Abstract
To better understand early steps in human breast carcinogenesis, we examined allele imbalance or loss of heterozygosity (LOH), in co-existing normal-appearing breast epithelium and cancers. We microdissected a total of 173 histologically normal ducts or terminal ductolobular units (TDLUs) and malignant epithelial samples from 18 breast cancer cases, and examined their DNA for LOH at 21 microsatellite markers on 10 chromosome arms. Fourteen of 109 (13%) normal ducts/TDLUs, from 8 of 18 (44%) cases, contained LOH. The location of these 14 ducts/TDLUs appeared unrelated to distance from the cancer. LOH in normal-appearing epithelium involved only single markers, whereas LOH in cancers commonly encompassed all informative markers on a chromosome arm. In only 1 of 14 (7%) ducts/TDLUs with LOH, was the same LOH seen in the co-existing cancer. Global differences in LOH per arm in normal-appearing tissue were not demonstrated, but less LOH was seen at 11q and 17p than at 1q (P = 0.002), 16q (P = 0.01), and possibly 17q (P = 0.06). These results indicate that in a large fraction of women with breast cancer, histologically normal breast epithelium harbors occult aberrant clones. Individual clones rarely are precursors of co-existing cancers. However, they might constitute a reservoir from which proliferative lesions or second cancers develop once additional genetic abnormalities occur, they could contribute to intratumoral genetic heterogeneity, and they are consistent with a role for genetic instability early in tumorigenesis.
Breast cancers, even carcinoma in situ (CIS), the earliest recognized breast malignancy, contain numerous genetic abnormalities. 1,2 No signature abnormality characterizes breast cancers, but allele imbalances (AI) or loss [commonly described as loss of heterozygosity (LOH)], are especially frequent. Recurrent sites of LOH are thought to identify the location of genes important to tumorigenesis. Cancer precursors must exist, and identification of these precursors and delineation of their genetic abnormalities is important to elucidate critical early steps in breast cancer development, to determine targets for chemopreventive agents, and to identify lesions destined to progress to invasive disease.
Candidate precursors include proliferative lesions; depending on their histology, varying proportions of these lesions demonstrate LOH, often at the same sites as breast carcinomas. 3 Given the frequency of LOH in proliferative lesions, it is not surprising that LOH has been detected recently in histologically normal epithelium, ie, ducts and terminal ductolobular units (TDLUs). 4-7 LOH, often at sites implicated in breast carcinogenesis, has been found in several small series examining normal-appearing breast epithelium from women with and without breast cancer, in tissue both adjacent to and distant from the primary tumor, and with an incidence possibly increasing with cancer risk. Reproducible LOH indicates that a substantial proportion of the sample’s cells contain an identical DNA abnormality, compared to the individual’s normal somatic pattern. Therefore, samples with LOH likely contain a genetically aberrant, clonal population. It is unclear whether histologically normal epithelial samples with LOH represent early precursors in breast cancer, markers of increased risk, or background abnormalities.
To begin to clarify this issue, we evaluated LOH in histologically normal human breast epithelium and co-existing cancers. We microdissected multiple normal epithelial samples (primarily TDLUs) and co-existing in situ and invasive malignant lesions from 18 breast cancer cases. We examined their DNA for LOH at 21 microsatellite loci selected primarily for their location at chromosome regions that undergo LOH frequently in breast cancer. We speculated that within a given case, we could identify normal and malignant samples with a possible precursor-product relationship, when the samples had LOH at the same locus (particularly if they were adjacent). In contrast, normal and malignant samples with different LOH should represent distinct clones. We also conjectured that determining the sites and extent of LOH in morphologically normal epithelium should help elucidate early genetic events contributing to the development of sporadic breast cancer.
Materials and Methods
Specimen Acquisition
Eighteen consecutive lumpectomy or mastectomy specimens were selected at random from the Department of Pathology, Boston Medical Center, archives. Demographic characteristics of the catchment area suggest few cases would represent familial breast cancer. Review of the pathology findings also suggested a nonselected population: 17 of 18 cases were ductal carcinomas, 1 case (case 2008) was lobular, 2 cases (cases 0071R and 0071L) represented synchronous bilateral disease (analyzed separately); 80% of CIS had some evidence of high-grade disease (high nuclear grade, comedo histology, or necrosis) and 66% of invasive cancers were grade III/III.
