Abstract
Oxidative tissue damage has been shown to be associated with carcinogenesis. In human cancers p16INK4A is one of the most frequently mutated tumor suppressor genes. The present study used the ferric nitrilotriacetate (Fe-NTA)-induced rat renal carcinogenesis model to determine whether oxidative damage can cause specific allelic loss of p16 INK4A. By the use of fluorescent in situ hybridization in combination with imprint cytology at single-cell resolution, we found that the number of renal tubular cells with aneuploidy (1 or 3 signals) at the p16INK4A locus was significantly and specifically increased (1 week, 37.2 ± 2.3%; 3 weeks, 37.8 ± 1.3% vs control, 22.5 ± 1.9%; mean ± SE, N = 8; P < 0.001 and P < 0.0001, respectively) after repeated intraperitoneal administration of 5 to10 mg of iron/kg in the form of Fe-NTA for 3 weeks. No increase in aneuploidy was observed at the loci of either the p53 or vhl tumor suppressor gene. Furthermore, the increase in the cells with 3 signals was followed by a continuous increase in those with 1 signal. Therefore, the p16 INK4A locus is specifically vulnerable to oxidative damage, leading to its allelic loss within weeks, presumably due to a deficiency in the replication of both the alleles.
Recently, studying the activation of oncogenes and/or inactivation of tumor suppressor genes by a variety of mutations has been a primary approach to elucidating the biological mechanisms of human neoplasia. 1 The tumor suppressor gene p16 (INK4A/MTS-1/CDKN2A), first described in 1994 2-4 is a major target in carcinogenesis: the frequency of p16 INK4A mutation in tumors is rivaled only by that of mutations of the p53 tumor suppressor gene. p16 INK4A is unique in that the major form of mutation is a homozygous deletion, 5 and that it not only inhibits the phosphorylation of retinoblastoma protein through inhibition of cyclin-dependent kinases 4 and 6 as a gatekeeper in the cell cycle, but also acts upstream of the p53 pathway via alternative splicing as p19 (p14)ARF, resulting in suppression of the expression of MDM2 protein, which promotes degradation of p53 protein. 6,7
Inactivation of an allele by intragenic mutation of tumor suppressor genes results in a genomic environment in which the probability of early inactivation of the remaining allele is increased, as demonstrated in various familial cancer syndromes including hereditary retinoblastoma (rb), the Li-Fraumeni syndrome (p53), von Hippel-Lindau syndrome (vhl), and familial melanoma kindreds (p16 INK4A). 1 On the other hand, it was recently shown that allelic loss is often the first hit in the biallelic inactivation of p53 and DPC4 genes during sporadic pancreatic carcinogenesis. 8,9 Despite these findings, the events responsible for allelic loss in vivo are still largely unknown.
The hydroxyl radical is the most reactive known species in biological systems and can induce DNA double-strand breaks. 10 Furthermore, free radical-associated tissue damage resulting from irradiation, 11 chronic inflammation, 12 or overload of transition metals 13 has been shown to be associated with carcinogenesis. We previously established a rat model of renal cell carcinoma induced by ferric nitrilotriacetate (Fe-NTA) 14 as a model of free radical-induced cancer. In this model, oxidative molecular damage such as formation of aldehydes and oxidative DNA base modifications has been well characterized. 15-17 Furthermore, one of the major target genes in this model has recently been identified as p16INK4A, and allelic loss of p16INK4A was observed in 38.5% of the induced renal cell carcinoma. 18 In the present study we attempted to determine when the allelic loss occurs during carcinogenesis and whether this loss is specific to the p16INK4A locus.
