become irrelevant as time approaches zero, and we therefore used nonlinear regression methods (see Materials and Methods) to approximate the hypothetical initial velocity at which BC1 RNA proceeds into dendrites. On the basis of an exponential function, this velocity was calculated as 388 ± 15 μm/hr. As a note of caution, it should be added that this value is an approximation that is predicated on the selection of a mathematical model that best fits the experimental data (see Materials and Methods). Nonetheless, we consider this a valuable estimate of the anterograde velocity of BC1 RNA in dendrites.
DISCUSSION
We have used a microinjection protocol with sympathetic neurons in primary culture to analyze dendritic delivery of neuronal BC1 RNA. Our results demonstrate that BC1 RNA is selectively transported to dendritic target sites in such neurons. This targeted transport is specific; control RNAs, including nuclear and irrelevant RNAs of similar size, were not observed to enter dendritic domains to significant extents. Experiments with chimeric RNAs demonstrate that BC1 RNA contains information sufficient to impart dendritic targeting competence on normally nondendritic mRNAs. Microinjection experiments with BC1 segments further indicate that a cis-acting element within the 5′ region of the RNA is responsible for its dendritic targeting. Our data thus demonstrate that mechanisms exist in neurons that mediate the targeted transport of select RNAs to dendritic domains.
The dendritic transport of BC1 RNA is rapid; the average dendritic transport rate of BC1 RNA during the first hour after injection was measured at 242 ± 25 μm/hr, and a hypothetical initial velocity was calculated at 388 ± 15 μm/hr. The delivery of total newly synthesized cellular RNA to dendrites has been analyzed previously by pulse labeling with [3H]uridine (Davis et al., 1987; 1990). [3H]RNA appeared in dendrites at an estimated rate of 11–21 μm/hr. The dendritic transport of BC1 RNA is thus significantly higher than the delivery rate of total newly synthesized RNA. We attribute this to the heterogeneous nature of newly synthesized endogenous RNA, which is likely to include rRNA (Kleiman et al., 1993, 1994), tRNA (Tiedge and Brosius, 1996), and other RNA species that may accumulate in dendrites by different and slower mechanisms. Although we have not yet analyzed whether BC1 transport is an active, energy-dependent process, the speed and specificity of this transport argue against passive dispersal followed by selective localization through mechanisms such as trapping. Rapid transport in dendrites (minimum transport rate, 200–300 μm/hr) has also been described in a preliminary report for the appearance of Arc mRNA in the molecular layer of the dentate gyrus after induction of expressionin vivo (Wallace et al., 1995). Granules containing myelin basic protein (MBP) mRNA are transported in oligodendrocyte processes at ∼720 μm/hr (Ainger et al., 1993), a rate that reflects sustained anterograde movement of single granules. Dye-labeled RNA-containing granules in dendrites have been shown to move in both anterograde and retrograde directions, with a transport velocity of 360 μm/hr in either direction (Knowles et al., 1996).
As Figures 3 and 4 illustrate, BC1 RNA contains, within a 5′ segment of 62 nucleotides or less, a cis-acting element that is responsible for its dendritic targeting. This observation, together with the fact that BC1 RNA is an RNA polymerase III transcript (Martignetti and Brosius, 1995), raises the question of whether the BC1-targeting element (BTE) is shared by mRNAs (i.e., RNA polymerase II transcripts) that have been localized to dendrites. To address this question, we compared the BC1 5′ sequence with sequences of dendritic and nondendritic mRNAs. The “prototypical” dendritic mRNAs, encoding high-Mr MAP2a/b isoforms (Garner et al., 1988), contain two motifs with sequence similarity to BC1 RNA (GaGGUUGGGGAU and GUAGAGCuCUU, upper case lettering indicating matches). In MAP2a/b mRNAs, both motifs are spaced closely (nt 3012–3023 and 3027–3037; numbering according to Kindler et al., 1990), whereas in BC1 RNA they are located (nt 1–12 and 24–34; for sequence see DeChiara and Brosius, 1987) such that they are flanking box A (nt 14–24) of the RNA polymerase III promoter. In MAP2a/b mRNAs, these motifs are contained within the central segment of the coding region that is lacking from nondendritic MAP2c mRNAs. Because the 5′ and 3′ untranslated regions (UTRs) of MAP2a/b and MAP2c mRNAs are identical, it has been predicted that a dendritic targeting element in MAP2a/b mRNAs would have to be contained within the part of the coding regions that is unique to the high-Mr isoforms (Kindler et al., 1996). Because this prediction has not yet been confirmed experimentally, the relevance of sequence similarities between BC1 RNA and MAP2a/b mRNAs will have to be ascertained in future work. Although RNA localization signals, in particular in theDrosophila system, have in several examples been associated with 3′ UTRs (for review, see St Johnston, 1995), it is unclear whether the relative position of such a signal within an mRNA is directly relevant for its function. In fact, our experiments with chimeric RNAs indicate that a sequence imparting dendritic targeting competence can also be introduced 5′ to a coding region.
