Abstract
Using a general strategy for evaluating clinical tissue specimens, we found that 70% ethanol fixation and paraffin embedding is a useful method for molecular profiling studies. Human prostate and kidney were used as test tissues. The protein content of the samples was analyzed by one-dimensional gel electrophoresis, immunoblot, two-dimensional gel electrophoresis, and layered expression scanning. In each case, the fixed and embedded tissues produced results similar to that obtained from snap-frozen specimens, although the protein quantity was somewhat decreased. Recovery of mRNA was reduced in both quantity and quality in the ethanol-fixed samples, but was superior to that obtained from formalin-fixed samples and sufficient to perform reverse transcription polymerase chain reactions. Recovery of DNA from ethanol-fixed specimens was superior to formalin-fixed samples as determined by one-dimensional gel electrophoresis and polymerase chain reaction. In conclusion, specimens fixed in 70% ethanol and embedded in paraffin produce good histology and permit recovery of DNA, mRNA, and proteins sufficient for several downstream molecular analyses. Complete protocols and additional discussion of relevant issues are available on an accompanying website (http://cgap-mf.nih.gov/).
The information from the Human Genome Project and new high-throughput expression technologies are permitting investigators to comprehensively measure mRNA and protein levels in biological samples. 1-8 These data sets are useful for determining biochemical pathways and regulatory elements that are active in cells of various phenotypes or those exhibiting a particular behavior. Moreover, they permit comparison of the temporal patterns of expression that occur during normal development or evolution of a disease process.
In humans, the most widely used approach to characterizing expression levels is to measure mRNA or protein abundance in cell lines in vitro. These models are powerful tools that have lead to novel discoveries and a mechanistic understanding of many cellular processes. However, work in our laboratory and that of others is raising significant questions regarding the validity of cultured cells as accurate global expression models of human cells in vivo. For example, Celis and co-workers 9 found that short-term culturing of bladder cancer cells leads to changes in expression of several proteins involved in key cellular activities. Our group used two-dimensional polyacrylamide gel electrophoresis (2D-PAGE) analysis to compare proteomic profiles of prostate epithelial cells that were microdissected directly from a human prostate gland with two widely used prostate epithelial cell lines (LNCAP, PC3) and two epithelial lines derived from the patient (1542-N, 1542-T). Even at the level of high abundance proteins, the primary tissue samples were significantly dissimilar from the cultured lines. 10 These data indicate that the research community must be cautious in the use of cell lines as representative expression models of cells in vivo, and further suggest that analysis of clinical tissue specimens will be an important component of efforts to completely characterize gene expression in humans.
There are several obstacles to high-throughput molecular analyses of tissue samples, starting with the methods used for fixation and embedding. At first glance, sample acquisition and processing may seem of little concern to investigators; however, these procedures impact heavily on subsequent studies. For example, in most countries, tissue specimens have been processed for the past several decades using aldehyde-based (eg, formalin) fixation which induces extensive protein cross-linking and makes recovery of biomolecules tenuous. 11,12 These samples are not satisfactory for high-throughput expression methodologies such as cDNA microarrays, serial analysis of gene expression (SAGE), or two-dimensional polyacrylamide gel electrophoresis (2D-PAGE). Therefore, a critical need exists for development of new tissue processing methods that produce high-quality histological detail and also permit recovery of mRNA and protein of sufficient quality for molecular profiling studies.
Many factors must be considered when developing and evaluating a new clinical methodology, including balancing diagnostic and research objectives, and protecting patient confidentiality. This process is accomplished by our group with a three-stage approach (see website, “Prostate Molecular Profiling” section). Using this strategy, we evaluated a series of fixatives and embedding compounds. Histological quality, preservation of biomolecules for subsequent molecular profiling studies, and ease-of-use in a clinical setting were each independently assessed.
Materials and Methods
Histology
Human prostate and kidney specimens from four patients were uniformly cut (2 to 5 mm diameter) and placed into one of eight fixatives, including two aldehyde-based and six alcohol-based (non-crosslinking) fixatives. The particular formulations were selected on the basis of a review of the literature and our previous work with murine tissues. 13-20 The samples were embedded in paraffin and sectioned onto glass slides using standard protocols. Five surgical pathologists from two separate institutions (National Cancer Institute (NCI) and John Hopkins University) evaluated the histology of the tissue sections without knowledge of the processing conditions. Rankings were based on nuclear morphology, cellular morphology, tissue architecture, and staining characteristics.
Prostatectomy Specimens
Prostatectomy specimens were placed immediately on ice after surgery, the margins were inked, and the specimens were transversely sectioned into 3- to 5-mm-thick sections. In a subset of cases, one transverse section was frozen in OCT, another was formalin-fixed and paraffin-embedded, another was 70% ethanol-fixed and polyester wax-embedded, and the remaining pieces of tissue were fixed in 70% ethanol and paraffin-embedded. All tissue fixation periods were for approximately 24 hours and ethanol fixation was performed at 4°C while formalin fixation was performed at room temperature.