Materials and Methods
Chemicals
Ferric nitrate enneahydrate, sodium hydrogen carbonate, sodium citrate monohydrate, ammonium acetate, formamide, dextran sulfate, salmon sperm DNA, and Escherichia coli tRNA were from Wako (Osaka, Japan); nitrilotriacetic acid disodium salt was from Nacalai Tesque, Inc. (Kyoto, Japan). Block Ace was from Dai-nihon-seiyaku (Osaka, Japan). All of the chemicals used were of analytical quality; deionized water was used throughout. Fe-NTA solution was prepared as previously described. 15
Animals and Imprint Cytology
Five-week-old specific-pathogen-free male Wistar rats were purchased from Shizuoka Laboratory Animal Center, Shizuoka, Japan. They were kept in stainless steel cages in a temperature-controlled room (22 to 24°C) with a light/dark cycle of 12 hours each. This experiment was approved by the Animal Research Committee, Graduate School of Medicine, Kyoto University, Kyoto, Japan. Twenty-four animals were divided into three groups of eight animals each: an untreated control group, a one-week treatment group, and a three-week treatment group. An established protocol for Fe-NTA-induced renal carcinogenesis 19 was used. Briefly, 5 mg of iron/kg in the form of Fe-NTA was injected intraperitoneally (i.p.) on days 1 to 3, 10 mg of iron/kg on days 4 and 5, and 10 mg of iron/kg five days a week after a two-day break. The animals were killed 48 hours after the final administration of Fe-NTA. Both kidneys were removed immediately. One was used for imprint cytology as described previously. 20 After fixation in 95% ethanol at 4°C, each glass slide was air-dried and then stored at −80°C until use. The other was fixed in 10% neutral formalin solution for histological examination with hematoxylin and eosin staining.
Screening of Genomic Library for Fluorescent in Situ Hybridization Probes
A λ DASH II rat genomic library (Stratagene, La Jolla, CA) was screened according to the manufacturer’s instructions to obtain phage clones that contained the coding sequences of the p16INK4A, p53, or vhl tumor suppressor genes. Probes used for library screening were obtained by PCR amplification, as summarized in Table 1A ▶ . 21-23 cDNA was obtained from the kidney of a male Wistar rat by RNA extraction using a modified acid guanidium/phenol/chloroform method (Isogen, Nippon Gene, Tokyo, Japan), poly(A)-rich RNA isolation with oligo(dT)-latex beads (Nippon Roche, Tokyo, Japan), and reverse transcription (First-strand cDNA synthesis kit, Amersham Pharmacia Biotech, Tokyo, Japan), and was used as a substrate for PCR amplification. The probes were labeled with α-[32P]dCTP by random priming (Megaprime, Amersham Pharmacia Biotech), and nylon membranes (Biodyne B, Nihon Pall, Tokyo, Japan) bearing the cloned phage DNA of the genome library described above were hybridized with the probes and then washed under stringent condition (60°C, 0.1 × standard saline citrate (SSC), 0.1% sodium dodecyl sulfate (SDS)). DNA of each phage clone was extracted using a standard procedure, 24 and analyzed by agarose gel electrophoresis after digestion with appropriate restriction enzymes (Takara, Shiga, Japan). Complete or partial sequencing of exons was performed after PCR amplification using phage DNA as a substrate and subsequent subcloning (TA cloning, Invitrogen, Groningen, the Netherlands) for the purpose of eliminating phage clones of p53 pseudogenes 25-27 or vhl pseudogenes. 28 So far no pseudogenes have been reported for p16 INK4A. The most appropriate clone was selected for each tumor suppressor gene. The size of the inserts in the phage clones and the conditions for PCR amplification are summarized in Table 1B ▶ . The exon 1β region (p19ARF) was not included in the fluorescent in situ hybridization (FISH) probe for p16INK4A based on PCR analysis by the use of rat kidney cDNA as a positive control (data not shown). Sequence analyses revealed a complete match with GenBank data in each exonic region.
Table 1.