Other dendritic mRNAs share the 5′-BC1 dodecamer motif (but not the hendecamer motif) to varying degrees. For example, the dodecamer motif is represented in InsP3R mRNA (Mignery et al., 1990) with 10 of 12 matches in the coding region, and in Ng/RC3 mRNA (Sato et al., 1995) with 10 of 12 matches in the 3′ UTR. A weaker degree of similarity was observed in other dendritic RNAs. The limited sequence overlap between BC1 RNA and some of these dendritic mRNAs raises two important issues. First, it is questionable whether the BC1 targeting element is used universally by other dendritic RNAs. Such RNAs may contain other and/or additional targeting elements. For example, because dendritic RNAs are distributed differentially in dendrites (Steward, 1995), one would assume that at least some of the elements that are responsible for targeting and localization should be distinctive. For the same reason, it seems likely that the differential delivery of dendritic RNAs to their respective target sites would require, in at least some RNAs, the concerted or sequential participation of multiple cis-acting elements, as has also been discussed for localized RNAs in other systems (St Johnston, 1995). It is hoped that the degree to which such elements are being shared between BC1 RNA and other dendritic RNAs will become clearer in the future as more dendritic targeting elements will be identified in dendritic mRNAs. Second, it should be emphasized that the sequence per se of a cis-acting element such as the one in BC1 RNA may not be the sole or even the most important determinant of dendritic RNA targeting competence. For example, secondary and higher-order structures may be of greater importance than the primary structure in mediating interactions of the RNA with a conjectured dendritic transport machinery. Secondary and tertiary RNA structures frequently serve as recognition motifs for RNA-binding proteins, and such structures have been shown to play important roles in the specificity of many protein–RNA interactions (for review, see Draper, 1995). In BC1 RNA, a 5′ segment of 75 nt has been predicted to form a hairpin structure or alternatively, although of much lower stability, a tRNA-like cloverleaf structure (Deininger et al., 1996). Although the 5′-BC1 segment that was used in this study was shorter (62 nt) and will therefore not display the full secondary structure conformation of the 75 nt segment, we presume that secondary structure motifs are likely to be relevant for the dendritic targeting of BC1 RNA.
In vivo, BC1 RNA has been shown to be complexed with proteins to form an RNP (Kobayashi et al., 1991; Cheng et al., 1996). The molecular mass of the core BC1 particle has been estimated at 188 kDa (BC1 RNA, 50 kDa; associated proteins, 138 kDa; Cheng et al., 1996). It is thus plausible to assume that BC1 RNA is being transported in dendrites as part of this minimal molecular unit. We do not know at this time whether some or all of these proteins also bind to microinjected BC1 RNA; obviously, however, injected BC1 RNA is being recognized by factors that mediate interactions with the cellular transport machinery. Such factors may or may not be part of the core BC1 particle, an issue to be resolved pending further purification of BC1 particle proteins. The high rate at which microinjected BC1 RNA moves into dendrites would seem to implicate interactions with cellular motors of the kinesin superfamily. This and the fact that translocation of RNA granules in dendrites has been shown to be dependent on intact microtubules (Knowles et al., 1996) leads us to hypothesize that BC1 transport may proceed along dendritic microtubules. Finally, we wish to suggest that at least transiently, for example en route or at its destination points, BC1 RNA or the BC1 core particle may be integrated within higher supramolecular assemblies that may play a role in RNA transport and/or translation in dendrites.