All of the paraffin-embedded tissue was processed in a V.I.P. tissue processor (Sakura Finetek, Inc., Torrance, CA). The formalin-fixed tissue was processed routinely as performed in a standard pathology department. The ethanol-fixed and paraffin-embedded tissue was processed at 40°C in 70% ethanol for 30 minutes, then 80% ethanol for 30 minutes, then twice in 95% ethanol (each time for 45 minutes), then four times in 100% ethanol (each time for 45 minutes). Finally, the tissue was infiltrated at 58°C four times in paraffin wax (Oxford Labware, St. Louis, MO) for 30 minutes each time, and embedded to form tissue blocks.
The tissue which was infiltrated in polyester wax was processed manually. Samples were processed at 4°C in 70% ethanol twice for 2 hours each, then 90% ethanol for 90 minutes, and then 99% ethanol for 90 minutes. The tissue was subsequently placed in 100% ethanol at room temperature for 150 minutes and then infiltrated in low-melt polyester wax (Gallard-Schlesinger Industries, Inc., Carle Place, NY). Infiltration was performed at 45°C with agitation first in 50:50 polyester wax:ethanol for 150 minutes, then in 90:10 polyester wax:ethanol overnight (approximately 15 hours). The tissue was then polyester wax-embedded to form tissue blocks.
Tissue Staining
Five-μm-thick sections of frozen, ethanol-fixed and paraffin-embedded, ethanol-fixed and polyester wax-embedded, and formalin-fixed and paraffin-embedded tissue from a whole-mount prostatectomy specimen were cut onto glass slides. The frozen tissue sections were stored at −80°C until use. After obtaining sections, the following protocol was used to stain tissue before mRNA and DNA analysis. Paraffin-embedded tissue sections were dewaxed in two consecutive baths of xylenes for 5 minutes each and polyester wax-embedded tissue sections were dewaxed in two consecutive baths of 100% ethanol for 5 minutes each. The sections were then placed in decreasing concentrations of ethanol (100%, 95%, then 70%) for approximately 10 seconds each. The tissue sections were eosin-stained (Sigma-Aldrich St. Louis, MO) for 5 seconds followed by immersion in increasing concentrations of ethanol (95%, 100%) for 10 seconds each. Finally, the sections were immersed in xylenes for 20 seconds. Snap frozen tissue sections were placed in 70% ethanol for 20 seconds followed by eosin for 5 seconds. The sections were then immersed in 95% and then 100% ethanol for 10 seconds each followed by xylenes for 20 seconds. The same protocol was used for staining tissue for protein analysis except one tablet of Complete, Mini protease inhibitor cocktail (Roche Diagnostics, Indianapolis, IN) was added per 10 ml of all of the staining reagents except xylenes. We have previously observed that the recovery of nucleic acids and proteins is superior from tissue which was stained only with eosin rather than hematoxylin and eosin. Since our interest is the analysis of the effect of fixation and embedding on the recovery of DNA, RNA, and proteins, we decided to analyze tissue which was stained only with eosin.
Immunohistochemistry
Prostate-specific antigen (PSA) protein expression was compared for ethanol-fixed, paraffin-embedded and formalin-fixed, paraffin-embedded prostate tissue using a polyclonal anti-PSA antibody (DAKO, Carpinteria, CA). The samples were run on a Ventana (Tucson, AZ) autostainer and the antibody-antigen complex was visualized using diaminobenzidine tetrahydrochloride (DAB) as the substrate. The sections were counterstained with Mayer’s hematoxylin and coverslipped using Permount (Fisher Scientific, Pittsburgh, PA). The intensity and specificity of epithelial staining were analyzed.
Protein Analysis by One-Dimensional PAGE
Equivalent volumes of tissue from each sample preparation were placed in 800 μl of a 1:1 mixture of Tissue Protein Extraction Reagent (T-PER, Pierce, Rockford IL) and 2X sodium dodecyl sulfate (SDS) sample buffer (4% SDS, 160 mmol/L Tris-HCl (pH 6.8) 20% glycerol, and 5% β-mercaptoethanol) and were incubated for 2 hours at 70°C, or at 80°C, or not at all. Following incubation, the lysates were heated to 95°C for 10 minutes, were briefly spun, and 10 μl of each of the supernatants run on a 4 to 20% denaturing Tris-glycine gel. Proteins were transferred onto a polyvinylidene difluoride (PVDF) membrane, stained with Sypro Ruby protein blot stain (Molecular Probes, Eugene, OR) according to the manufacturer’s procedure, and visualized using a Fluorimager SI (Molecular Dynamics, Sunnyvale, CA).
