Abstract
We investigated DNA copy-number aberrations in 22 cell lines derived from small cell lung cancers (SCLCs) using comparative genomic hybridization. A minimal common region at 5p13, within the 5p11-p13 amplicon that was most frequently involved, harbored the CDH6, PC4, and SKP2 genes. These three genes showed amplification and consequent overexpression in the SCLC cell lines. SKP2 positively regulates progression of cell cycle by targeting several regulators, such as the cell-cycle inhibitor p27KIP1, for ubiquitin-mediated degradation. SKP2 was amplified in 7 (44%) of 16 primary SCLC tumors, and consequently overexpressed in 10 (83%) of the 12 of those tumors we examined. Expression levels of SKP2 protein were cell cycle-dependent in SCLC cells as well as in normal cells, and were correlated with the DNA copy-number of the gene. There was an inverse correlation between the expression of SKP2 and p27KIP1 proteins. Down-regulation of SKP2 using an anti-sense oligonucleotide remarkably suppressed the growth of SCLC cells. Our results indicate that SKP2 is likely to be a target of the 5p13 amplification and to play an important role in the growth of SCLC cells.
Small cell lung cancer (SCLC) is one of the most malignant tumors among human populations worldwide and accounts for ∼15 to 25% of all lung cancers. 1-5 Although it initially shows good response to chemo- and radiation therapy, the prognosis is quite poor. An increased understanding of the genetics of SCLC may lead to development of better protocols for its clinical management.
Accumulated evidence suggests that multiple genetic alterations occurring sequentially in a cell lineage, at the nucleotide levels as well as at the chromosome levels, underlie the carcinogenetic process in solid tumors. Amplification of chromosomal DNA is one of the mechanisms capable of activating genes that are implicated in developing tumors. Oncogenes such as MYC (8q24), MYCN (2p24), and MYCL1 (1p34) are known to be activated by amplification in SCLC, 6-8 and comparative genomic hybridization (CGH) studies have revealed additional regions of amplification. 9-15 Those amplified regions are likely to contain unidentified genes associated with the pathogenesis of SCLC. Indeed, our own CGH studies have revealed a number of novel regions of amplification in various types of tumors, including esophageal squamous cell carcinoma, 16,17 gastric cancer, 18,19 and malignant fibrous histiocytoma. 20 Furthermore, we have identified several novel target genes in these amplicons, including GASC1 at 9p23-p24 in esophageal squamous cell carcinoma 19 and MASL1 at 8p23.1 in malignant fibrous histiocytoma. 20 Our studies have also indicated possible involvement of CD44 in an 11p13 amplicon detected in gastric cancers, 18 and recently we identified cIAP1, an apoptosis inhibitor, as a probable target of 11q22 amplification in esophageal squamous cell carcinomas. 21
In the work reported here we examined 22 SCLC cell lines by CGH to explore genomic alterations that might affect initiation and progression of this type of tumor. Among losses and gains involving several chromosomal regions, we found the most frequent high-level gain (HLG), indicative of gene amplification, at 5p11-p13. HLG at 5p had been detected in 14% of SCLCs examined in previous CGH studies. 22 Gains in DNA copy-number within 5p have also been reported in 13.2% of solid tumors from 27 tissues, and in 40.7% of all types of lung cancer. 23 Taken together, these lines of evidence strongly suggested that the 5p11-p13 region harbors one or more target genes whose amplification renders them oncogenic in tumors of various types, and led us to carry out molecular definition of the 5p13 amplicon. Within this amplicon we identified a probable target gene, SKP2, whose product promotes the ubiquitin-mediated proteolysis of p27KIP1; 24-26 this gene exerts its oncogenic effect by cooperating with other oncogenes. 27,28
Materials and Methods
Cell Lines and Primary Samples
The 22 SCLC cell lines chosen for this study had been established and described previously (Table 1) ▶ . All lines except Lu-165 were established from Japanese patients; Lu-165 was derived from the tumor of an Indonesian patient. ACC-LC-5, -80, -170, -172, and -173 were maintained in RPMI 1640 supplemented with 5% fetal calf serum and penicillin-streptomycin. SBC-3 and SBC-5 were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum and penicillin-streptomycin. All others were maintained in RPMI 1640 supplemented with 10% fetal calf serum and penicillin-streptomycin. We obtained primary samples from 16 patients with SCLC at the Aichi Cancer Center Hospital. Before initiation of the present study, informed consent was obtained in the formal style approved by the Ethics Committee.
Table 1.
Summary of 22 SCLC Cell Lines
| No. | Cell line | Amplification sites | Reference |
|---|---|---|---|
| 1 | S-1 | 3q27-q29 | 50 |
| 2 | S-2 | None | 50 |
| 3 | 87-5 | None | 51 |
| 4 | SBC-3 | None | 52 |
| 5 | SBC-5 | 2p23-p25.3, 6q15-q16.2, 8q11.1-q12 | 53 |
| 6 | PC-6 | 5p15.2-p15.3 | 54 |
| 7 | ACC-LC-5* | 5p11-p13.3, 13q33-q34, 18q12.1-q21.1 | 55 |
| 8 | ACC-LC-80 | 20q13.1-q13.3 | 56 |
| 9 | ACC-LC-170 | 8q21.3-q23 | 57 |
| 10 | ACC-LC-172* | 5p11-p13.3, 5q11.1-q11.2, 8q22.1-q23, 12p11.1-p13.3, 12q11-q13.1, 13q31-q34 | 55 |
| 11 | ACC-LC-173 | 8p11.1-p12, 8q11.1-q24.3, 21q21-q22.3 | 58 |
| 12 | Lu-24 | 3q22-q29 | 59 |
| 13 | Lu-130 | 5p11-p15.3, 5q11.1-q11.2 | 59 |
| 14 | Lu-134A† | 5p11-p15.3, 5q11.1-q11.2 | 59 |
| 15 | Lu-134B† | 5p11-p15.3, 5q11.1-q11.2 | 59 |
| 16 | Lu-135 | None | 59 |
| 17 | Lu-138 | 19q11-q13.2, Xq21.1-q28 | 59 |
| 18 | Lu-139 | None | 59 |
| 19 | Lu-140 | 3q26.1-q29, Xq21.1-q28 | 59 |
| 20 | Lu-141 | 19q11-q13.2 | 60 |
| 21 | Lu-143 | 18q21.1-q21.2, 19q11-q13.2, Xp11.1-p22.3, Xq11.1-q13 | 61 |
| 22 | Lu-165 | 4p15.2-p16 | 61 |
*Established from the same patient after an interval of 9 months of chemotherapy and radiotherapy.
†Established from the same patient using a different method.