Probes for Library Screening and FISH
| A. PCR primers and conditions for production of probes for library screening | ||||
|---|---|---|---|---|
| Gene | Primer | Annealing temperature (cycle) | Product size | GenBank accession no. |
| p16INK4A | F-ATCTCCGAGAGGAAGGCGAACTCG | 57°C (35) | 667 bp | Swafford et al 21 L81167 |
| R-TCTGTCCCTCCCTCCCTCTGCTAAC | ||||
| p53 | F-TCACAGTCGGATATGAGCATCG | 58°C (35) | 1519 bp | Hulla et al 22 L07781 |
| R-GCTTTGCAGAGTGGAGGAAATG | ||||
| vhl | F-AACGAGCGTCCGGTTCCAAT | 58°C (35) | 622 bp | Kikuchi et al 23 S80345 |
| R-TCAATTTCAGACCATCAAGG | ||||
| B. Analysis of FISH probes | |||||
|---|---|---|---|---|---|
| Gene | Size of insert | Exon | Primer | Annealing temperature (no. of cycles) | Product size |
| p16INK4A | ∼16 kb | 1α | F-TCCGAGAGGAAGGCGAACTC | 55°C (35) | 160 bp |
| R-GGGTACGACCGAAAGTGTTCG | |||||
| 2 | F-AACGTCAAAGTGGCAGCTCTCCTG | 55°C (35) | 237 bp | ||
| R-GAGTAGATACCGCAAATACCGCAC | |||||
| (p19ARF) | 1β* | F1-GAGCATGGGTCGCAGGTTC | 55°C (35) | 193 bp | |
| F2-CGTGGTCACTGTGAGGATTCG | 55°C (35) | 175 bp | |||
| R-TGGTCCAGGATGTGGGTGC | |||||
| p53 | ∼20 kb | 2–4 | F-ATGGAGGATTCACAGTCGGATAT | 56°C (35) | 719 bp (intron 2, 260 bp; intron 3, 90 bp) |
| R-CGTGCACATAACAGACTTGG | |||||
| 9–10 | F-ATCCGTGGGCGTGAGCGCTTC | 56°C (35) | 889 bp (intron 9, 700 bp) | ||
| R-TCAGTCTGAGTCAGGCCCCA | |||||
| vhl | ∼18 kb | 1 | F-GCGTCGTGCTGCCTTTGTGG | 58°C (35) | 178 bp |
| R-tgagtagggacctggtgctc† (intron 1) | |||||
| 2 | F-actgctgttgccttgctcag (intron 1) | 55°C (35) | 302 bp | ||
| R-tcctcagccccaaggtctta (intron 2) | |||||
| 3 | F-tgactggagcctgcctcaga (intron 2) | 55°C (35) | 284 bp | ||
| R-TCAATTTCAGACCATCAAGG | |||||
FISH Analysis
Extracted DNA from each phage clone was used for FISH analysis by labeling with biotin-16-dUTP via nick translation (Roche, Tokyo, Japan). The size of the labeled probe was confirmed by agarose gel electrophoresis to be 300 to 500 base pairs. After ethanol precipitation (20 μl of products, 2.4 μl of 8 mol/L ammonium acetate, 2.0 μl of 10 μg/μl salmon sperm DNA, 2.0 μl of 10 μg/ml E. coli tRNA, and 79 μl of ethanol) at −80°C for 30 minutes, the labeled probe was dissolved in ULTRAhyb hybridization buffer (Ambion, Austin, TX). Mouse Cot-1 DNA was added (20 μg/ml; Life Technologies, Tokyo, Japan) to the hybridization mixture to minimize the background signal. Stock slides were completely air dried followed by 5 minutes of microwave treatment in 10 mmol/L citrate buffer, pH 6.0, and proteinase K treatment (4 μg/ml phosphate-buffered saline) at room temperature. Hybridization was done at 35°C overnight. After washing with 2 × SSC at 37°C and at room temperature sequentially, the tyramide signal amplification (TSA) system (NEN Life Science Products, Inc., Boston, MA) was applied to amplify signals according to the manufacturer’s instructions. Briefly, blocking solution was applied after washing, followed by avidin-biotin-peroxidase complex (Vector Laboratories, Burlingame, CA) and biotin tyramide, sequentially. Finally, avidin-FITC (Vector; 1:100 dilution with phosphate-buffered saline containing 1% Block Ace) and then propidium iodide was applied for nuclear counterstaining. The images were analyzed with a confocal laser microscope (Fluoview, Olympus, Osaka, Japan). Sequential sectioning at 0.5 μm along the Z-axis was used when necessary to confirm the number of signals. When DNA of an empty phage clone was used as a probe, no signal was obtained under the experimental conditions used. The number of cells with 1 to 4 signals was counted on at least 100 cells obtained from each animal. Cells showing signals of 5 or more were not included since the whole nuclear area showed ambiguous positivity in most of the cases.