Protein Analysis by Immunoblot
Approximately 35,000 prostate epithelial cells were microdissected using laser capture microdissection (LCM) from histological sections of ethanol-fixed, paraffin-embedded tissue, and frozen tissue. Two separate microdissections from the ethanol-fixed, paraffin-embedded sample were performed to assess reproducibility. The samples were lysed for 2 hours at 80°C in 30 μl of a 1:1 mixture of T-PER and 2X SDS sample buffer. Proteins were resolved on a 4 to 20% denaturing Tris-glycine gel. After transfer onto a PVDF membrane, standard immunoblotting was performed using a monoclonal anti-PSA antibody from Scripps Laboratories (San Diego, CA) (MP007) at a dilution of 1:1000 and the Western-Star chemiluminescence detection system (Tropix, Inc., Bedford MA).
Two-Dimensional PAGE Analysis
An ethanol-fixed, paraffin-embedded tissue section was dewaxed then lysed in 400 μl of isoelectric focusing buffer containing 7 mol/L urea, 2 mol/L thiourea, 4% CHAPS, 1% Mega-10, 1% OBG, 0.5% Triton X-100, 40 mmol/L Tris-HCl, 50 mmol/L dithiothreitol (DTT), 1% IPG buffer (pH 3–10), 1% β-mercaptoethanol, and 2 mmol/L tributylphosphine. The lysed sample was absorbed into a Pharmacia Immobiline IPG DryStrip system (Amersham Pharmacia, Piscataway, NJ) using pH 3–10 nonlinear gradient strips. Proteins were equilibrated for 15 minutes in buffer (50 mmol/L Tris-HCl (pH 6.9), 2% SDS, 7 mol/L urea, and 10% glycerol) reduced in 0.4% DTT, and then alkylated in 5% iodoacetamide. The first dimensional focusing of proteins was performed for 48 hours. Separation along the second dimension was performed on a 9 to 18% SDS-PAGE gel and the proteins were visualized by staining with ammoniacal silver.
Layered Expression Scanning
The proteins from an ethanol-fixed, paraffin-embedded, whole-mount prostate tissue section were transferred through ten membranes using capillary action and one liter of transfer buffer (Bio-Rad, Hercules, CA), and captured onto a nitrocellulose membrane. After transfer, the nitrocellulose membrane was stained with 0.25% Coomassie blue (Pierce, Boston, MA). Additional information on this technology is available in the “Protocols” section of the website (see “Protocols in Development”) and reference 28.
RNA Analysis by Denaturing Agarose Gel Electrophoresis
Equivalent volumes of tissue from the different preparations were lysed in 400 μl of guanidinium isothiocyanate:3.2 μl of β-mercaptoethanol (Stratagene, La Jolla, CA) either immediately at 4°C, or after 20 minutes at 60°C or 2 hours at 80°C. The RNA was subsequently isolated by phenol chloroform extraction, and the samples were electrophoresed on a denaturing 1% agarose gel and visualized by ethidium bromide staining.
RNA Analysis by Reverse Transcriptase Polymerase Chain Reaction
Approximately 15,000 epithelial cells were microdissected from frozen or ethanol-fixed, paraffin-embedded prostate sections. The cells were lysed in 200 μl of guanidinium isothiocyanate:1.6 μl of β-mercaptoethanol at 60°C for 20 minutes. The RNA was isolated by phenol chloroform extraction with DNase (Gen Hunter, Nashville, TN) treatment followed by reverse transcription polymerase chain reaction (RT-PCR) of β-actin (220-bp product). Since such a small amount of RNA was present, the sample was analyzed using incorporation of [32P]dCTP (NEN Dupont, Boston, MA). The products were electrophoresed on a 6% denaturing acrylamide gel and visualized by autoradiography.
Analysis of DNA Quality by Agarose Gel Electrophoresis
The quality of total DNA from tissue that was either ethanol-fixed and paraffin-embedded, ethanol-fixed and polyester wax-embedded, or formalin-fixed and paraffin-embedded was compared. Equivalent volumes of tissue were placed into 1 ml of proteinase K (Sigma-Aldrich, St. Louis, MO) solution (20 mg/ml) and digested overnight at 55°C followed by boiling at 94°C for 7 minutes. The DNA was isolated by phenol chloroform extraction and a 10-μl aliquot of each sample was loaded onto a 1% agarose gel. DNA was visualized with ethidium bromide staining.
Analysis of DNA Quality by PCR
The quality of DNA from all of the tissue preparations was compared by PCR amplification. DNA was prepared as described in the “Analysis of Total DNA Quality” section. A 3 μl aliquot of each of the samples was amplified by 25 cycles of PCR using primers for microsatellite marker D17S926 (Research Genetics, Huntsville, AL). Comparisons were made using incorporation of [32P]dCTP since we have extensive experience using this method for amplification of microsatellite markers using microdissected samples. The product was electrophoresed on a 6% denaturing acrylamide gel (Life Technologies, Gaithersburg, MD) and visualized by autoradiography. All samples were analyzed in duplicate.