Existing slides were reviewed by a single experienced breast pathologist (Adelas M) who identified multiple examples of normal ducts/TDLUs, and in situ and invasive carcinoma. Stroma or nodal tissue was available in 7 of 18 cases.
Microdissection and DNA Extraction and Quantitation
After identifying areas of interest on the hematoxylin and eosin (H&E)-stained slides, seven serial sections were cut from the corresponding blocks, and the top and bottom sections were stained with H&E. After reconfirming histology in these stained slides, they were used to guide microdissection of areas of interest from the unstained sections. Microdissection was performed using a laser capture microdissection apparatus (Arcturus Engineering, Mountain View, CA). 8 By counting nuclei and considering a cell to be 20 μm in diameter, we estimated that we obtained 200 to 1000 cells per normal-appearing sample, and considerably more cells per tumor sample. DNA was extracted using standard techniques that we have described previously. 5,9 DNA for control reactions was quantitated fluorimetrically (PicoGreen dsDNA Quantitation Kit; Molecular Probes, Eugene, OR).
Microsatellite Selection
Twenty-one microsatellite markers, located on 10 chromosomal arms, were selected for utility in fixed breast tissue based on the following criteria: 1) location at regions relevant to breast tumorigenesis (ie, regions of LOH in early-stage carcinomas, or at sites of identified or putative tumor suppressor genes). Markers at regions not believed relevant to breast tumorigenesis were also included; 2) size of amplified fragment <200 bp for reliable use in fixed tissue that produces fragmented template DNA; 3) highly polymorphic (ideally >75% heterozygosity); 4) ability to be multiplexed together without adverse interaction. Chromosomal regions and markers used were as follows: 1p: D1s468; 1q32-42: D1s549, D1s213; 3p24: D3s1283; 7q31: D7s486; 11p15: THO1, D11s2071; 11q13: PYGM; 11q23: D11s1818, D11s1819; 16q22-24: D16s265, D16s402, D16s413, D16s512; 17p13.1: TP53, D17s796, D17s525; 17q12-21: D17s1290, D17s579, D17s855; Xq11-12: AR. Primers were purchased from Research Genetics (Huntsville, AL) or synthesized commercially.
Polymerase Chain Reaction (PCR)/Electrophoresis
Six multiplexed PCRs were performed using ∼1/10 volume of the DNA solution as a template in a 50-μl reaction, 30 to 35 cycles of amplification, incorporation of α-32P-dCTP, and annealing temperatures between 55 to 60°C. One-fifth of the amplified products was electrophoresed through 7% denaturing gels that were then exposed to autoradiography film.
Determination of AI
The normal pattern at each microsatellite in each individual was defined as the pattern in stroma and nodes, or the predominant pattern in normal epithelium. LOH was defined at heterozygous loci as an imbalance of allele intensities >25%, ie, when (n1)(t2)/(n2)(t1) >1.33 or <0.75, where n1 = normal samples’ larger allele, n2 = normal samples’ smaller allele, t1 = test sample’s larger allele, t2 = test sample’s smaller allele. This degree of AI indicates that a substantial proportion of the cells within a sample contains the same DNA abnormality and likely represents the presence of a clonal population. Abnormal results were demonstrated at least twice with equivalent results. At certain loci AI probably reflects increased copy number rather than loss of an allele. Distinguishing between these possibilities is important conceptually, but would not change data analysis. Therefore, all AIs were labeled as LOH.
The proportion of LOH for each arm, or fractional allele loss (FAL), was calculated in each case as: number of LOH/number of observations per arm. This adjusts for multiple samples that may have separate patterns of LOH, which was common in normal-appearing samples. For each histology, overall FAL was calculated as the mean of all 10 arms’ FAL.
Statistical Tests
The exact Wilcoxon test was used to assess differences in median sample number between cases with versus without LOH in normal-appearing tissue. A t-test assessed differences in mean AI in control reactions versus abnormal duct/TDLUs. To assess global differences in FAL across arms within each histological type of tissue, we used analysis of variance methods that accounted for the correlated multiple observations coming from the same individual and weighted for the number of observations on which the calculation of FAL was based, using Proc Mixed in SAS. 10 We also used this strategy to evaluate specific hypotheses, ie, that different levels of LOH existed between specific arms in normal-appearing samples. Because of the weighting strategy, this approach adjusts for different numbers of samples per case and observations per sample.