Comparative Genomic Hybridization
CGH was performed using directly fluorochrome-conjugated DNA as described by Kallioniemi and colleagues, 29 with minor modification. 17,30 Briefly, tumor DNAs and normal DNAs were labeled respectively with Spectrum Green-dUTP and Spectrum Orange-dUTP (Vysis, Chicago, IL) by nick translation. Labeled tumor and normal DNAs (250 ng each), together with 10 μg of Cot-1 DNA (Life Technologies, Inc., Gaithersburg, MD), were denatured and hybridized to normal male metaphase chromosome spreads. The slides were washed and counterstained with 4′,6′-diamidino-2-phenylindole. Shifts in CGH profiles were rated as gains and losses if they reached at least the 1.2 and 0.8 thresholds. Overrepresentations were considered to be HLGs when the fluorescence ratio exceeded 1.5, as described elsewhere. 16-20,29
Fluorescence in Situ Hybridization (FISH)
We performed FISH experiments using as probes a total of 10 bacterial artificial chromosomes (BACs). We selected eight of them (RP11-313B16, 116O11, 16A6, 318D22, 36A10, 2A8, 309K8, and 313P15), containing respectively the CDH12, CDH10, CDH6, PC4, SKP2, RAD1, ZNF131, and GHR genes, on the basis of their locations according to the GeneMap’99 (http://www. ncbi.nlm.nih.gov/genemap99/) and Map View (http://www. ncbi.nlm.nih.gov/cgi-bin/Entrez/maps) databases. The other two, RP11-5N11 and 85N3, were selected on the basis of the BAC map published by the University of Washington (http://www.biotech.washington.edu/). Briefly, each probe was labeled by nick translation with biotin-16-dUTP or digoxigenin-11-dUTP (Boehringer Mannheim, Tokyo, Japan) and hybridized to metaphase chromosomes. Hybridization signals for multicolor FISH were detected as described elsewhere. 31,32
Southern and Northern Blot Analyses
cDNA sequences of PC4 (IMAGE clone ID, 265592) were purchased from Incyte Genomics (Palo Alto, CA) and used as probes for Southern and Northern blotting. For SKP2 and CHD6 we designed specific polymerase chain reaction (PCR) primers on the basis of their respective cDNA sequences 33,34 and used the PCR products as probes. The primer sequences were for SKP2: F, 5′-TGGAATCGTGCCAGATGGTA-3′ and R, 5′-GCTCCCT-AGTATACTTGCAG-3′; and for CHD6: F, 5′-TCAGCATGAGAACTTACCGC-3′ and R, 5′-ACTGATTCCACATCC-AGCTC-3′. Southern and Northern hybridization experiments were performed as described elsewhere. 35
Semiquantitative Reverse-Transcriptase (RT)-PCR of CDH6
cDNAs were synthesized from 5 μg of total RNA by means of the SuperScript Preamplification System (Life Technologies, Inc.). The RT-PCR exponential phase was determined on 15 to 35 cycles to allow semiquantitative comparisons among cDNAs. Each PCR regime involved an initial denaturation step of 4 minutes at 94°C, followed by 30 cycles for CDH6 and 25 cycles for GAPDH at 94°C for 30 seconds, 59°C for 30 seconds, and 72°C for 30 seconds on a thermocycler (GeneAmp PCR System 9700; Perkin Elmer, Branchburg, NJ). The primer sequences were mentioned above for CDH6, and previously described for GAPDH. 36
Western Blotting
Western blots were prepared according to published methods. 35 Anti-SKP2 polyclonal antibody was obtained from Zymed Laboratories Inc. (San Francisco, CA); anti-KIP1/p27 (clone 57) monoclonal antibody was from Transduction Laboratories (Lexington, KY); and anti-cyclin E (M-20) polyclonal antibody was from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-SKP2, anti-KIP1/p27, and anti-cyclin E served as primary antibodies at dilutions of 1:1000, 1:2500, and 1:2000, respectively. We used anti-rabbit or anti-mouse Ig (Amersham Pharmacia Biotech, Tokyo, Japan) diluted 1:2000 as secondary antibodies.
Cell-Cycle Analysis
Cells were blocked in G1/S by incubation in medium containing 2.5 mmol/L of thymidine (Sigma Chemical Co., St. Louis, MO) for 24 hours. 37 Four hours after release from the thymidine block, cells were synchronized in G2/M by incubation in medium containing 50 ng/ml of nocodazole (SIGMA) for 12 hours. 38 Then cells were shaken-off, 39 released from the mitotic block in fresh medium, and harvested at selected times. The synchronization and progression of the cell cycle were monitored by flow cytometry using a FACSCaliber scanner and Cell Quest software (Becton Dickinson Pharmingen, San Diego, CA). Suspensions of nuclei for determination of DNA content were prepared as previously described. 40
Anti-Sense Experiments
Anti-sense experiments were performed as described previously with minor modifications. 26,41 Briefly, two oligonucleotides that contain a phosphorothioate backbone were synthesized (Espec Oligo Service Corp., Tsukuba, Japan) as follows: oligonucleotide 1; 5′-CCTGGGGGATGTTCTCA (the anti-sense direction of human SKP2 cDNA nucleotides 180 to 196); oligonucleotide 2, 5′-GGCTTCCGGGCATTTAG (the scramble control for oligonucleotide 1). The oligonucleotides were delivered into ACC-LC-172 cells using Oligofectamine reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instruction. Briefly, cells were seeded in a 96-well plate at 1000 cells/well the day before transfection. Cells were transfected using 0.6 μl of Oligofectamine per well in 100 μl of Opti-MEM I Reduced Serum Medium (Invitrogen). Cells were treated with the oligonucleotides (anti-sense or scrambled) at a final concentration of 50, 200, and 1000 nmol/L, or Oligofectamine alone as a control. After incubation for 4 hours, 50 μl of growth medium containing three times the normal concentration of serum were added. Viable cells were counted by the MTT assay (see later) at 2, 4, and 6 days after transfection. For Western blot analysis, cells were plated in a 15-cm dish, transfected in the same way, and then harvested at 48 hours after transfection.
Measurement of Cell Viability
Cell growth was assessed using the MTT colorimetric assay (cell-counting kit-8; Dojindo Laboratories, Kumamoto, Japan), which measures the ability of viable cells to cleave a tetrazolium salt (WST-8) to a water-soluble formazan. WST-8 was added 4 hours before the end of culture, and absorbance was measured at 450 nm using a microplate reader (Benchmark; Bio-Rad Laboratories, Hercules, CA). Each determination was performed in quadruplicate.
Results
Genomic Changes in SCLC Cell Lines
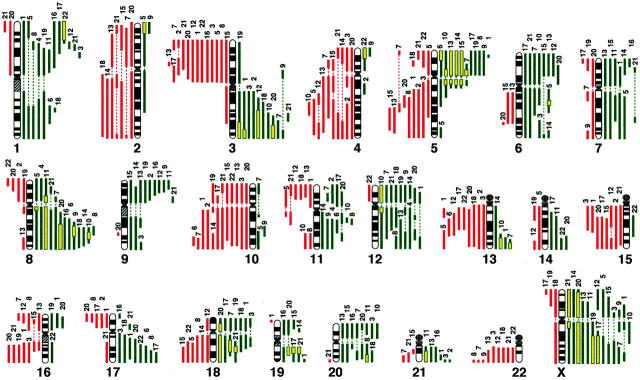
An overview of the genetic changes we detected among 22 SCLC cell lines is shown in Figure 1 ▶ . All of the lines (100%) showed copy-number aberrations, and gains predominated over losses in a ratio of 1.21:1. The average number of aberrations per cell line was 14.7 (range, 4 to 25); average numbers of gains and losses were 8.0 (range, 4 to 12) and 6.6 (range, 0 to 14), respectively.
Figure 1.
Summary of genetic imbalances detected by CGH in 22 SCLC cell lines. The 22 autosomes and X chromosome are represented by ideograms showing G-banding patterns. As judged by the computerized green-to-red profiles, vertical lines on the left (red) of each chromosome ideogram show losses of genomic material in cell lines, whereas those on the right (green) correspond to copy-number gains. HLGs are represented as yellow rectangles. The number at the top of each vertical line corresponds to the cell line in which each change was recorded (see Table 1 ▶ ).