Results and Discussion
Histology
There are several published articles that assess histological characteristics, immunohistochemical staining, 14,21,22 and the recovery of DNA 11,23,24 and RNA 11,25,26 from tissues that have been processed using alcohol-based fixation. The aim of the present study was to use a systematic approach to more fully evaluate the biomolecular status of a large number of clinical tissue specimens processed through a non-formalin fixation method. As an initial screen, we evaluated the histology of tissues processed in the pathology department at Johns Hopkins University with several different fixatives to determine whether these methods were sufficient for clinical diagnosis. An overall ranking for the fixatives was determined by averaging the scores of each criterion (Table 1) ▶ . Based on these findings, 70% ethanol and two embedding compounds (standard paraffin and low-melt polyester wax) were selected for in-depth clinical and molecular analysis.
Table 1.
Fixatives and Overall Rank
| Fixatives | Overall rank* |
|---|---|
| Alcohol-based | |
| 70% ethanol | 2 |
| 95% ethanol | 3 |
| 70% ethanol: 100% methanol (3:1) | 1 |
| 95% ethanol: 100% methanol (3:1) | 8 |
| SafeFix | 7 |
| Streck molecular biology fixative | 6 |
| Aldehyde-based | |
| 10% neutral buffered formalin | 5 |
| Omnifix | 4 |
*Ranking was based on the evaluation of nuclear morphology, cellular morphology, tissue architecture, and staining characteristics.
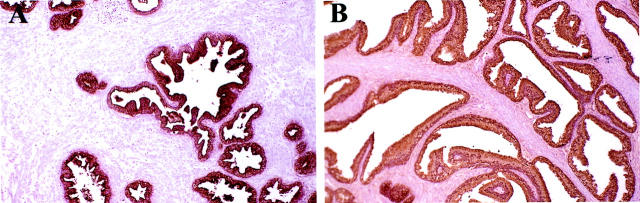
In the second phase of the study, fifty radical prostatectomies from patients with prostate cancer were fixed in 70% ethanol and studied at the National Cancer Institute over a two-year period. In five of the cases, the specimens were subdivided and processed through four separate methods (snap freezing; ethanol fixation, paraffin embedding; ethanol fixation, polyester embedding; formalin fixation, paraffin embedding) to permit direct comparison between the procedures. After completing the clinical and histological evaluation, three major conclusions were evident. First, there were no difficulties in making a clinical diagnosis in any of the cases. Second, ethanol fixation was consistently comparable to formalin fixation and superior to snap frozen for tissue architectural and staining qualities (Figure 1,A) ▶ . Ethanol fixation was also consistently superior to standard formalin fixation for visualizing nuclear detail of prostate epithelial cells (Figure 1B) ▶ , permitting more accurate grading of hyperplastic, premalignant, and tumor cell nuclei. Third, the polyester wax was technically difficult to use for embedding large tissue specimens (>1 cm), thus this method likely will be limited to use with small tissue biopsies only.
Figure 1.

Comparison of the histological quality of 5-μm-thick sections of prostate tissue stained with H&E. A: Normal prostate. Original magnification, ×100. A: Formalin-fixed, paraffin-embedded. B: 70% ethanol-fixed, paraffin-embedded. C: 70% ethanol-fixed, polyester wax-embedded. D: Frozen. Note the comparable staining quality and architecture for the formalin-fixed and ethanol-fixed tissues, which are superior to the frozen. B: High-grade prostatic intraepithelial neoplasia from ethanol-fixed, paraffin-embedded prostate tissue showing nuclear overlap and fine nuclear detail, including prominence of nucleoli. H&E stained, original magnification, ×200.
There are a few issues regarding 70% ethanol fixation that warrant special attention. First, it should be noted that 70% ethanol penetrates prostate tissue slower than 10% normal buffered formalin, thus it is essential that the tissue is grossly cut into thin sections no thicker than 3 to 5 mm. Second, even though immunohistochemical staining for PSA gave comparable results for both 70% ethanol and normal buffered formalin-fixed prostate tissue in our studies (Figure 2, A and B) ▶ , many commercially available antibodies have been selected for use on formalin-fixed tissue. Therefore, investigators need to assess the performance of their antibodies of interest on ethanol-fixed tissue before settling on a single method of fixation. Finally, even though 70% ethanol does not give the same level of nuclear detail as buffered formalin containing zinc (which is used in many pathology laboratories), the utility of tissues fixed in formalin containing zinc in molecular analysis is limited due to the adverse effects of heavy metals.
Figure 2.
Immunohistochemical stain of normal prostate tissue for PSA. Original magnification, ×100, hematoxylin counterstain. A: Ethanol-fixed, paraffin-embedded. B: Formalin-fixed, paraffin-embedded. Both preparations show comparable staining with positive glandular epithelial cells and negative stroma.