Statistics
The unpaired Student’s t-test and one-way analysis of variance was used for statistical analysis. P values < 0.05 were considered statistically significant.
Results
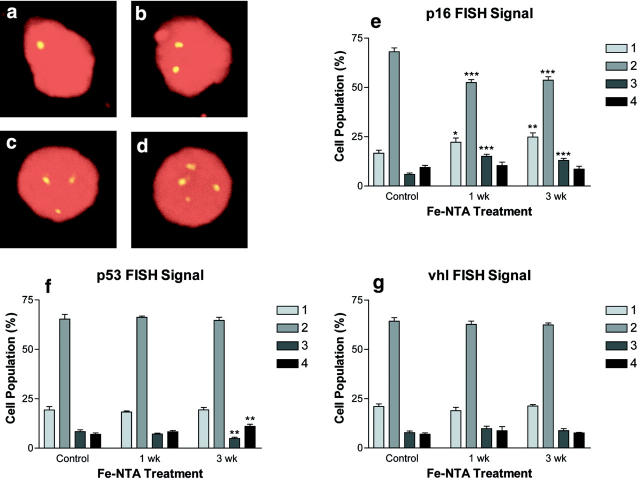
Histological analysis of the kidney after 1 or 3 weeks of treatment of Fe-NTA showed no necrosis, but revealed many regenerative tubular cells with an enlarged nucleus and prominent nucleoli as previously reported. 16 Representative FISH signals at single-cell resolution are shown in Figure 1,a–d ▶ . We found that the fraction of renal tubular cells with aneuploidy (1 signal or 3 signals) at the locus of p16 INK4A was significantly increased after Fe-NTA treatment for 1 or 3 weeks, whereas no increase in aneuploidy was observed at the loci of either the p53 or vhl tumor suppressor gene (Figure 1, e–g) ▶ . When fractions with 1 signal and 3 signals were combined, the difference was more significant (1 week, 37.2 ± 2.3%; 3 weeks, 37.8 ± 1.3% vs control 22.5 ± 1.9%; mean ± SE, n = 8; P = 0.0003 and P = 0.00002, respectively).
Figure 1.
FISH analysis in imprint cytology from rat kidney after a ferric nitrilotriacetate-induced renal carcinogenesis protocol. More than 800 cells from eight animals for each group were classified according to the signal number (FITC-labeled, propidium iodide nuclear counterstaining). Refer to Materials and Methods for details. a, 1 signal; b, 2 signals; c, 3 signals; d, 4 signals; e–g, cell population (%) with each copy number (1 to 4, left to right) of allele for p16INK4A, p53, and vhl tumor suppressor genes (mean ± SE). *, P < 0.05; **, P < 0.01; ***, P < 0.001 vs control group, Student’s t-test.
A one-way analysis of variance was also performed. The analysis was done among the three groups with different treatments (control, 1-week treatment, and 3-week treatment) from the viewpoint of signal number. The P values of p16 INK4A for each signal number (1, 2, 3, and 4 signals) were 0.022, <0.0001, <0.0001, and 0.67, respectively. The P values of p53 for each signal number (1, 2, 3, and 4 signals) was 0.77, 0.80, 0.0079, and 0.010, respectively. The P values of vhl for each signal number (1, 2, 3, and 4 signals) was 0.40, 0.65, 0.44, and 0.66, respectively.