Common Regions Involved in DNA Copy-Number Aberrations
Table 2 ▶ indicates that the minimal common regions of loss that occurred most frequently were at 3p and 10q (13 of 22 each, 59.1%); 22q (9 of 22, 40.3%); 4q31.2-q34, 5q21-q23.1, 13q14.1-q21, and 15q15-q22.1 (8 of 22 each, 36.4%); 16q12.2-q21 (7 of 22, 31.8%). The minimal regions involving copy-number gains were identified at 8q21.1-q24.3, and Xq21.1-q28 (12 of 22 each, 54.5%); 3q23-q29 and 5p11-p15.3 (11 of 22 each, 50.0%); 9p21-p24 and 12p11-p13.3 (9 of 22 each, 40.9%); 6p21.3-p22.2 and 17q23-q25 (8 of 22 each, 36.4%); 11q13-q14.2, 18q12.1-q12.3, and 20p12-p13 (7 of 22 each, 31.8%). The smallest regions of HLGs were seen at 5p11-p13 (five cases); 3q26-q29 and 8q22.2-q24.3 (four cases each); 18q12.1-q21.3, 19q12-q13.2, and Xq21.1-q28 (three cases each); 13q33-q34 and Xp22.3-q13 (two cases each).
Table 2.
Minimal Overlapping Regions of Common DNA Copy Number Changes in SCLC Cell Lines
| DNA copy number | Chromosome regions | Frequency (%) |
|---|---|---|
| Gains | 8q21.1-q24.3 | 54.5 (12/22) |
| Xq21.1-q28 | 54.5 (12/22) | |
| 3q23-q29 | 50.0 (11/22) | |
| 5p11-p15.3 | 50.0 (11/22) | |
| 9p21-p24 | 40.9 (9/22) | |
| 12p11-p13.3 | 40.9 (9/22) | |
| 6p21.3-p22.2 | 36.4 (8/22) | |
| 17q23-q25 | 36.4 (8/22) | |
| 1p31.3-p32.3 | 31.8 (7/22) | |
| 11q13-q14.2 | 31.8 (7/22) | |
| 12q23-q24.1 | 31.8 (7/22) | |
| 18q12.1-q12.3 | 31.8 (7/22) | |
| 20p12-p13 | 31.8 (7/22) | |
| High level gains (amplifications) | 5p11-p13 | 22.7 (5/22) |
| 3q26-q29 | 18.2 (4/22) | |
| 8q22.2-q24.3 | 18.2 (4/22) | |
| 18q12.1-q21.3 | 13.6 (3/22) | |
| 19q12-q13.2 | 13.6 (3/22) | |
| Xq21.1-q28 | 13.6 (3/22) | |
| 13q33-q34 | 9.1 (2/22) | |
| Xp11.1-p22.3, Xq11.1-q13 | 9.1 (2/22) | |
| Losses | 3p | 59.1 (13/22) |
| 10q | 59.1 (13/22) | |
| 22q | 40.9 (9/22) | |
| 4q31.2-q34 | 36.4 (8/22) | |
| 5q21-q23.1 | 36.4 (8/22) | |
| 13q14.1-q21 | 36.4 (8/22) | |
| 15q15-q22.1 | 36.4 (8/22) | |
| 16q12.2-q21 | 31.8 (7/22) |
Definition of the 5p13 Amplicon by FISH
We focused further examination on amplification at 5p11-p13, because this was the region most frequently involved. To delineate a map of the 5p11-p13 amplicon, we performed FISH on 11 SCLC cell lines that had exhibited copy-number gains in this region by CGH. We selected 10 BACs covering the amplified region as probes for the FISH experiments (Figure 2A) ▶ . Four BACs (5N11, 16A6, 318D22, and 36A10) produced strong signals as homogeneous staining regions (HSRs) in ACC-LC-5 and ACC-LC-172, indicating that these BACs represented part of the amplicon. In contrast, the six other BACs did not show HSR patterns in the same cell lines, reflecting positions outside the 5p13 amplicon (Figure 2E) ▶ . On the other hand, when we used BACs 5N11, 16A6, 318D22, or 36A10 as probes the numbers of FISH signals in the amplified region ranged from seven to nine in three cell lines (Lu-130, -134A, and -134B; Figure 2D ▶ ), amounted to three or four in four lines (ACC-LC-80, -170, -173, and PC-6; Figure 2C ▶ ) and two in two cell lines (SBC-3 and SBC-5; Figure 2B ▶ ). In this way we defined the smallest commonly affected region in the 5p13 amplicon between BACs 5N11 and 36A10 (markers AFM238ZA9 and WI12996), a region harboring the CDH6, PC4, and SKP2 genes.
Figure 2.
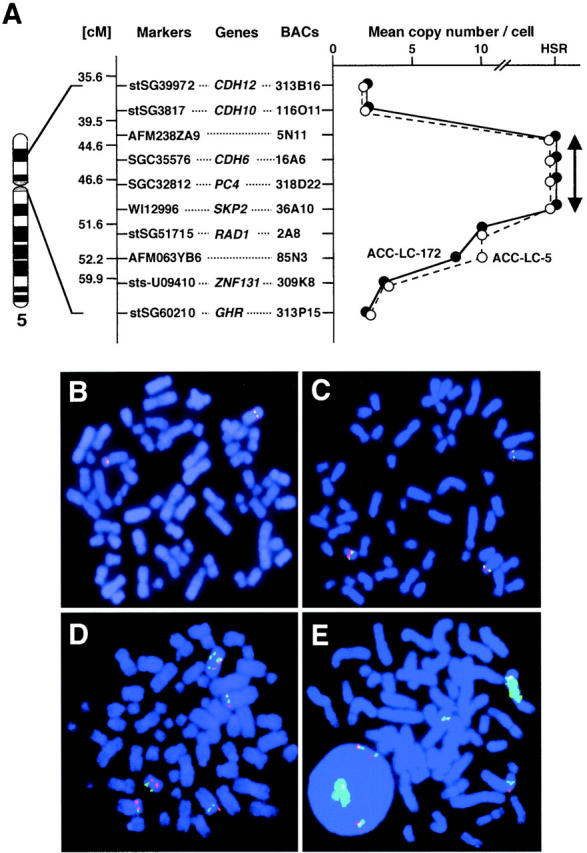
Map of the amplicon at 5p13 and representative images of two-color FISH in SCLC cell lines. A: Represented to the right of the chromosome 5 ideogram are STS markers selected according to the GeneMap 1999 database (http://www.ncbi.nlm.nih.gov/genemap99/), the positions of eight genes (CDH12, CDH10, CDH6, PC4, SKP2, RAD1, ZNF131, and GHR), and the 10 BACs used as probes for FISH experiments. The RP11-prefix is omitted here for all BACs. The graph at far right represents the copy number per cell and the extent of HSR determined by FISH on metaphase chromosomes from two SCLC cell lines, ACC-LC-5 (open circles) and ACC-LC-172 (filled circles). B–E: Two-color FISH using BAC RP11–36A10 (green) and RP11–309K8 (red) on metaphase chromosomes from cell lines SBC-5 (B), ACC-LC-173 (C), Lu-134A (D), and ACC-LC-172 (E). BAC RP11–36A10 generated strong signals as a HSR on a marker chromosome in ACC-LC-172 (E); BAC RP11–309K8 generated three signals in the same cell line. In other lines both RP11–36A10 and RP11–309K8 generated two signals in SBC-5 (B), three in ACC-LC-173 (C), and eight in Lu-134A (D).
Amplification and Overexpression of CDH6, PC4, and SKP2 in SCLC Cell Lines
To determine whether CDH6, PC4, and SKP2 were individual targets of amplification, we examined amplification status and expression levels for each of these genes in all of the SCLC cell lines except Lu-141 and S-1, by Southern and Northern blotting and by semiquantitative RT-PCR. As Figure 3A ▶ represents, all three genes were amplified in comparison to normal genomic DNA in five (ACC-LC-5, ACC-LC-172, Lu-130, Lu-134A, and Lu-134B) of the 20 cell lines examined. In Northern blot analyses, SKP2 and PC4 showed overexpression in 15 (75%) and nine (45%) of the 20 cell lines, respectively (Figure 3B) ▶ . Semiquantitative RT-PCR analysis demonstrated that expression levels of CDH6 were elevated in 13 (65%) of the 20 cell lines (Figure 3C) ▶ . Taken together, these findings indicated that CDH6, PC4, and SKP2 were co-amplified and consequently overexpressed in all five cell lines that had exhibited HLG at 5p13. Among the three genes, we focused further investigation on SKP2, because the gene was most frequently overexpressed among the three.