Protein Analysis
Total protein in the specimens was initially analyzed by one-dimensional gel electrophoresis (Figure 3,A) ▶ . To maximize protein recovery, each sample was subdivided and processed through one of three incubation steps (immediate analysis, 70°C for 2 hours, and 80°C for 2 hours). As can be seen in the figure, the general quality and quantity of the proteins in the ethanol-fixed samples is similar to that of snap-frozen material and superior to formalin-fixed tissue. Incubation of formalin-fixed tissue at 80°C for 2 hours resulted in somewhat improved protein recovery. However, this effect was variable from case to case and the protein yield was consistently less than from the ethanol-fixed and frozen samples.
Figure 3.

A: Protein recovery from prostate tissue sections. Samples were either snap frozen (Froz), ethanol-fixed and paraffin-embedded (Et/Para), ethanol-fixed and polyester wax-embedded (Et/PEW), or formalin-fixed and paraffin-embedded (Form/Para). B: Immunoblot for PSA. Approximately 35,000 prostate epithelial cells were microdissected from histological sections using LCM. Two separate microdissections from the ethanol-fixed, paraffin-embedded sample were performed to assess reproducibility.
We microdissected cells from histological prostate sections using LCM 27 and performed an immunoblot for PSA (Figure 3B) ▶ . A band representing PSA is visible in the ethanol-fixed, paraffin-embedded samples and is of similar intensity to the snap-frozen tissue. Studies in our group have now established that immunoblots can be used to measure the levels of many different proteins in ethanol- fixed samples, including several phosphoproteins.
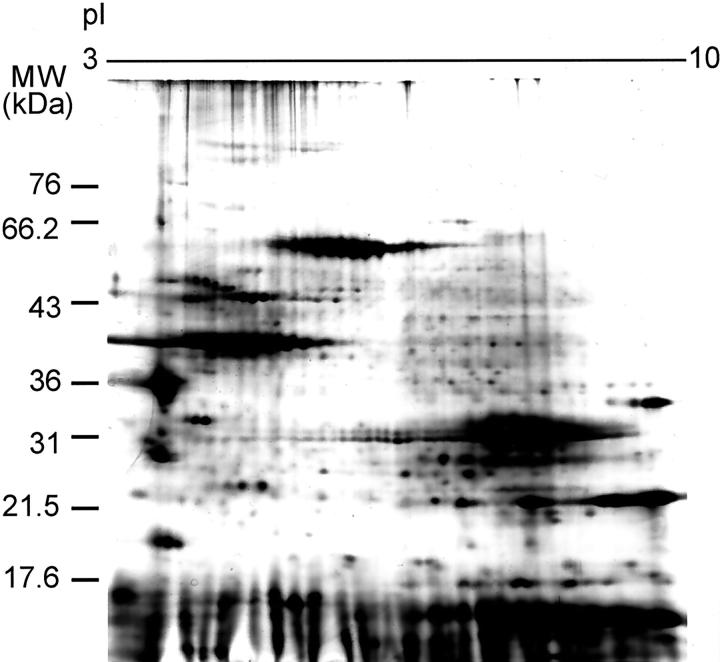
We also analyzed total protein using 2D-PAGE (Figure 4) ▶ . The proteins on the gel are from an ethanol-fixed, paraffin-embedded prostate and share 98% identity with a matched sample from the same patient that was snap-frozen (data not shown), indicating that the molecular weights and isoelectric points of the proteins are not significantly affected by the tissue processing method. However, in general, we did observe an approximate 50% decrease in the amount of protein that was observed by 2D-PAGE from the ethanol-fixed samples as compared to snap-frozen specimens.
Figure 4.
Two-dimensional PAGE analysis of ethanol-fixed, paraffin-embedded prostate tissue.
Layered expression scanning is a high-throughput array method that is under development at the NCI. 28 We were able to successfully transfer total protein from a transverse histological section of an ethanol-fixed, paraffin-embedded prostate specimen through 10 capture layers, indicating that a fully processed sample can be used for this new technology (Figure 5) ▶ . Taken together, these results indicate that ethanol-fixed tissue specimens can be successfully used for a variety of high-throughput proteomic technologies.
Figure 5.
Layered expression scanning using an ethanol-fixed, paraffin-embedded prostate which has been transferred onto nitrocellulose and stained with Coomassie blue. The labeled regions demonstrate that the protein staining pattern retains characteristic prostate structures.