Conclusions
The results presented here demonstrate that a spatial determinant, specifying the dendritic localization of a neuronal RNA, is encoded within a specific segment of the RNA itself. Thus, genetic information defines a topological parameter in nerve cells. In a number of other eukaryotic cell systems, cis-acting elements have been shown to mediate RNA localization (reviewed by Kislauskis and Singer, 1992;Lehmann, 1995; Lipshitz, 1995; St Johnston, 1995). In theDrosophila and Xenopus oocyte–embryo systems, RNA localization is instrumental in the establishment of embryonic polarity (for review, see Ding and Lipshitz, 1993). In neurons, functional correlates of RNA localization have not yet been as well defined but are likely to include the regulation of mosaic postsynaptic protein repertoires (Steward, 1995). Converging evidence is thus suggesting that RNA targeting may play an important role in the management of microgeometry in various eukaryotic cell systems.
Footnotes
This work was supported in part by National Science Foundation Grant IBN-9210149, Human Frontier Science Program Organization Grant RG-84/94 B, and National Institutes of Health Grant NS34158 (H.T.). I.A.M. is an Aaron Diamond Foundation Fellow. E.S. was supported by a fellowship from the Istituto Pasteur-Fondazione Cenci Bolognetti. We thank the following colleagues for plasmids: J. Brosius, C. Garner, R. Müller, R. Heim, R. Tsien, and A. Weiner. We further thank W. Makalowski for GenBank/EMBL database searches for ID elements and J. Brosius for comments on this manuscript.
Correspondence should be addressed to Henri Tiedge, Department of Pharmacology, State University of New York, Health Science Center at Brooklyn, 450 Clarkson Avenue, Brooklyn, NY 11203.
Sean Perini’s present address: Department of Radiology, University of California at San Francisco, San Francisco, CA 94143.
REFERENCES
- 1.Ainger K, Avossa D, Morgan F, Hill SJ, Barry C, Barbarese E, Carson JC. Transport and localization of exogenous myelin basic protein mRNA microinjected into oligodendrocytes. J Cell Biol. 1993;123:431–441. doi: 10.1083/jcb.123.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banker G, Goslin K. Characterizing and studying neuronal cultures. In: Banker G, Goslin K, editors. Culturing nerve cells. MIT; Cambridge, MA: 1991. pp. 76–109. [Google Scholar]
- 3.Berleth T, Burri M, Thoma G, Bopp D, Richstein S, Frigerio G, Noll M, Nüsslein-Volhard C. The role of localization of bicoid RNA in organizing the anterior pattern of the Drosophila embryo. EMBO J. 1988;7:1749–1756. doi: 10.1002/j.1460-2075.1988.tb03004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bringmann P, Appel B, Rinke J, Reuter H, Theissen H, Lührmann R. Evidence for the existence of snRNAs U4 and U6 in a single ribonucleoprotein complex and their association by intermolecular base-pairing. EMBO J. 1984;3:1357–1363. doi: 10.1002/j.1460-2075.1984.tb01977.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brosius J. Superpolylinkers in cloning and expression vectors. DNA. 1989;8:759–777. doi: 10.1089/dna.1989.8.759. [DOI] [PubMed] [Google Scholar]
- 6.Brosius J, Tiedge H. Neural BC1 RNA: dendritic localization and transport. In: Lipshitz HD, editor. Localized RNAs. RG Landes; Austin, TX: 1995. pp. 289–300. [Google Scholar]
- 7.Bruckenstein DA, Lein PJ, Higgins D, Fremeau RT. Distinct spatial localization of specific mRNAs in cultured sympathetic neurons. Neuron. 1990;5:809–819. doi: 10.1016/0896-6273(90)90340-l. [DOI] [PubMed] [Google Scholar]