Discussion
Studying the activation of oncogenes and/or inactivation of tumor suppressor genes by a variety of mutations has been a major approach to elucidating the biological mechanisms of human neoplasia. 1 Despite the insights obtained thereby, there is little data available on the events responsible for the mutations in vivo. Using the Fe-NTA-induced rat renal carcinogenesis model, we have focused on understanding free radical-induced carcinogenesis, and recently found that the p16 INK4A tumor suppressor gene is one of the major target genes in this model. Since allelic loss is observed frequently, 18 we undertook to determine when this event occurs during carcinogenesis and whether it is specific for the p16 INK4A loci. We chose p53 and vhl tumor suppressor genes as control genes since we previously analyzed mutations of these two genes in Fe-NTA-induced renal cancer; only one case of G to T transversion mutation in p53 (exon 7) was observed out of 12 cases of Fe-NTA-induced renal cell carcinoma, 19 and no genetic change of vhl was observed in 34 cases. 29
We obtained phage clones as FISH probes by screening a rat genomic library. The majority of exons in each gene were sequenced and pseudogenes were eliminated. To reproducibly perform FISH analyses at the single-cell level via imprint cytology with touch preparation, the use of mouse Cot-1 DNA at hybridization and tyramide signal amplification (TSA) were essential. The use of Cot-1 DNA decreased the background staining, while the use of TSA increased the sensitivity. The analysis we have undertaken in the present study is technically more difficult than that of tumor cells since individual cells need to be evaluated, and thus, optimal conditions are required. Even in untreated control animals, 17 to 20% of cells showed one signal and 6 to 10% showed 3 signals. We believe on the basis of repeated experiments that this is due to either insufficient nuclear transfer to glass slides in the touch preparation or insufficient hybridization in the deeper part of the nuclei. For this reason we did not perform double staining with the two probes, which could further complicate this issue.
The results clearly indicate that the population of cells with allelic loss of the p16 INK4A locus (aneuploidy) is significantly increased within weeks (Figure 1) ▶ . In this study we defined signals of 1 and 3 as representing aneuploidy. Analysis of variance analyses also revealed significant changes in the fraction of 1, 2, and 3 signals for p16 INK4A among each treatment group of the animals. The population with 3 signals reached a plateau at 1 week whereas the population with one signal continuously increased until 3 weeks. This may indicate that allelic loss is induced by an error of replication at one of the alleles, possibly by DNA double-strand breaks, which ultimately leads to allelic loss in one of the cells after mitosis. Fe-NTA can induce DNA double-strand breaks via a Fenton-like reaction, as well as single-strand breaks 10 and oxidative DNA base modifications. 16 We previously showed that the number of double-strand breaks catalyzed by Fe-NTA is proportional to that of single-strand breaks in a simple supercoiled plasmid model. 10 The decrease in the 3-signal population and concomitant increase in the 4-signal population at the p53 locus may mean that these loci are replicated earlier than the p16 INK4A and vhl loci after Fe-NTA treatment for 3 weeks. Increased aneuploidy at the p16INK4A locus was not observed in the liver, a non-target organ, in this carcinogenesis model (data not shown).
It is notable that human mesothelioma, which is closely associated with iron-mediated oxidative damage, 13,30 has an incidence of 22 to 73% homozygous deletion of p16 INK4A in primary tumors. 31-33 This, in combination with our data, strongly suggests that Fenton chemistry-associated DNA damage is one of the causes of p16 INK4A deletion. The most frequent mutation induced by the hydroxyl radical in plasmid vectors is G to T transversions. 34 However, in this kind of simple plasmid system, it is not possible to detect deletion of a large fragment (>20 kb) such as seen in our rat renal cancer model. Furthermore, free radical damage is not so simple as the direct attack of hydroxyl radicals on DNA since secondary reactions associated with a variety of aldehydes may be involved. 17 Thus, further studies are necessary to determine whether this concept is applicable to other types of oxidative damage such as inflammation and radiation.