RNA Analysis
We compared the recovery of total RNA from the ethanol-fixed, frozen and formalin-fixed tissue samples (Figure 6,A) ▶ . Each specimen was subdivided and processed through one of three incubation steps (4°C, 60°C, and 80°C) before RNA purification. Incubation of the frozen and ethanol-fixed samples at 60°C resulted in improved RNA yield. However, incubation of the RNA at 80°C or higher resulted in significant RNA hydrolysis and fragmentation. The sample that was snap frozen immediately after surgical resection provided high-quality RNA as judged by intact 28S and 18S ribosomal RNA bands. RNA could be recovered from ethanol-fixed tissue that was either paraffin- or polyester-embedded, although the quality was reduced, as observed in the loss of the 28S and 18S bands and the appearance of an RNA smear on the gel. However, the RNA was sufficient to perform a number of molecular techniques such as gene-specific RT-PCR (Figure 6B) ▶ and cDNA library production.
Figure 6.

A: Denaturing agarose gel of total RNA from prostate tissue sections that were either frozen, ethanol-fixed, polyester wax-embedded (EtOH/PEW), ethanol-fixed, paraffin-embedded (EtOH/Pf), or formalin-fixed, paraffin-embedded (Form/Pf). B: RT-PCR for actin. Approximately 15,000 epithelial cells were microdissected from frozen or ethanol-fixed, paraffin-embedded (EtOH/Pf) prostate sections. Actin cDNA and prostate RNA from cell culture are included as positive controls. The arrow indicates the 220-bp band corresponding to actin.
We have found that transcriptome amplification for cDNA microarray analysis is possible using ethanol-fixed and paraffin-embedded specimens. However, further experimentation is needed to more fully assess this method as the effect of RNA hydrolysis observed in the ethanol-fixed samples on subsequent array experiments is not yet clear. Certainly, this raises the concern that experimental artifact could be introduced into a study. For example, cDNA-based microarray analysis comparing samples with RNA of significantly differing quality could produce misleading results based on the length of the labeled cDNA and subsequent hybridization characteristics. Therefore, at present, our group focuses on intrapatient specimen comparisons (ie, normal versus tumor from the same tissue section) to “normalize” RNA quality and minimize this effect. 29,30 Continued efforts to refine the fixation and embedding methodology are yet needed to improve the RNA quality obtained from processed samples.
DNA Analysis
The DNA in the samples was assessed by gel electrophoresis and PCR amplification. Although DNA analysis is not a part of expression profiling per se, determination of the epigenetic events that occur during normal development and in disease processes will be valuable information that can be integrated with mRNA and protein data sets. 31 The DNA from the ethanol-fixed samples migrates on an agarose gel as a smear of fragments, ranging in size from several hundred base pairs (bp) to few kbp (Figure 7,A) ▶ . The quality of the DNA is superior to that recovered from formalin-fixed tissue. Experiments using PCR showed that DNA from the ethanol-fixed tissue consistently amplified more robustly than DNA from formalin-fixed tissue (as an example, see Figure 7B ▶ ).
Figure 7.

A: Analysis of total DNA quality in prostate tissue that was either ethanol-fixed and paraffin-embedded (Et/Para), ethanol-fixed and polyester wax-embedded (Et/PEW), or formalin-fixed and paraffin-embedded (Form/Para). Each sample was loaded onto a 1% agarose gel. DNA was visualized with ethidium bromide staining. B: PCR-based analysis of DNA quality in prostate tissue that was either snap frozen, ethanol-fixed and paraffin-embedded (Et/Para), ethanol-fixed and polyester wax-embedded (Et/PEW), or formalin-fixed and paraffin-embedded (Form/Para). The PCR product from amplification of microsatellite marker D17S926 was electrophoresed on a 6% denaturing acrylamide gel and visualized by autoradiography. All samples were analyzed in duplicate.
The better quality DNA in the ethanol-fixed samples has two important advantages. First, techniques that require relatively large fragments of DNA can be used. These approaches are difficult if not impossible to perform using DNA from formalin-fixed archival samples. Second, the number of successful PCR amplifications that can be performed per cell number is significantly increased. Thus, investigators can generate substantial data (eg, mutation analysis, gene promoter methylation status, allelic loss patterns, and single nucleotide polymorphism profiles) from limited amounts of tissue such as small biopsies or microdissected cells.
Future Directions
We are currently evaluating three additional tissue processing parameters. The first is the long-term stability of biomolecules in ethanol-fixed and embedded samples. To date, we have not detected significant quantitative or qualitative changes in nucleic acid or protein content after extended storage (>18 months). The second is a low-melt embedding compound or methodology that preserves proteins, RNA, and DNA in tissue specimens, but is also easy for histologists to use in a clinical setting. This will be particularly important for mRNA-based studies since we consistently observed increased RNA hydrolysis in the paraffin-embedded tissues as compared to the tissues embedded in low-melt polyester, likely due to the elevated temperature that occurs during the paraffin infiltration process. Finally, we are testing the use of reversible protein cross-linking agents during the tissue fixation process. This constricts movement of biomolecules (inhibiting RNase and proteinase activity), yet permits the DNA, mRNA, and proteins to be recovered for subsequent molecular studies after reversal of cross-linking. This may have particular utility for technologies such as layered expression scanning that are designed to permit investigators to capture and measure individual proteins as well as their in vivo binding partners.