- 8.Burgin KE, Waxman MN, Rickling S, Westgate SA, Mobley WC, Kelly PT. In situ hybridization histochemistry of Ca2+/calmodulin-dependent protein kinase in developing rat brain. J Neurosci. 1990;10:1788–1798. doi: 10.1523/JNEUROSCI.10-06-01788.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng JG, Tiedge H, Brosius J. Identification of BC1 RNP particles. DNA Cell Biol. 1996;15:549–559. doi: 10.1089/dna.1996.15.549. [DOI] [PubMed] [Google Scholar]
- 10.Chicurel ME, Terrian DM, Potter H. mRNA at the synapse: analysis of a synaptosomal preparation enriched in hippocampal dendritic spines. J Neurosci. 1993;13:4054–4063. doi: 10.1523/JNEUROSCI.13-09-04054.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crino PB, Eberwine J. Molecular characterization of the dendritic growth cone: regulated mRNA transport and local protein synthesis. Neuron. 1996;17:1173–1187. doi: 10.1016/s0896-6273(00)80248-2. [DOI] [PubMed] [Google Scholar]
- 12.Davis L, Banker G, Steward O. Selective dendritic transport of RNA in hippocampal neurons in culture. Nature. 1987;330:477–479. doi: 10.1038/330477a0. [DOI] [PubMed] [Google Scholar]
- 13.Davis L, Burger B, Banker GA, Steward O. Dendritic transport: quantitative analysis of the time course of somatodendritic transport of recently synthesized RNA. J Neurosci. 1990;10:3056–3068. doi: 10.1523/JNEUROSCI.10-09-03056.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeChiara TM, Brosius J. Neural BC1 RNA: cDNA clones reveal nonrepetitive sequence content. Proc Natl Acad Sci USA. 1987;84:2624–2628. doi: 10.1073/pnas.84.9.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deininger PL, Tiedge H, Kim J, Brosius J. The BC1 RNA gene as a master gene for ID amplification: evolution, expression and function. In: Cohn WE, Moldave K, editors. Progress in nucleic acid research and molecular biology, Vol 52. Academic; San Diego: 1996. pp. 67–88. [DOI] [PubMed] [Google Scholar]
- 16.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ding D, Lipshitz HD. Localized RNA and their functions. BioEssays. 1993;15:651–658. doi: 10.1002/bies.950151004. [DOI] [PubMed] [Google Scholar]
- 18.Draper DE. Protein-RNA recognition. Annu Rev Biochem. 1995;64:593–620. doi: 10.1146/annurev.bi.64.070195.003113. [DOI] [PubMed] [Google Scholar]
- 19.Furuichi T, Simon-Chazottes D, Fujino I, Yamada N, Hasegawa M, Miyawaki A, Yoshikawa S, Guénet JL, Mikoshiba K. Widespread expression of inositol 1,4,5-trisphosphate receptor type 1 gene (Insp 3r 1) in the mouse central nervous system. Receptors Channels. 1993;1:11–24. [PubMed] [Google Scholar]
- 20.Garner CC, Tucker RP, Matus A. Selective localization of messenger RNA for cytoskeletal protein MAP2 in dendrites. Nature. 1988;336:674–677. doi: 10.1038/336674a0. [DOI] [PubMed] [Google Scholar]
- 21.Hashimoto C, Steitz JA. U4 and U6 RNAs coexist in a single small ribonucleoprotein particle. Nucleic Acids Res. 1984;12:3283–3293. doi: 10.1093/nar/12.7.3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hausner TP, Giglio LM, Weiner AM. Evidence for base-pairing between mammalian U2 and U6 small nuclear ribonucleoprotein particles. Genes Dev. 1990;4:2146–2156. doi: 10.1101/gad.4.12a.2146. [DOI] [PubMed] [Google Scholar]
- 23.Heim R, Cubitt A, Tsien R. Improved green fluorescence. Nature. 1995;373:663–664. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
- 24.Higgins D, Lein PJ, Osterhout DJ, Johnson MI. Tissue culture of mammalian autonomic neurons. In: Banker G, Goslin K, editors. Culturing nerve cells. MIT; Cambridge, MA: 1991. pp. 177–205. [Google Scholar]
- 25.Kang H, Schuman EM. A requirement for local protein synthesis in neurotrophin-induced hippocampal synaptic plasticity. Science. 1996;273:1402–1406. doi: 10.1126/science.273.5280.1402. [DOI] [PubMed] [Google Scholar]