After we found that p16 INK4A is one of the target genes in Fe-NTA-induced renal cancer, the next question has been whether this is due to natural selection or targeted disruption. The present findings also demonstrate the presence of “fragile sites in the genome to free radical attack” since no increase in aneuploidy was observed at the p53 and vhl loci. Though we cannot completely rule out the possibility of the selection mechanism, at present we believe that hemizygous deletion of the tumor suppressor genes investigated does not alter the expression of these genes or the phenotype. This concept is supported by a recent finding that formation of 8-oxoguanine, an oxidatively modified DNA base, is different among three different gene loci after Fe-NTA treatment. 35 In further studies it will be important to determine specific chromosomal or more localized areas vulnerable to free radical attack in the genome.
On the other hand, our results may help explain the recent findings that homozygous deletion of p16INK4A but not methylation of the p16INK4A promoter is a genetic target in the pathogenesis of smoking-induced human lung cancer 36 and that a higher incidence of p16 INK4A deletion is observed in cultured cell lines than in primary tumors. 37 In both of these conditions, cells have been exposed to unusually high levels of reactive oxygen species.
In conclusion, oxidative tissue damage can cause specific allelic loss of the p16 INK4A tumor suppressor gene within a few weeks in animal experiments. Therefore, p16INK4A should be reevaluated as a potential target for the prevention and therapy of free radical-associated cancer.
Acknowledgments
We thank Dr. James E. Strickland (Rockville, MD) for critically reviewing the manuscript and Ms. Yoshie Fujiwara for excellent technical assistance.
Footnotes
Address reprint requests to Shinya Toyokuni, M.D./Ph.D., Department of Pathology and Biology of Diseases, Graduate School of Medicine, Kyoto University, Yoshida-Konoe-cho, Sakyo-ku, Kyoto 606-8501, Japan. E-mail: toyokuni@path1.med.kyoto-u.ac.jp.
Supported in part by a Grant-in-Aid from the Ministry of Education, Science, Sports, and Culture of Japan, a Grant-in-Aid for Cancer Research from the Ministry of Health, Labour and Welfare of Japan, and a grant from the Program for Promotion of Basic Research Activities for Innovative Bioscience (PROBRAIN).
References
- 1.Vogelstein B, Kinzler KW: The genetic basis of human cancer. 1998. McGraw-Hill, New York
- 2.Nobori T, Miura K, Wu D, Lois A, Takabayashi K, Carson D: Deletions of the cyclin-dependent kinase-4 inhibitor gene in multiple human cancers. Nature 1994, 368:753-756 [DOI] [PubMed] [Google Scholar]
- 3.Kamb A, Gruis NA, Weaver-Feldhaus J, Liu Q, Harshman K, Tavtigian SV, Stockert E, Day RS, Johnson BE, Skolnick MH: A cell cycle regulator potentially involved in genesis of many tumor types. Science 1994, 264:436-440 [DOI] [PubMed] [Google Scholar]
- 4.Cairns P, Mao L, Merlo A, Lee DJ, Schwab D, Eby Y, Tokino K, van der Riet P, Blaugrund JE, Sidransky D: Rates of p16 (MTS1) mutations in primary tumors with 9p loss. Science 1994, 265:415-417 [DOI] [PubMed] [Google Scholar]
- 5.Liggett WH, Jr, Sidransky D: Role of the p16 tumor suppressor gene in cancer. J Clin Oncol 1998, 16:1197-1206 [DOI] [PubMed] [Google Scholar]