Figure 3.
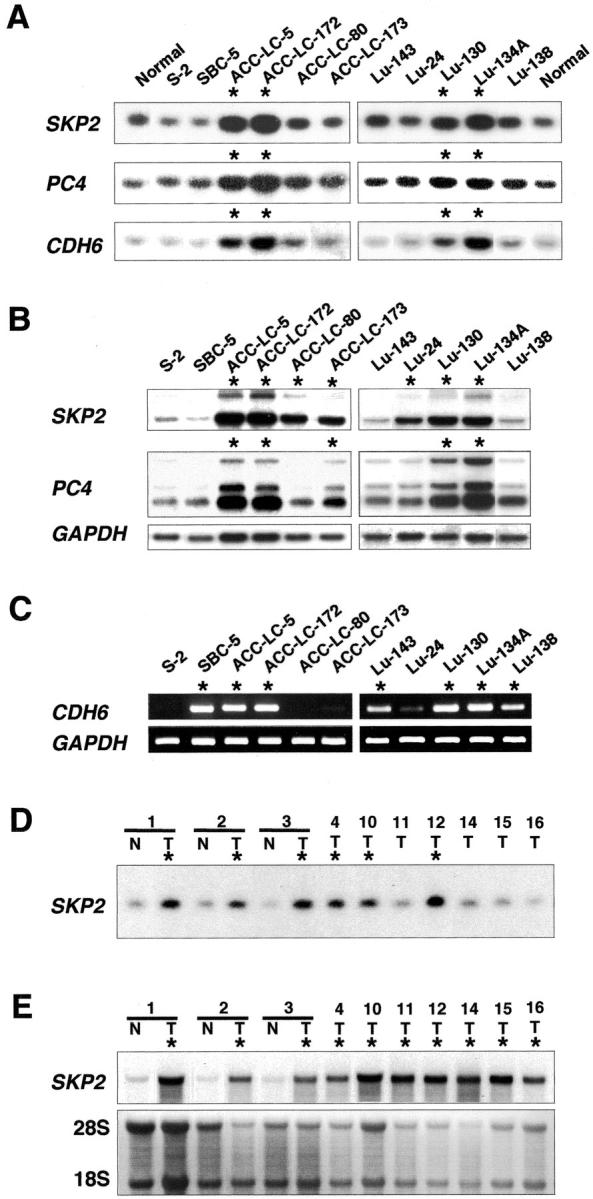
Amplification and overexpression of three genes (SKP2, PC4, and CDH6) lying within the 5p13 amplicon in SCLC cell lines and primary tumors. Asterisks indicate cell lines and primary tumors bearing amplification or overexpression of these genes. A: Representative Southern blots containing 5 μg of genomic DNA derived from cell lines and normal peripheral lymphocytes. B: Northern-blot analyses of SKP2 and PC4 using 10 μg of total RNA from the same SCLC cell lines; GAPDH served as a quantity-control probe. C: Semiquantitative RT-PCR analyses of CDH6 in the same cell lines, using GAPDH as control. D: Representative Southern blots analysis of SKP2 containing 5 μg of genomic DNA derived from 10 primary SCLC tumors (T) and nontumorous counterparts (N) of three of them. E: Northern-blot analysis of SKP2 using 10 μg of total RNA from each of the same 10 primary SCLC tumors and three nontumorous counterparts. Two rRNA species served as controls.
Amplification and Overexpression of SKP2 in Primary SCLC Tumors
To ascertain whether SKP2 was amplified and overexpressed in primary tumors, we examined 16 primary SCLCs by Southern blotting and 12 of those 16 by Northern blotting. As shown in Figure 3D ▶ , SKP2 was amplified in 7 (43.8%) of the 16 tumors compared with their nontumorous counterparts. In Northern blots, 10 (83.3%) of 12 tumors showed high expression of this gene (Figure 3E) ▶ . All of the tumors with amplification of SKP2, except for one whose RNA was not available, showed consequent overexpression.
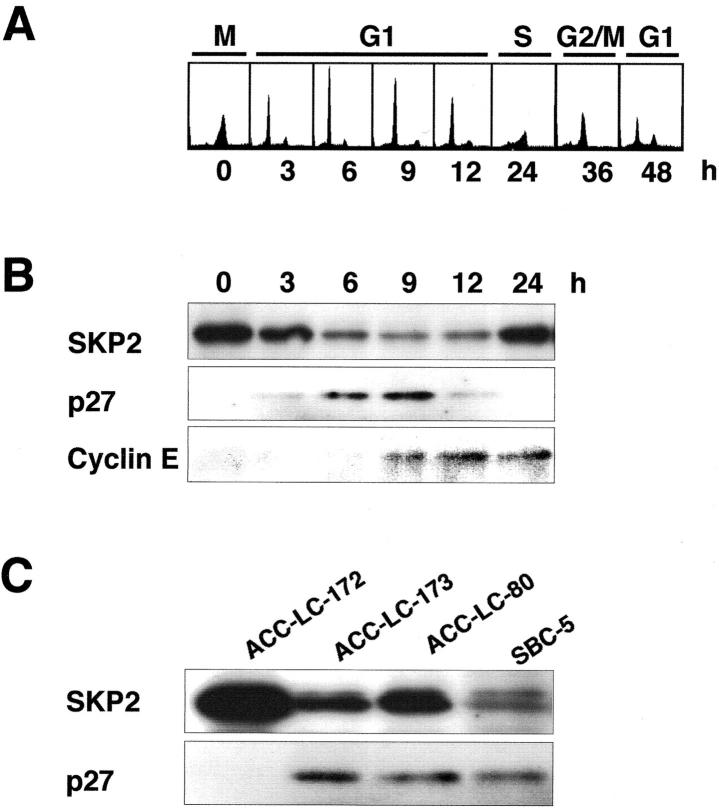
Expression of SKP2 During Cell-Cycle Progression
To confirm whether expression levels of SKP2 protein correlated with amplification levels of the gene, we conducted immunoblotting experiments. Because expression of SKP2 protein was cell cycle-dependent in both normal and transformed cells, 26,42 we determined its levels during cell-cycle progression in ACC-LC-172, a cell line showing amplification of SKP2. We examined the expression of p27KIP1 and cyclin E proteins as well. Cells were synchronized in the M phase with nocodazole. We confirmed the synchrony and monitored the progression of the cell cycle after removal of nocodazole, by flow-cytometric analysis (Figure 4A) ▶ . As shown in Figure 4B ▶ , immunoblots revealed that expression levels of SKP2 were high in the M phase (0 hours) and gradually decreased during transition from the M to G1 phases (6 hours). This decrease was inversely proportional to the increase in levels of p27KIP1. As the cell cycle progressed, SKP2 levels increased again in S phase (24 hours). Expression levels of p27KIP1 decreased during transition from the mid-G1 phase (12 hours) to S phase. Cyclin E was expressed during the transition from G1 to S. These results verified that expression of SKP2 was regulated by the cell cycle even in SCLC lines where the gene was amplified and inversely proportional to that of p27KIP1.
Figure 4.