In summary, using a general strategy for evaluating clinical tissue specimens, we have found that ethanol fixation and paraffin embedding of clinical tissue specimens is a useful method for molecular profiling studies. This approach allows investigators to perform high-throughput molecular analyses on all of the cell populations in a sample, including those that are required for clinical diagnosis. Continued improvement of tissue processing methodologies will be a critical step toward ultimately determining the complete molecular anatomy of normal and diseased human cell types.
Footnotes
Address reprints requests J.W. Gillespie, Pathogenetics Unit, Laboratory of Pathology, National Cancer Institute, Rm. 2A33, Bldg. 10, 9000 Rockville Pike, Bethesda, MD 20892.
Carolyn J.M. Best, Verena E. Bichsel, Kristina A. Cole, and Susan F. Greenhut contributed substantially to this work.
Research was performed at the Pathogenetics Unit, Laboratory of Pathology, National Cancer Institute, Rm. 2C500, Bldg. 10, 9000 Rockville Pike, Bethesda, MD 20892.
References
- 1.Schena M, Shalon D, Davis RW, Brown PO: Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 1995, 270:467-469 [DOI] [PubMed] [Google Scholar]
- 2.Velculescu V, Zhang L, Vogelstein B, Kinzler KW: Serial analysis of gene expression. Science 1995, 270:484-487 [DOI] [PubMed] [Google Scholar]
- 3.Anderson N, Anderson NG: Proteome and proteomics: new technologies, new concepts, and new words. Electrophoresis 1998, 19:1853-1861 [DOI] [PubMed] [Google Scholar]
- 4.Duggan D, Bittner M, Chen Y, Meltzer P, Trent JM: Expression profiling using cDNA microarrays. Nat Genet 1999, 21:10-14 [DOI] [PubMed] [Google Scholar]
- 5.Lipshutz R, Fodor SPA, Gingeras TR, Lockhart DJ: High density synthetic oligonucleotide arrays. Nat Genet 1999, 21:20-24 [DOI] [PubMed] [Google Scholar]
- 6.Lander E: Array of hope. Nat Genet 1999, 21:3-4 [DOI] [PubMed] [Google Scholar]
- 7.Emmert-Buck MR, Gillespie JW, Paweletz CP, Ornstein DK, Basrur V, Appella E, Wang Q, Huang J, Hu N, Taylor P, Petricoin EF: A strategic approach for proteomic analysis of human tumors. Mol Carcinog 1999, 27:158-165 [PubMed] [Google Scholar]
- 8.DeRisi J, Penland L, Brown PO, Bittner ML, Meltzer PS, Ray M, Chen Y, Su YA, Trent JM: Use of a cDNA microarray to analyze gene expression patterns in human cancer. Nat Genet 1996, 14:457-460 [DOI] [PubMed] [Google Scholar]
- 9.Celis A, Rasmussen HH, Celis P, Bodil B, Lauridsen JB, Ratz G, Hein B, Ostergaard M, Wolf H, Orntoft T, Celis J: Short-term culturing of low-grade superficial bladder transitional cell carcinomas leads to changes in the expression levels of several proteins involved in key cellular activities. Electrophoresis 1999, 20:355-361 [DOI] [PubMed] [Google Scholar]
- 10.Ornstein DK, Gillespie JW, Paweletz CP, Duray PH, Herring J, Vocke CD, Topalian SL, Bostwick DG, Linehan WM, Petricoin EF, Emmert-Buck MR: Proteomic analysis of laser capture microdissected human prostate cancer and in vitro prostate cell lines. Electrophoresis 2000, 21:2235-2242 [DOI] [PubMed] [Google Scholar]
- 11.Ben-Ezra J, Johnson DA, Rossi J, Cook N, Wu A: Effect of fixation on the amplification of nucleic acids from paraffin-embedded material by the polymerase chain reaction. J Histochem Cytochem 1991, 39:351-354 [DOI] [PubMed] [Google Scholar]
- 12.Foss R, Guha-Thakurta N, Conran RM, Gutman P: Effects of fixative and fixation time on the extraction and polymerase chain reaction amplification of RNA from paraffin-embedded tissue: comparison of two housekeeping gene mRNA controls. Diagn Mol Pathol 1994, 3:148-155 [DOI] [PubMed] [Google Scholar]
- 13.Klimecki W, Futscher BW, Dalton WS: Effects of ethanol and paraformaldehyde on RNA yield and quality. Biotechniques 1994, 16:1021-1023 [PubMed] [Google Scholar]
- 14.Hudock J, Hanau CA, Christen R, Bibbo M: Expression of estrogen and progesterone receptors in cytologic specimens using various fixatives. Diagn Cytopathol 1996, 15:78-83 [DOI] [PubMed] [Google Scholar]