- 26.Kindler S, Schwanke B, Schulz B, Garner CC. Complete cDNA sequence encoding rat high and low molecular weight MAP2. Nucleic Acid Res. 1990;18:2822. doi: 10.1093/nar/18.9.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kindler S, Müller R, Chung WJ, Garner CC. Molecular characterization of dendritically localized transcripts encoding MAP2. Mol Brain Res. 1996;36:63–69. doi: 10.1016/0169-328x(95)00237-m. [DOI] [PubMed] [Google Scholar]
- 28.Kislauskis EH, Singer RH. Determinants of mRNA localization. Curr Opin Cell Biol. 1992;4:975–978. doi: 10.1016/0955-0674(92)90128-y. [DOI] [PubMed] [Google Scholar]
- 29.Kleiman R, Banker G, Steward O. Differential subcellular localization of particular mRNAs in hippocampal neurons in culture. Neuron. 1990;5:821–830. doi: 10.1016/0896-6273(90)90341-c. [DOI] [PubMed] [Google Scholar]
- 30.Kleiman R, Banker G, Steward O. Subcellular distribution of rRNA and poly (A) RNA in hippocampal neurons in culture. Mol Brain Res. 1993;20:305–312. doi: 10.1016/0169-328x(93)90057-v. [DOI] [PubMed] [Google Scholar]
- 31.Kleiman R, Banker G, Steward O. Development of subcellular mRNA compartmentation in hippocampal neurons in culture. J Neurosci. 1994;14:1130–1140. doi: 10.1523/JNEUROSCI.14-03-01130.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knowles RB, Sabry JH, Martone ME, Deerinck TF, Ellisman MH, Bassell GJ, Kosik KS. Translocation of RNA granules in living neurons. J Neurosci. 1996;16:7812–7820. doi: 10.1523/JNEUROSCI.16-24-07812.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kobayashi S, Goto S, Anzai K. Brain-specific small RNA transcript of the identifier sequences is present as a 10 S ribonucleoprotein particle. J Biol Chem. 1991;66:4726–4730. [PubMed] [Google Scholar]
- 34.Landry CF, Watson JB, Kashima T, Campagnoni AT. Cellular influences on RNA sorting in neurons and glia: an in situ hybridization histochemical study. Mol Brain Res. 1994;27:1–11. doi: 10.1016/0169-328x(94)90178-3. [DOI] [PubMed] [Google Scholar]
- 35.Lehmann R. Cell-cell signaling, microtubules, and the loss of symmetry in the Drosophila oocyte. Cell. 1995;83:353–356. doi: 10.1016/0092-8674(95)90111-6. [DOI] [PubMed] [Google Scholar]
- 36.Link W, Konietzko U, Kauselmann G, Krug M, Schwanke B, Frey U, Kuhl D. Somatodendritic expression of an immediate early gene is regulated by synaptic activity. Proc Natl Acad Sci USA. 1995;92:5734–5738. doi: 10.1073/pnas.92.12.5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lipshitz HD. Localized RNAs. RG Landes; Austin, TX: 1995. [Google Scholar]
- 38.Lührmann R. Function of U-snRNPs. Mol Biol Rep. 1990;14:183–192. doi: 10.1007/BF00360468. [DOI] [PubMed] [Google Scholar]
- 39.Lyford GL, Yamagata K, Kaufmann WE, Barnes CA, Sanders LK, Copeland NG, Gilbert DJ, Jenkins NA, Lanahan AA, Worley PF. Arc, a growth factor and activity-regulated gene, encodes a novel cytoskeleton-associated protein that is enriched in neuronal dendrites. Neuron. 1995;14:433–445. doi: 10.1016/0896-6273(95)90299-6. [DOI] [PubMed] [Google Scholar]
- 40.Martignetti JA, Brosius J. BC1 RNA: transcriptional analysis of a neural cell-specific RNA polymerase III transcript. Mol Cell Biol. 1995;15:1642–1650. doi: 10.1128/mcb.15.3.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mayford M, Baranes D, Podsypanina K, Kandel ER. The 3′-untranslated region of CamKIIα is a cis-acting signal for the localization and translation of mRNA in dendrites. Proc Natl Acad Sci USA. 1996;93:13250–13255. doi: 10.1073/pnas.93.23.13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mignery GA, Newton CL, Archer BT, Südhof TC., III Structure and expression of the rat inositol 1,4,5-trisphosphate receptor. J Biol Chem. 1990;265:12679–12685. [PubMed] [Google Scholar]