Expression of SKP2 protein during cell-cycle progression. A: Flow-cytometric analysis. ACC-LC-172 cells were synchronized to the M phase with nocodazole as described in Materials and Methods, and harvested at the indicated times after release from the nocodazole block. The x axis shows DNA content and the y axis shows the number of cells. B: Protein extracts of ACC-LC-172 cells analyzed by Western blotting with antibodies to SKP2, p27, and cyclin E during the M phase (0 hours) and the S phase (24 hours). C: Immunoblots of protein extracts from M phase showing expression of SKP2 and p27 in four SCLC cell lines that had shown different SKP2 DNA copy-numbers in FISH. Experiments using the SKP2-specific BAC 36A10 (Figure 2) ▶ had shown the gene amplification as HSR in ACC-LC-172, three-copy signals in ACC-LC-173 and ACC-LC-80, and two-copy signals in SBC-5. The results of FISH analyses also agreed with those of Southern blotting (Figure 3A) ▶ .
We next compared expression levels of SKP2 protein in four SCLC cell lines (ACC-LC-172, ACC-LC-173, ACC-LC-80, and SBC-5) that had different status of SKP2 DNA copy-number, by synchronizing these cells to the M phase when its levels were high. As shown in Figure 4C ▶ , we found the greatest amount of SKP2 in ACC-LC-172 and the least in SBC-5, a cell line not showing amplification of the gene. Expression levels of SKP2 protein were concordant with levels of the amplification of this gene. By contrast, p27KIP1 was expressed at the lowest level in ACC-LC-172, an inverse correlation with SKP2.
SKP2 Anti-Sense Oligonucleotide Inhibits the Growth of SCLC Cells
To investigate the effect of SKP2 overexpression on cell growth, SKP2 was down-regulated in ACC-LC-172 cells using an anti-sense oligonucleotide targeting the same site on SKP2 mRNA as described previously. 26,41 The anti-sense oligonucleotide, but not the control oligonucleotide or the transfecting agent alone, induced a decrease in SKP2 protein (Figure 5A) ▶ . Down-regulation of SKP2 resulted in remarkable growth inhibition in ACC-LC-172 cells (Figure 5, B and C) ▶ . The most striking anti-sense effects were obtained when ACC-LC-172 cells were treated with 200 nmol/L of anti-sense oligonucleotides (Figure 5B) ▶ and cell viability was determined at 4 days after transfection (Figure 5C) ▶ . These findings strongly suggested that SKP2 overexpression led to promoting the growth of SCLC cells.
Figure 5.
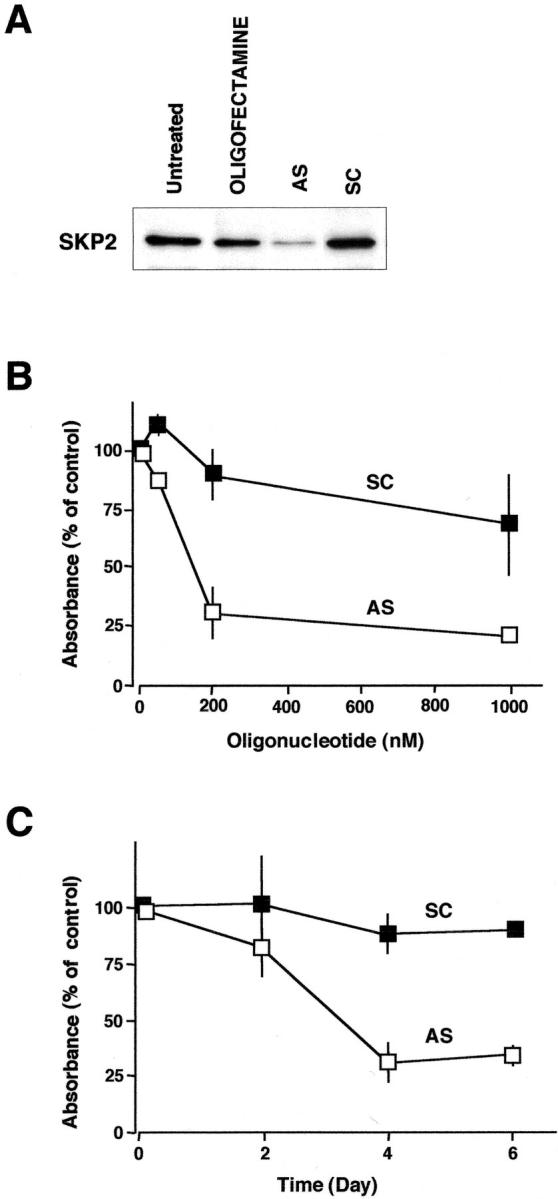
Growth inhibition by the anti-sense oligonucleotide targeting SKP2 mRNA. ACC-LC-172 cells, exhibiting amplification and overexpression of SKP2, were treated with an anti-sense oligonucleotide targeting SKP2 (AS) or a scramble oligonucleotide (SC) by using Oligofectamine. A: Levels of SKP2 determined by Western blotting. Cells were treated with 200 nmol/L of oligonucleotides (anti-sense or scrambled) or Oligofectamine alone, and then harvested at 48 hours after transfection. Untreated cells were maintained under identical experimental conditions. Six μg of whole cell lysate were separated by polyacrylamide gel electrophoresis and analyzed by immunoblotting. B and C: The effects of anti-sense oligonucleotide on the viability of ACC-LC-172 cells. Cells were treated with the indicated concentrations of oligonucleotides (anti-sense or scrambled) and cell viability was determined by MTT assay at 4 days after transfection (B). Cells were treated with 200 nmol/L of oligonucleotides (anti-sense or scrambled) and cell viability was determined at the indicated times after transfection (C). The percentage was calculated against the absorbance of the control cells treated with Oligofectamine alone. The values were shown on the plot represent the means ± SD of four independent experiments.
Discussion
Results from our CGH analysis of 22 SCLC cell lines, summarized in Table 2 ▶ and Figure 1 ▶ , mostly were in accord with previous CGH data, 9-15 except that we more often found gains at 9p and 6p, and observed loss at 17p less frequently. HLG at 5p11-p13 was detected in five (22.7%) of the 22 cell lines. In two previous CGH studies involving primary SCLCs, amplification of 5p occurred in 15% in one (2 of 13) 12 and 14% in the other (3 of 22). 10 Amplification of 5p has been reported also in non-small cell lung cancer (NSCLC), squamous cell carcinoma of the head and neck, carcinoma of the uterine cervix, osteosarcoma, and malignant fibrous histiocytoma. 22 Although in some SCLCs examined the entire 5p arm was amplified, in others the commonly amplified regions were restricted to the region spanning 5p11-p13, 9-11 strongly suggesting the existence there of one or more genes whose amplification could contribute to tumorigenesis of SCLC. Thus we chose to perform molecular cytogenetic characterization of 5p13 amplification in this type of cancer cell.
First, by FISH analysis we successfully narrowed the region of overlap within the 5p13 amplification and observed that BACs containing CDH6, PC4, and SKP2 were localized in this limited region (Figure 2) ▶ . We focused further examination on SKP2. Our experiments clearly demonstrated that SKP2 was amplified and consequently overexpressed not only in multiple SCLC cell lines, but also in primary tumors (Figure 3) ▶ . Moreover, the gene was always overexpressed in both the cell lines and primary tumors where it was amplified, as well as in some lines and tumors not showing amplification. The findings supported the idea that it is a specific target within the 5p13 amplicon, because some mechanism other than amplification should be operating for a gene to be important for tumorigenesis.