- 6.Kamijo T, Weber JD, Zambetti G, Zindy F, Roussel MF, Sherr CJ: Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc Natl Acad Sci USA 1998, 95:8292-8297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Honda R, Yasuda H: Association of p19 (ARF) with Mdm2 inhibits ubiquitin ligase activity of Mdm2 for tumor suppressor p53. EMBO J 1999, 18:22-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luttges J, Galehdari H, Brocker V, Schwarte-Waldhoff I, Henne-Bruns D, Kloppel G, Schmiegel W, Hahn S: Allelic loss is often the first hit in the biallelic inactivation of the p53 and DPC4 genes during pancreatic carcinogenesis. Am J Pathol 2001, 158:1677-1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilentz R, Argani P, Hruban R: Loss of heterozygosity or intragenic mutation, which comes first? Am J Pathol 2001, 158:1561-1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toyokuni S, Sagripanti J-L: DNA single- and double-strand breaks produced by ferric nitrilotriacetate in relation to renal tubular carcinogenesis. Carcinogenesis 1993, 14:223-227 [DOI] [PubMed] [Google Scholar]
- 11.Boice JJ, Lubin J: Occupational and environmental radiation and cancer. Cancer Causes Control 1997, 8:309-322 [DOI] [PubMed] [Google Scholar]
- 12.Weitzman SA, Gordon LI: Inflammation and cancer: role of phagocyte-generated oxidants in carcinogenesis. Blood 1990, 76:655-663 [PubMed] [Google Scholar]
- 13.Toyokuni S: Iron-induced carcinogenesis: the role of redox regulation. Free Radic Biol Med 1996, 20:553-566 [DOI] [PubMed] [Google Scholar]
- 14.Ebina Y, Okada S, Hamazaki S, Ogino F, Li JL, Midorikawa O: Nephrotoxicity and renal cell carcinoma after use of iron- and aluminum-nitrilotriacetate complexes in rats. J Natl Cancer Inst 1986, 76:107-113 [PubMed] [Google Scholar]
- 15.Toyokuni S, Uchida K, Okamoto K, Hattori-Nakakuki Y, Hiai H, Stadtman ER: Formation of 4-hydroxy-2-nonenal-modified proteins in the renal proximal tubules of rats treated with a renal carcinogen, ferric nitrilotriacetate. Proc Natl Acad Sci USA 1994, 91:2616-2620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toyokuni S, Mori T, Dizdaroglu M: DNA base modifications in renal chromatin of Wistar rats treated with a renal carcinogen, ferric nitrilotriacetate. Int J Cancer 1994, 57:123-128 [DOI] [PubMed] [Google Scholar]
- 17.Toyokuni S, Luo XP, Tanaka T, Uchida K, Hiai H, Lehotay DC: Induction of a wide range of C2–12 aldehydes and C7–12 acyloins in the kidney of Wistar rats after treatment with a renal carcinogen, ferric nitrilotriacetate. Free Radic Biol Med 1997, 22:1019-1027 [DOI] [PubMed] [Google Scholar]
- 18.Tanaka T, Iwasa Y, Kondo S, Hiai H, Toyokuni S: High incidence of allelic loss on chromosome 5 and inactivation of p15 INK4B and p16 INK4A tumor suppressor genes in oxystress-induced renal cell carcinoma of rats. Oncogene 1999, 18:3793-3797 [DOI] [PubMed] [Google Scholar]
- 19.Nishiyama Y, Suwa H, Okamoto K, Fukumoto M, Hiai H, Toyokuni S: Low incidence of point mutations in H-, K-, and N-ras oncogenes and p53 tumor suppressor gene in renal cell carcinoma and peritoneal mesothelioma of Wistar rats induced by ferric nitrilotriacetate. Jpn J Cancer Res 1995, 86:1150-1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toyokuni S, Iwasa Y, Kondo S, Tanaka T, Ochi H, Hiai H: Intranuclear distribution of 8-hydroxy-2′-deoxyguanosine: an immunocytochemical study. J Histochem Cytochem 1999, 47:833-836 [DOI] [PubMed] [Google Scholar]