- 43.Miyashiro K, Dichter M, Eberwine J. On the nature and differential distribution of mRNAs in hippocampal neurites: implications for neuronal functioning. Proc Natl Acad Sci USA. 1994;91:10800–10804. doi: 10.1073/pnas.91.23.10800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Racca C, Gardiol A, Triller A. Dendritic and postsynaptic localizations of glycine receptor subunit mRNAs. J Neurosci. 1997;17:1691–1700. doi: 10.1523/JNEUROSCI.17-05-01691.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rao A, Steward O. Evaluation of RNAs present in synapodendrosomes: dendritic, glial, and neuronal cell body contribution. J Neurochem. 1993;61:835–844. doi: 10.1111/j.1471-4159.1993.tb03594.x. [DOI] [PubMed] [Google Scholar]
- 46.Sato T, Xiao DM, Li H, Huang FL, Huang KP. Structure and regulation of the gene encoding the neuron-specific protein kinase C substrate neurogranine (RC3 protein). J Biol Chem. 1995;270:10314–10322. doi: 10.1074/jbc.270.17.10314. [DOI] [PubMed] [Google Scholar]
- 47.St Johnston D. The intracellular localization of messenger RNA. Cell. 1995;81:161–170. doi: 10.1016/0092-8674(95)90324-0. [DOI] [PubMed] [Google Scholar]
- 48.Steward O. Dendrites as compartments for macromolecular synthesis. Proc Natl Acad Sci USA. 1994;91:10766–10768. doi: 10.1073/pnas.91.23.10766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Steward O. Targeting of mRNAs to subsynaptic microdomains in dendrites. Curr Opin Neurobiol. 1995;5:55–61. doi: 10.1016/0959-4388(95)80087-5. [DOI] [PubMed] [Google Scholar]
- 50.Steward O. mRNA localization in neurons: a multipurpose mechanism? Neuron. 1997;18:9–12. doi: 10.1016/s0896-6273(01)80041-6. [DOI] [PubMed] [Google Scholar]
- 51.Steward O, Banker GA. Getting the message from the gene to the synapse: sorting and intracellular transport of RNA in neurons. Trends Neurosci. 1992;15:180–186. doi: 10.1016/0166-2236(92)90170-d. [DOI] [PubMed] [Google Scholar]
- 52.Steward O, Levy WB. Preferential localization of polyribosomes under the base of dendritic spines in granule cells of the dentate gyrus. J Neurosci. 1982;2:284–291. doi: 10.1523/JNEUROSCI.02-03-00284.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Steward O, Reeves TM. Protein-synthetic machinery beneath postsynaptic sites on CNS neurons: association between polyribosomes and other organelles at the synaptic site. J Neurosci. 1988;8:176–184. doi: 10.1523/JNEUROSCI.08-01-00176.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tiedge H. The use of UV light as a cross-linking agent for cells and tissue sections in in situ hybridization. DNA Cell Biol. 1991;10:143–147. doi: 10.1089/dna.1991.10.143. [DOI] [PubMed] [Google Scholar]
- 55.Tiedge H, Fremeau RT, Weinstock PH, Jr, Arancio O, Brosius J. Dendritic location of neural BC1 RNA. Proc Natl Acad Sci USA. 1991;88:2093–2097. doi: 10.1073/pnas.88.6.2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tiedge H, Brosius J. Translational machinery in hippocampal neurons in culture. J Neurosci. 1996;16:7171–7181. doi: 10.1523/JNEUROSCI.16-22-07171.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Torre ER, Steward O. Demonstration of local protein synthesis within dendrites using a new cell culture system which permits the isolation of living axons and dendrites from their cell bodies. J Neurosci. 1992;12:762–772. doi: 10.1523/JNEUROSCI.12-03-00762.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wallace CS, Lyford GL, Worley PF, Steward O. Rapid translocation of Arc mRNA in the rat dentate gyrus following ECS. Soc Neurosci Abstr. 1995;21:600. [Google Scholar]
- 59.Zuker M. Computer prediction of RNA structure. Methods Enzymol. 1989;180:261–288. doi: 10.1016/0076-6879(89)80106-5. [DOI] [PubMed] [Google Scholar]