SKP2 is an F-box substrate-recognition subunit of the SCF ubiquitin-protein ligase complex. It regulates the progression of the cell cycle by targeting regulators, such as the cell-cycle inhibitor p27KIP1, for ubiquitin-mediated degradation. 24-26,33 This molecule is also required for ubiquitination of cyclin E, but only in the latter’s free, non-CDK2 bound form. 43 Decreased levels of p27KIP1 seem to be associated with high aggressiveness and poor prognosis in a variety of cancers. 44 Skp2-deficient mice grow more slowly and have smaller organs than littermate controls. 43 Moreover, cells in the mutant mice contain markedly enlarged nuclei with polyploidy and multiple centrosomes, and show a reduced growth rate and increased apoptosis. Overexpression of SKP2 has been observed in transformed cells 33 and in human hepatocellular carcinomas, 45 colorectal carcinomas, 46 lymphomas, 28 and oral carcinomas. 27,47 In a direct evaluation of the potential of SKP2 to deregulate growth in vivo, Latres and colleagues 28 generated transgenic mice expressing the SKP2 gene targeted to the T-lymphoid lineage, and also generated double-transgenic mice co-expressing SKP2 and activated N-Ras. Their experiments demonstrated that the double-transgenic mice developed T cell lymphoma, suggesting that SKP2 had oncogenic potential.
In the present study, our results clearly showed that expression of SKP2 protein was cell cycle-dependent, even in SCLC cell lines with amplification of the gene, as it is in normal cells 26,33 (Figure 4, A and B) ▶ . Moreover, we ascertained that expression levels of SKP2 protein were closely correlated with the DNA copy numbers of SKP2 (Figure 4C) ▶ . We also examined the relation between SKP2 and p27KIP1 expression levels during cell-cycle progression. During the transition from M to G1 phase (0 to 6 hours in Figure 4B ▶ ), an inverse correlation appeared between expression of SKP2 and p27KIP1; after that, however, p27KIP1 was reduced in the mid-G1 phase (9 to 12 hours) before SKP2 levels were elevated again in the S phase (24 hours; Figure 4B ▶ ). These findings, together with evidence presented recently by Malek and colleagues, 48 suggested that down-regulation of p27KIP1 in the G1 phase might reflect an SKP2-independent mechanism. Yatabe and colleagues 49 previously observed that SCLC tumors, in contrast to NSCLC, showed increased expression of p27KIP1, a fact seemingly inconsistent with our results that SKP2 was frequently overexpressed in SCLCs. However, an SKP2-independent pathway for controlling p27KiP1 could account for this discrepancy. Indeed, recently Kudo and colleagues 47 reported high expression of SKP2 protein despite high p27KIP1 expression in some primary oral cancers and one oral cancer cell line. Therefore we suggest that amplification and consequent overexpression of SKP2 might play an important role in SCLC tumorigenesis because of not only mediated reduction of p27KIP1, but also because of a dominantly acting function that has not yet been clarified. The involvement of SKP2 or p27KIP1 in tumorigenesis at least should be discussed with a view to their joint dependency on the cell cycle.
To elucidate the functional role of SKP2 in SCLC cells, we down-regulated the expression levels of SKP2 protein using the anti-sense oligonucleotide (Figure 5A) ▶ . We demonstrated that down-regulation of SKP2 induced growth inhibition in SCLC cells (Figure 5, B and C) ▶ . This result provided direct evidence for the role of SKP2 overexpression in the growth of tumor cells.
In summary, the CGH studies reported here revealed that amplification in SCLC cell lines was most frequent in the 5p11-p13 region. We found that SKP2, lying within the minimal common region of the amplicon at 5p13, often showed amplification and consequent overexpression in not only cell lines, but also primary SCLCs. Expression levels of SKP2 protein were correlated with amplification levels of the gene, in a cell cycle-dependent pattern. The anti-sense oligonucleotide targeting the SKP2 gene reduces the growth rate of SCLC cells. These findings indicate that SKP2 is likely to be a target of the amplification mechanism, and to promote the growth of SCLC cells. Thus, SKP2 may provide an excellent therapeutic target for SCLCs.
Acknowledgments
We thank Professor Yusuke Nakamura for his continuous encouragement.
Footnotes
Address reprint requests to Dr. Johji Inazawa, Department of Molecular Cytogenetics, Medical Research Institute, Tokyo Medical and Dental University, 1-5-45 Yushima, Bunkyo-ku, Tokyo 113-8519, Japan. E-mail: johinaz.cgen@mri.tmd.ac.jp.
Supported by Grants-in-Aid for Cancer Research from the Ministry of Education, Culture, Sports, Science, and Technology, Japan; and from the Ministry of Health, Labour, and Welfare, Japan.
References
- 1.Shimosato Y, Nakajima T, Hirohashi S, Morinaga S, Terasaki T, Yamaguchi K, Saijo N, Suemasu K: Biological, pathological and clinical features of small cell lung cancer. Cancer Lett 1986, 33:241-258 [DOI] [PubMed] [Google Scholar]
- 2.Vincent RG, Pickren JW, Lane WW, Bross I, Takita H, Houten L, Gutierrez AC, Rzepk T: The changing histopathology of lung cancer: a review of 1682 cases. Cancer 1977, 39:1647-1655 [DOI] [PubMed] [Google Scholar]
- 3.Landis SH, Murray T, Bolden S, Wingo PA: Cancer statistics, 1999. CA Cancer J Clin 1999, 49:8-31 [DOI] [PubMed] [Google Scholar]
- 4.Yoshimura K, Yamashita N: Clinical statistical observation of 4,931 cases with lung cancer by histological types; results of field study in Japan. Haigan 1982, 22:1-17 [Google Scholar]
- 5.: The Editorial Board of the Cancer Center Statistics in Japan:: Cancer Statistics in Japan 1999 1999:12-47 Foundation for Promotion of Cancer Research, Tokyo
- 6.Little C, Nau MM, Carney DN, Gazdar AF, Minna JD: Amplification and expression of the c-myc oncogene in human lung cancer cell lines. Nature 1983, 306:194-196 [DOI] [PubMed] [Google Scholar]
- 7.Nau MM, Brooks BJ, Jr, Carney DN, Gazdar AF, Battey JF, Sausville EA, Minna JD: Human small-cell lung cancers show amplification and expression of the N-myc gene. Proc Natl Acad Sci USA 1986, 83:1092-1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nau MM, Brooks BJ, Battey J, Sausville E, Gazdar AF, Kirsch IR, McBride OW, Bertness V, Hollis GF, Minna JD: L-myc, a new myc-related gene amplified and expressed in human small cell lung cancer. Nature 1985, 318:69-73 [DOI] [PubMed] [Google Scholar]
- 9.Levin NA, Brzoska PM, Warnock ML, Gray JW, Christman MF: Identification of novel regions of altered DNA copy number in small cell lung tumors. Genes Chromosom Cancer 1995, 13:175-185 [DOI] [PubMed] [Google Scholar]