- 21.Swafford DS, Middleton SK, Palmisano WA, Nikula KJ, Tesfaigzi J, Baylin SB, Herman JG, Belinsky SA: Frequent aberrant methylation of p16INK4a in primary rat lung tumors. Mol Cell Biol 1997, 17:1366-1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hulla J, Schneider R: Structure of the rat p53 tumor suppressor gene. Nucleic Acids Res 1993, 21:713-717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kikuchi Y, Kobayashi E, Nishizawa M, Hamazaki S, Okada S, Hino O: Cloning of the rat homologue of the von Hippel-Lindau tumor suppressor gene and its non-somatic mutation in rat renal cell carcinomas. Jpn J Cancer Res 1995, 86:905-909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch EF, Maniatis T: Molecular Cloning: A Laboratory Manual. 1989. NY, Cold Spring Harbor Laboratory Press, Cold Spring Harbor
- 25.Lin Y, Chan S: Cloning and characterization of two processed p53 pseudogenes from the rat genome. Gene 1995, 156:183-189 [DOI] [PubMed] [Google Scholar]
- 26.Weghorst C, Buzard G, Calvert R, Hulla J, Rice J: Cloning and sequence of a processed p53 pseudogene from rat: a potential source of false “mutations” in PCR fragments of tumor DNA. Gene 1995, 166:317-322 [DOI] [PubMed] [Google Scholar]
- 27.Ciotta C, Dogliotti E, Bignami M: Mutation analysis in two newly identified rat p53 pseudogenes. Mutagenesis 1995, 10:123-128 [DOI] [PubMed] [Google Scholar]
- 28.Bradley J, Rothberg P: Processed pseudogene from the von Hippel-Lindau disease gene is located on human chromosome 1. Diagn Mol Pathol 1999, 8:101-106 [DOI] [PubMed] [Google Scholar]
- 29.Toyokuni S, Okada K, Kondo S, Nishioka H, Tanaka T, Nishiyama Y, Hino O, Hiai H: Development of high-grade renal cell carcinomas in rats independent of somatic mutations in the Tsc2 and VHL tumor suppressor genes. Jpn J Cancer Res 1998, 89:814-820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murthy S, Testa J: Asbestos, chromosomal deletions, and tumor suppressor gene alterations in human malignant mesothelioma. J Cell Physiol 1999, 180:150-157 [DOI] [PubMed] [Google Scholar]
- 31.Cheng J, Jhanwar S, Klein W, Bell D, Lee W, Altomare D, Nobori T, Olopade O, Buckler A, Testa J: p16 alterations and deletion mapping of 9p21–p22 in malignant mesothelioma. Cancer Res 1994, 54:5547-5551 [PubMed] [Google Scholar]
- 32.Xio S, Li D, Vijg J, Sugarbaker D, Corson J, Fletcher J: Co-deletion of p15 and p16 in primary malignant mesothelioma. Oncogene 1995, 11:511-515 [PubMed] [Google Scholar]
- 33.Prins J, Williamson K, Kamp M, Van HE, Van dKT, Hagemeijer A, Versnel M: The gene for the cyclin-dependent-kinase-4 inhibitor, CDKN2A, is preferentially deleted in malignant mesothelioma. Int J Cancer 1998, 75:649-653 [DOI] [PubMed] [Google Scholar]
- 34.Jeong J, Juedes M, Wogan G: Mutations induced in the supF gene of pSP189 by hydroxyl radical and singlet oxygen: relevance to peroxynitrite mutagenesis. Chem Res Toxicol 1998, 11:550-556 [DOI] [PubMed] [Google Scholar]
- 35.Nomoto M, Yamaguchi R, Kawamura M, Kohno K, Kasai H: Analysis of 8-hydroxyguanine in rat kidney genomic DNA after administration of a renal carcinogen, ferric nitrilotriacetate. Carcinogenesis 1999, 20:837-841 [DOI] [PubMed] [Google Scholar]
- 36.Sanchez-Cespedes M, Decker P, Doffek K, Esteller M, Westra W, Alawi E, Herman J, Demeure M, Sidransky D, Ahrendt S: Increased loss of chromosome 9p21 but not p16 inactivation in primary non-small cell lung cancer from smokers. Cancer Res 2001, 61:2092-2096 [PubMed] [Google Scholar]
- 37.Zhang S, Klein-Szanto A, Sauter E, Shafarenko M, Mitsunaga S, Nobori T, Carson D, Ridge J, Goodrow T: Higher frequency of alterations in the p16/CDKN2 gene in squamous cell carcinoma cell lines than in primary tumors of the head and neck. Cancer Res 1994, 54:5050-5053 [PubMed] [Google Scholar]