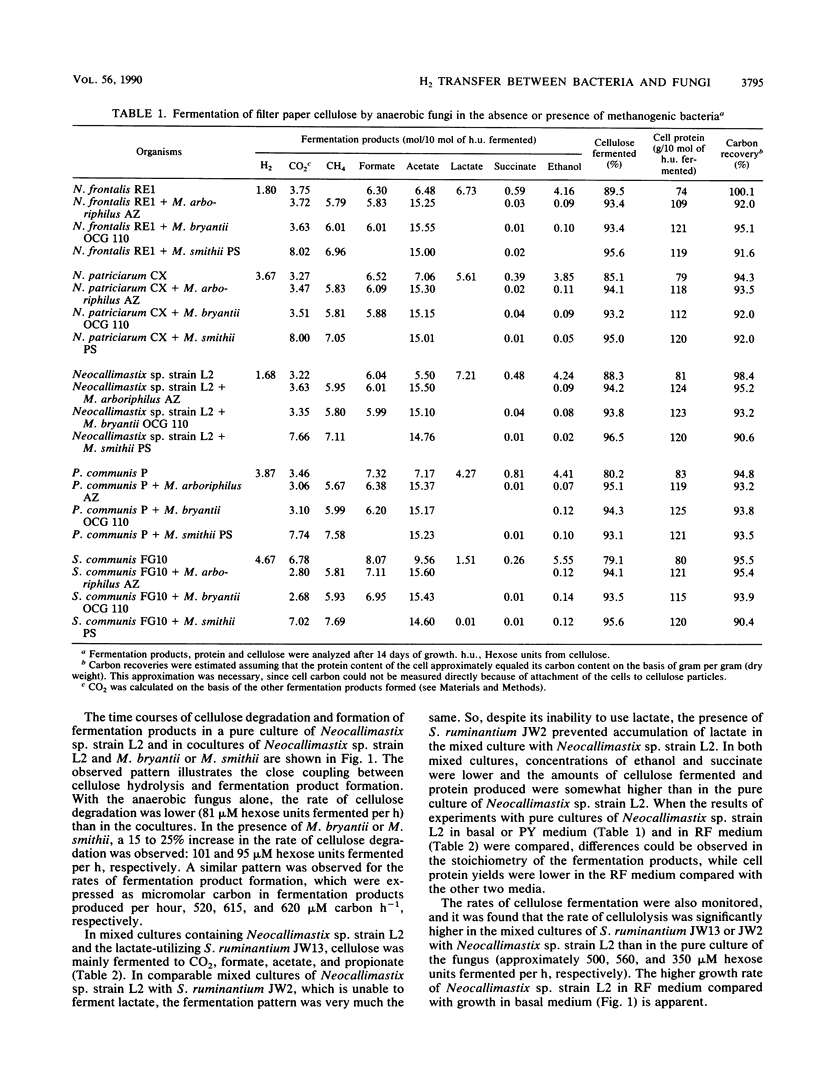
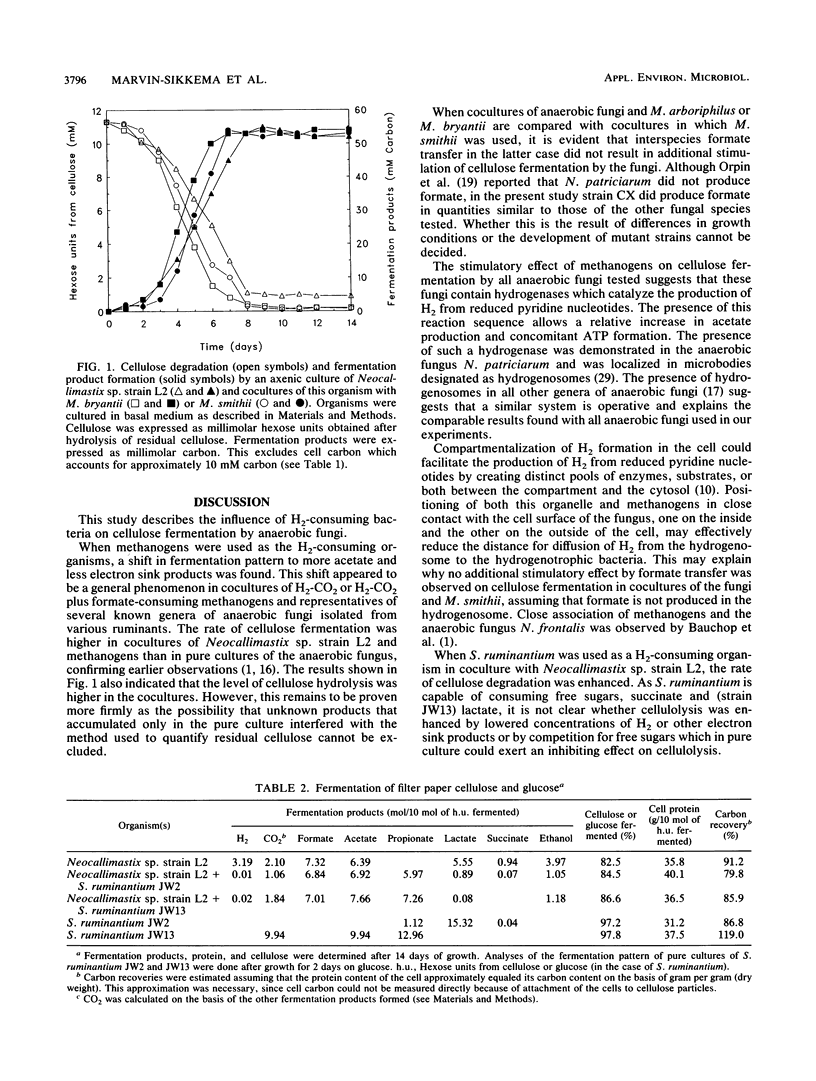
Abstract
The presence of methanogens Methanobacterium arboriphilus, Methanobacterium bryantii, or Methanobrevibacter smithii increased the level of cellulose fermentation by 5 to 10% in cultures of several genera of anaerobic fungi. When Neocallimastix sp. strain L2 was grown in coculture with methanogens the rate of cellulose fermentation also increased relative to that for pure cultures of the fungus. Methanogens caused a shift in the fermentation products to more acetate and less lactate, succinate, and ethanol. Formate transfer in cocultures of anaerobic fungi and M. smithii did not result in further stimulation of cellulolysis above the level caused by H2 transfer. When Selenomonas ruminatium was used as a H2-consuming organism in coculture with Neocallimastix sp. strain L2, both the rate and level of cellulolysis increased. The observed influence of the presence of methanogens is interpreted to indicate a shift of electrons from the formation of electron sink carbon products to H2 via reduced pyridine nucleotides, favoring the production of additional acetate and probably ATP. It is not known how S. ruminantium exerts its influence. It might result from a lowered production of electron sink products by the fungus, from consumption of electron sink products or H2 by S. ruminantium, or from competition for free sugars which in pure culture could exert an inhibiting effect on cellulolysis.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bauchop T., Mountfort D. O. Cellulose fermentation by a rumen anaerobic fungus in both the absence and the presence of rumen methanogens. Appl Environ Microbiol. 1981 Dec;42(6):1103–1110. doi: 10.1128/aem.42.6.1103-1110.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone D. R., Johnson R. L., Liu Y. Diffusion of the Interspecies Electron Carriers H(2) and Formate in Methanogenic Ecosystems and Its Implications in the Measurement of K(m) for H(2) or Formate Uptake. Appl Environ Microbiol. 1989 Jul;55(7):1735–1741. doi: 10.1128/aem.55.7.1735-1741.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson C. The influence of extracellular hydrogen on the metabolism of Bacteroides ruminicola, Anaerovibrio lipolytica and Selenomonas ruminantium. J Gen Microbiol. 1980 Aug;119(2):485–491. doi: 10.1099/00221287-119-2-485. [DOI] [PubMed] [Google Scholar]