- 10.Petersen I, Langreck H, Wolf G, Schwende A, Psille R, Vogt P, Reichel MB, Ried T, Dietel M: Small-cell lung cancer is characterized by a high incidence of deletions on chromosomes 3p, 4q, 5q, 10q, 13q and 17p. Br J Cancer 1997, 75:79-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwendel A, Langreck H, Reichel M, Schrock E, Ried T, Dietel M, Petersen I: Primary small-cell lung carcinomas and their metastases are characterized by a recurrent pattern of genetic alterations. Int J Cancer 1997, 74:86-93 [DOI] [PubMed] [Google Scholar]
- 12.Levin NA, Brzoska P, Gupta N, Minna JD, Gray JW, Christman MF: Identification of frequent novel genetic alterations in small cell lung carcinoma. Cancer Res 1994, 54:5086-5091 [PubMed] [Google Scholar]
- 13.Lui WO, Tanenbaum DM, Larsson C: High level amplification of 1p32-33 and 2p22-24 in small cell lung carcinomas. Int J Oncol 2001, 19:451-457 [DOI] [PubMed] [Google Scholar]
- 14.Ried T, Petersen I, Holtgreve-Grez H, Speicher MR, Schrock E, du Manoir S, Cremer T: Mapping of multiple DNA gains and losses in primary small cell lung carcinomas by comparative genomic hybridization. Cancer Res 1994, 54:1801-1806 [PubMed] [Google Scholar]
- 15.Walch AK, Zitzelsberger HF, Aubele MM, Mattis AE, Bauchinger M, Candidus S, Prauer HW, Werner M, Hofler H: Typical and atypical carcinoid tumors of the lung are characterized by 11q deletions as detected by comparative genomic hybridization. Am J Pathol 1998, 153:1089-1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pimkhaokham A, Shimada Y, Fukuda Y, Kurihara N, Imoto I, Yang ZQ, Imamura M, Nakamura Y, Amagasa T, Inazawa J: Nonrandom chromosomal imbalances in esophageal squamous cell carcinoma cell lines: possible involvement of the ATF3 and CENPF genes in the 1q32 amplicon. Jpn J Cancer Res 2000, 91:1126-1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shinomiya T, Mori T, Ariyama Y, Sakabe T, Fukuda Y, Murakami Y, Nakamura Y, Inazawa J: Comparative genomic hybridization of squamous cell carcinoma of the esophagus: the possible involvement of the DP1 gene in the 13q34 amplicon. Genes Chromosom Cancer 1999, 24:337-344 [PubMed] [Google Scholar]
- 18.Fukuda Y, Kurihara N, Imoto I, Yasui K, Yoshida M, Yanagihara K, Park JG, Nakamura Y, Inazawa J: CD44 is a potential target of amplification within the 11p13 amplicon detected in gastric cancer cell lines. Genes Chromosom Cancer 2000, 29:315-324 [DOI] [PubMed] [Google Scholar]
- 19.Yang ZQ, Imoto I, Fukuda Y, Pimkhaokham A, Shimada Y, Imamura M, Sugano S, Nakamura Y, Inazawa J: Identification of a novel gene, GASC1, within an amplicon at 9p23-24 frequently detected in esophageal cancer cell lines. Cancer Res 2000, 60:4735-4739 [PubMed] [Google Scholar]
- 20.Sakabe T, Shinomiya T, Mori T, Ariyama Y, Fukuda Y, Fujiwara T, Nakamura Y, Inazawa J: Identification of a novel gene, MASL1, within an amplicon at 8p23.1 detected in malignant fibrous histiocytomas by comparative genomic hybridization. Cancer Res 1999, 59:511-515 [PubMed] [Google Scholar]
- 21.Imoto I, Yang ZQ, Pimkhaokham A, Tsuda H, Shimada Y, Imamura M, Ohki M, Inazawa J: Identification of cIAP1 as a candidate target gene within an amplicon at 11q22 in esophageal squamous cell carcinomas. Cancer Res 2001, 61:6629-6634 [PubMed] [Google Scholar]
- 22.Knuutila S, Björkvist AM, Autio K, Tarkkanen M, Wolf M, Monni O, Szymanska J, Larramendy ML, Tapper J, Pere H, El-Rifai W, Hemmer S, Wasenius VM, Vidgren V, Zhu Y: DNA copy number amplifications in human neoplasms: review of comparative genomic hybridization studies. Am J Pathol 1998, 152:1107-1123 [PMC free article] [PubMed] [Google Scholar]
- 23.Rooney PH, Murray GI, Stevenson DA, Haites NE, Cassidy J, McLeod HL: Comparative genomic hybridization and chromosomal instability in solid tumours. Br J Cancer 1999, 80:862-873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sutterluty H, Chatelain E, Marti A, Wirbelauer C, Senften M, Muller U, Krek W: p45(SKP2) promotes p27(Kip1) degradation and induces S phase in quiescent cells. Nat Cell Biol 1999, 1:207-214 [DOI] [PubMed] [Google Scholar]
- 25.Tsvetkov LM, Yeh KH, Lee SJ, Sun H, Zhang H: p27(Kip1) ubiquitination and degradation is regulated by the SCFSkp2 complex through phosphorylated Thr187 in p27. Curr Biol 1999, 9:661-664 [DOI] [PubMed] [Google Scholar]
- 26.Carrano AC, Eytan E, Hershko A, Pagano M: SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat Cell Biol 1999, 1:193-199 [DOI] [PubMed] [Google Scholar]
- 27.Gstaigert M, Jordan R, Lim M, Catzavelos C, Mestan J, Slingerland J, Krek W: Skp2 is oncogenic and overexpressed in human cancers. Proc Natl Acad Sci USA 2001, 98:5043-5048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Latres E, Chiarle R, Schulman BA, Pavletich NP, Pellicer A, Inghirami G, Pagano M: Role of the F-box protein Skp2 in lymphomagenesis. Proc Natl Acad Sci USA 2001, 98:2515-2520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kallioniemi A, Kallioniemi OP, Sudar D, Rutovitz D, Gray JW, Waldman F, Pinkel D: Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science 1992, 258:818-821 [DOI] [PubMed] [Google Scholar]
- 30.Ariyama Y, Sakabe T, Shinomiya T, Mori T, Fukuda Y, Inazawa J: Identification of amplified DNA sequences on double minute chromosomes in a leukemic cell line KY821 by means of spectral karyotyping and comparative genomic hybridization. J Hum Genet 1998, 43:187-190 [DOI] [PubMed] [Google Scholar]
- 31.Inazawa J, Ariyama T, Abe T: Physical ordering of three polymorphic DNA markers spanning the regions containing a tumor suppressor gene of renal cell carcinoma by three-color fluorescent in situ hybridization. Jpn J Cancer Res 1992, 83:1248-1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ariyama T, Inazawa J, Uemura Y, Kakazu N, Maekawa T, Urase F, Irimajiri K, Horiuchi A, Nakamura Y, Abe T: Clonal origin of Philadelphia chromosome negative cells with trisomy 8 appearing during the course of alpha-interferon therapy for pH-positive chronic myelocytic leukemia. Cancer Genet Cytogenet 1995, 81:20-23 [DOI] [PubMed] [Google Scholar]
- 33.Zhang H, Kobayashi R, Galaktionov K, Beach D: p19Skp1 and p45Skp2 are essential elements of the cyclin A-CDK2 S phase kinase. Cell 1995, 82:915-925 [DOI] [PubMed] [Google Scholar]
- 34.Shimoyama Y, Gotoh M, Terasaki T, Kitajima M, Hirohashi S: Isolation and sequence analysis of human cadherin-6 complementary DNA for the full coding sequence and its expression in human carcinoma cells. Cancer Res 1995, 55:2206-2211 [PubMed] [Google Scholar]
- 35.Yasui K, Imoto I, Fukuda Y, Pimkhaokham A, Yang ZQ, Naruto T, Shimada Y, Nakamura Y, Inazawa J: Identification of target genes within an amplicon at 14q12–q13 in esophageal squamous cell carcinoma. Genes Chromosom Cancer 2001, 32:112-118 [DOI] [PubMed] [Google Scholar]