- 15.Monger LJ, Nagabhushan M, Pretlow TG, Pretlow TP: A novel approach to the characterization of whole prostate pathology in glycol methacrylate. Am J Pathol 1994, 145:54-60 [PMC free article] [PubMed] [Google Scholar]
- 16.Finke J, Fritzen R, Ternes P, Lange W, Dolken G: An improved strategy and a useful housekeeping gene for RNA analysis from formalin-fixed, paraffin-embedded tissues by PCR. Biotechniques 1993, 14:448-453 [PubMed] [Google Scholar]
- 17.Mies C: Molecular biological analysis of paraffin-embedded tissues. Hum Pathol 1994, 25:555-560 [DOI] [PubMed] [Google Scholar]
- 18.Oke B, Suarez-Quian CA: Localization of secretory, membrane-associated, and cytoskeletal proteins in rat testis using an improved immunocytochemical protocol that employs polyester wax. Biol Reprod 1993, 48:621-631 [DOI] [PubMed] [Google Scholar]
- 19.Kraft A, Duncan BW, Bijwaard KE, Taubenberger JK, Lichy JH: Optimization of the isolation and amplification of RNA from formalin-fixed, paraffin-embedded tissue: the Armed Forces Institute of Pathology experience and literature review. Mol Diagn 1997, 2:217-230 [DOI] [PubMed] [Google Scholar]
- 20.Masuda N, Ohnishi T, Kawamoto S, Monden M, Okubo K: Analysis of chemical modification of RNA from formalin-fixed samples and optimization of molecular biology applications for such samples. Nucleic Acids Res 1999, 27:4436-4443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Battifora H, Kopinski M: The influence of protease digestion and duration of fixation on the immunostaining of keratins: a comparison of formalin and ethanol fixation. J Histochem Cytochem 1986, 34:1095-1100 [DOI] [PubMed] [Google Scholar]
- 22.Perez-Martin C, Garcia-Fernandez A, Ferreras-Estrada MC, Escudero-Diez A, Espinosa-Alvarez J, Garcias-Iglesias MJ: Optimization of the immunohistochemical demonstration of keratins in paraffin wax-embedded mouse skin. J Comp Pathol 1998, 119:177-181 [DOI] [PubMed] [Google Scholar]
- 23.Giannella C, Zito FA, Colonna F, Paradiso A, Marzullo F, Alaibac M, Schittulli F: Comparison of formalin, ethanol, and histochoice fixation on the PCR amplification from paraffin-embedded breast cancer tissue. Eur J Clin Chem Clin Biochem 1997, 35:633-635 [PubMed] [Google Scholar]
- 24.Alaibac M, Folitico R, Giannella C, Paradiso A, Labriola A, Marzullo F: The effect of fixation type on DNA extracted from paraffin-embedded tissue for PCR studies in dermatopathology. Dermatology 1997, 195:105-107 [DOI] [PubMed] [Google Scholar]
- 25.Tyrrell L, Elias J, Longley J: Detection of specific mRNAs in routinely processed dermatopathology specimens. Am J Dermatopathol 1995, 17:476-483 [DOI] [PubMed] [Google Scholar]
- 26.Goldsworthy SM, Stockton PS, Trempus CS, Foley JF, Maronpot RR: Effects of fixation on RNA extraction and amplification from laser microdissected tissue. Mol Carcinog 1999, 25:86-91 [PubMed] [Google Scholar]
- 27.Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zhang Z, Goldstein SR, Weiss RA, Liotta LA: Laser capture microdissection. Science 1996, 274:998-1001 [DOI] [PubMed] [Google Scholar]
- 28.Englert C, Baibakov GV, Emmert-Buck MR: Layered expression scanning: rapid molecular profiling of tumor samples. Cancer Res 2000, 60:1526-1530 [PubMed] [Google Scholar]
- 29.Cole K, Krizman DB, Emmert-Buck MR: The genetics of cancer: a 3D model. Nat Genet 1999, 21:38-41 [DOI] [PubMed] [Google Scholar]
- 30.Emmert-Buck MR, Strausberg RL, Krizman DB, Bonaldo MF, Bonner RF, Buetow KH, Chuaqui RF, Cole KA, Duray PH, Englert CR, Gillespie JW, Greenhut S, Grouse L, Hillier LW, Katz KS, Klausner RD, Kuznetzov V, Lash AE, Lennon G, Linehan WM, Liotta LA, Marra MA, Munson PJ, Ornstein DK, Prabhu VV, Prange C, Schuler GD, Soares MB, Tolstoshev CM, Vocke CD, Waterston RH: Molecular profiling of clinical tissue specimens: feasibility and applications. Am J Pathol 2000, 156:1109-1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goelz S, Hamilton SR, Vogelstein B: Purification of DNA from formaldehyde-fixed and paraffin-embedded human tissue. Biochem Biophys Res Commun 1985, 130:118-126 [DOI] [PubMed] [Google Scholar]