- Joblin K. N. Isolation, enumeration, and maintenance of rumen anaerobic fungi in roll tubes. Appl Environ Microbiol. 1981 Dec;42(6):1119–1122. doi: 10.1128/aem.42.6.1119-1122.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungermann K., Thauer R. K., Leimenstoll G., Decker K. Function of reduced pyridine nucleotide-ferredoxin oxidoreductases in saccharolytic Clostridia. Biochim Biophys Acta. 1973 May 30;305(2):268–280. doi: 10.1016/0005-2728(73)90175-8. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laanbroek H. J., Geerligs H. J., Sijtsma L., Veldkamp H. Competition for sulfate and ethanol among desulfobacter, desulfobulbus, and desulfovibrio species isolated from intertidal sediments. Appl Environ Microbiol. 1984 Feb;47(2):329–334. doi: 10.1128/aem.47.2.329-334.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mountfort D. O., Asher R. A., Bauchop T. Fermentation of Cellulose to Methane and Carbon Dioxide by a Rumen Anaerobic Fungus in a Triculture with Methanobrevibacter sp. Strain RA1 and Methanosarcina barkeri. Appl Environ Microbiol. 1982 Jul;44(1):128–134. doi: 10.1128/aem.44.1.128-134.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn E. A., Orpin C. G., Greenwood C. A. The ultrastructure and possible relationships of four obligate anaerobic chytridiomycete fungi from the rumen of sheep. Biosystems. 1988;22(1):67–81. doi: 10.1016/0303-2647(88)90051-2. [DOI] [PubMed] [Google Scholar]
- Relationship of lactate dehydrogenase specificity and growth rate to lactate metabolism by Selenomonas ruminantium. Appl Microbiol. 1975 Dec;30(6):916–921. doi: 10.1128/am.30.6.916-921.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheifinger C. C., Wolin M. J. Propionate formation from cellulose and soluble sugars by combined cultures of Bacteroides succinogenes and Selenomonas ruminantium. Appl Microbiol. 1973 Nov;26(5):789–795. doi: 10.1128/am.26.5.789-795.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updegraff D. M. Semimicro determination of cellulose in biological materials. Anal Biochem. 1969 Dec;32(3):420–424. doi: 10.1016/s0003-2697(69)80009-6. [DOI] [PubMed] [Google Scholar]
- Wolin M. J. Metabolic interactions among intestinal microorganisms. Am J Clin Nutr. 1974 Nov;27(11):1320–1328. doi: 10.1093/ajcn/27.11.1320. [DOI] [PubMed] [Google Scholar]
- Yarlett N., Orpin C. G., Munn E. A., Yarlett N. C., Greenwood C. A. Hydrogenosomes in the rumen fungus Neocallimastix patriciarum. Biochem J. 1986 Jun 15;236(3):729–739. doi: 10.1042/bj2360729. [DOI] [PMC free article] [PubMed] [Google Scholar]