- 36.Mahotka C, Wenzel M, Springer E, Gabbert HE, Gerharz CD: Survivin-deltaEx3 and survivin-2B: two novel splice variants of the apoptosis inhibitor survivin with different antiapoptotic properties. Cancer Res 1999, 59:6097-6102 [PubMed] [Google Scholar]
- 37.Bootsma D, Budke L, Vas O: Studies on synchronous division of tissue culture cells initiated by excess thymidine. Exp Cell Res 1964, 33:301-309 [DOI] [PubMed] [Google Scholar]
- 38.Zieve GW, Turnbull D, Mullins JM, McIntosh JR: Production of large numbers of mitotic mammalian cells by use of the reversible microtubule inhibitor nocodazole. Nocodazole accumulated mitotic cells. Exp Cell Res 1980, 126:397-405 [DOI] [PubMed] [Google Scholar]
- 39.Terashima T, Tolmach LJ: Changes in X-ray sensitivity of HeLa cells during the division cycle. Nature 1961, 190:1210-1211 [DOI] [PubMed] [Google Scholar]
- 40.Block AL, Bauer KD, Williams TJ, Seidenfeld J: Experimental parameters and a biological standard for acridine orange detection of drug-induced alterations in chromatin condensation. Cytometry 1987, 8:163-169 [DOI] [PubMed] [Google Scholar]
- 41.Yu ZK, Gervais JL, Zhang H: Human CUL-1 associates with the SKP1/SKP2 complex and regulates p21CIP1/WAF1 and cyclin D proteins. Proc Natl Acad Sci USA 1998, 95:11324-11329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lisztwan J, Marti A, Sutterluty H, Gstaiger M, Wirbelauer C, Krek W: Association of human CUL-1 and ubiquitin-conjugating enzyme CDC34 with the F-box protein p45SKP2: evidence for evolutionary conservation in the subunit composition of the CDC34-SCF pathway. EMBO J 1998, 17:368-383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakayama K, Nagahama H, Minamishima YA, Matsumoto M, Nakamichi I, Kitagawa K, Shirane M, Tsunematsu R, Tsukiyama T, Ishida N, Kitagawa M, Nakayama K, Hatakeyama S: Targeted disruption of Skp2 results in accumulation of cyclin E and p27Kip1, polyploidy and centrosome overduplication. EMBO J 2000, 19:2069-2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsihlias J, Kapusta LR, DeBoer G, Morava-Protzner I, Zbieranowski I, Bhattacharya N, Catzavelos GC, Klotz LH, Slingerland JM: Loss of cyclin dependent kinase inhibitor p27Kip1 is a novel prognostic factor in localized human prostate adenocarcinoma. Cancer Res 1998, 58:542-548 [PubMed] [Google Scholar]
- 45.Chao Y, Shih YL, Chiu JH, Chau GY, Lui WY, Yang WK, Lee SD, Huang TS: Overexpression of cyclin A but not Skp 2 correlates with the tumor relapse of human hepatocellular carcinoma. Cancer Res 1998, 58:985-990 [PubMed] [Google Scholar]
- 46.Hershko D, Bornstein G, Ben-Izhak O, Carrano A, Pagano M, Krausz MM, Hershko A: Inverse relation between levels of p27Kip1 and of its ubiquitin ligase subunit Skp2 in colorectal carcinomas. Cancer 2001, 91:1745-1751 [DOI] [PubMed] [Google Scholar]
- 47.Kudo Y, Kitajima S, Sato S, Miyauchi M, Ogawa I, Takata T: High expression of S-phase kinase-interacting protein 2, human F-box protein, correlates with poor prognosis in oral squamous cell carcinomas. Cancer Res 2001, 61:7044-7047 [PubMed] [Google Scholar]
- 48.Malek NP, Sundberg H, McGrew S, Nakayama K, Kyriakidis TR, Roberts JM: A mouse knock-in model exposes sequential proteolytic pathways that regulate p27Kip1 in G1 and S phase. Nature 2001, 413:323-327 [DOI] [PubMed] [Google Scholar]
- 49.Yatabe Y, Masuda A, Nakamura S, Kuroishi T, Osada H, Takahashi T, Mitsudomi T: P27Kip1 in human lung cancers: differential changes in small cell and non-small cell carcinomas. Cancer Res 1998, 58:1042-1047 [PubMed] [Google Scholar]
- 50.Kobayashi S, Okada S, Inaba H, Hashimoto K, Nakada T: Study on growth characteristics and morphological findings of human small cell carcinoma in vitro. Kokenshi 1986, 38:103-111 [Google Scholar]
- 51.Kobayashi S, Okada S, Inaba H, Syoji W, Hasumi T, Sato N, Fujimura S: Culture and growth characteristics of human non-small cell lung cancer cells. Kokenshi 1990, 42:73-83 [Google Scholar]
- 52.Kishimoto N, Ohnoshi T, Hiraki S, Miyake K, Ozawa S, Tamura T, Numata K, Kawahara N, Tamura R, Miyamoto H, Nishii K, Kimura I: Establishment and characterization of five cell lines from human small cell carcinoma. Haigan 1984, 24:644 [Google Scholar]
- 53.Mitsuhashi Y, Inaba M, Sugiyama Y, Kobayashi T: In vitro measurement of chemosensitivity of human small cell lung and gastric cancer cell lines toward cell cycle phase-nonspecific agents under the clinically equivalent area under the curve. Cancer 1992, 70:2540-2546 [DOI] [PubMed] [Google Scholar]
- 54.Tsuji K, Hayata Y, Sato M, Shimosato Y, Fukushima Y: Neuronal differentiation of oat cell carcinoma in vitro by dibutyryl cyclic adenosine 3′, 5′-monophosphate. Cancer Lett 1976, 1:311-318 [Google Scholar]
- 55.Takahashi T, Obata Y, Sekido Y, Hida T, Ueda R, Watanabe H, Ariyoshi Y, Sugiura T, Takahashi T: Expression and amplification of myc gene family in small cell lung cancer and its relation to biological characteristics. Cancer Res 1989, 49:2683-2688 [PubMed] [Google Scholar]
- 56.Hibi K, Takahashi T, Sekido Y, Ueda R, Hida T, Ariyoshi Y, Takagi H, Takahashi T: Coexpression of the stem cell factor and the c-kit genes in small-cell lung cancer. Oncogene 1991, 6:2291-2296 [PubMed] [Google Scholar]
- 57.Suyama M, Saito T, Hara K, Nakamura W, Tateno W, Hosoda S, Kito H, Ariyoshi H, Kato K: Successive transplantation of 2 small cell (oat-cell) carcinoma of the lung and the thymus containing neuron-specific enolase into nude mice. Igaku no Ayumi 1984, 131:161-162 [Google Scholar]
- 58.Hida T, Ariyoshi Y, Kuwabara M, Sugiura T, Takahashi T, Takahashi T, Hosoda K, Niitsu Y, Ueda R: Glutathione S-transferase levels in a panel of lung cancer cell lines and its relation to chemo-radiosensitivity. Jpn J Clin Oncol 1993, 23:14-19 [PubMed] [Google Scholar]
- 59.Terasaki T, Shimosato Y, Nakajima T, Tsumuraya M, Morinaga S, Hirohashi S, Yamaguchi K, Kato K, Ichinose H, Nagatsu T: Changes in cell characteristics due to culture conditions in cell lines from human small cell lung cancer. Jpn J Clin Oncol 1986, 16:203-212 [PubMed] [Google Scholar]
- 60.Matsumoto T, Terasaki T, Mukai K, Wada M, Okamoto A, Yokota J, Yamaguchi K, Kato K, Nagatsu T, Shimosato Y: Relation between nucleolar size and growth characteristics in small cell lung cancer cell lines. Jpn J Cancer Res 1991, 82:820-828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Terasaki T, Matsuno Y, Shimosato Y, Yamaguchi K, Ichinose H, Nagatsu T, Kato K: Establishment of a human small cell lung cancer cell line producing a large amount of anti-diuretic hormone. Jpn J Cancer Res 1994, 85:718-722 [DOI] [PMC free article] [PubMed] [Google Scholar]