Results
Samples
From 18 independent breast cancer-containing lumpectomy or mastectomy specimens, we microdissected 173 lesions including normal-appearing terminal ductal-lobular units (TDLUs), or rarely simple ducts, CIS, invasive carcinomas (Inv), and when available, uninvolved lymphoid tissue or stroma. Figure 1 ▶ illustrates a representative microdissection. Table 1 ▶ lists the number of samples per histology and specimen. Samples were taken from all available blocks: 28 of 109 ducts/TDLUs located on the same block as the cancer, the remainder came from elsewhere in the specimen.
Figure 1.

Laser capture microdissection (LCM) of fixed, unstained morphologically normal breast epithelium. A: H&E-stained section showing a normal-appearing TDLUs; B, adjacent unstained section; C, after microdissection; D, tissue on cap.
Table 1.
Microdissected Samples
| Case no. | No. of samples | |||
|---|---|---|---|---|
| Stroma, lymph node | Normal | CIS | Inv | |
| 2004 | – | 5 | 3 | – |
| 2008 | – | 4 | 2 | 2 |
| 2012 | – | 4 | – | 2 |
| 2014 | 1 | 1 | 1 | – |
| 2028 | 3 | 7 | 1 | – |
| 2031 | 2 | 5 | 3 | 2 |
| 2032 | 3 | 6 | – | 2 |
| 2034 | – | 9 | 1 | – |
| 2044 | – | 8 | – | 3 |
| 0038 | – | 5 | 3 | 5 |
| 0039 | – | 4 | 4 | – |
| 0052 | 1 | 8 | 2 | 1 |
| 0053 | 1 | 9 | 2 | 1 |
| 0070 | – | 4 | 3 | – |
| 0071R | – | 5 | 2 | 1 |
| 0071L | – | 4 | – | 1 |
| 0072 | 1 | 10 | 3 | – |
| 0074 | – | 11 | 2 | – |
| Total | 12 | 109 | 32 | 20 |
LOH in Controls
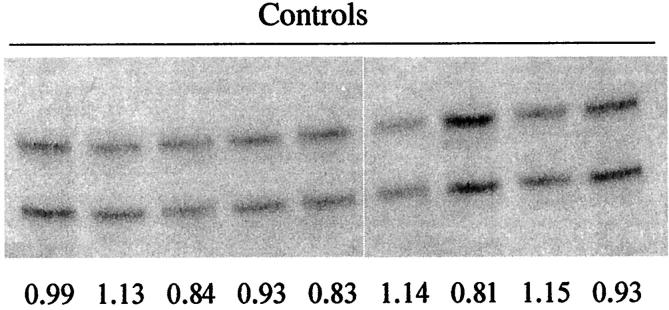
To establish the rate at which LOH might mistakenly be identified in normal-appearing ducts/TDLUs (ie, a false-positive rate), we performed 40 independent control PCRs. Each reaction contained 125 pg of lymph node DNA that had been microdissected and extracted using the same conditions as the breast samples. This template (reflecting the DNA content of ∼20 cells, our lowest estimated cell number per reaction), was amplified at two markers (D1s549 and D17s579). We found limited variation in allele ratios among these 40 reactions, with the mean allele ratio = 1.06, SD = 0.25 (Figure 2) ▶ . Eight of 40 (20%) reactions had allele ratios outside our predetermined cutoff values (>1.33, <0.75). Because our criteria for LOH require at least two independent demonstrations of abnormal allele ratios, this indicates that our estimated rate of false-positives is, at most, (0.2) × (0.2) = 0.04, or (4%). It is probably closer to 2% because artifactual imbalances should affect each allele half the time.
Figure 2.
Reproducibility of PCR. Representative examples from 40 independent control reactions, each amplifying marker D1s549 from 125 pg of template DNA. Normalized allele ratios, indicated below each lane, all fall within normal limits.
LOH in Breast Samples
Using multiplex PCR, DNA from each microdissected breast sample was analyzed at 21 microsatellite markers on 10 chromosome arms. On average, 11 markers were informative and interpretable per case. Some LOH could have been missed because of admixture of truly normal cells. Table 2 ▶ summarizes these data.
Table 2.
LOH in Histologically Normal Ducts/TDLUs versus Co-Existing Cancers
| Samples | No. (%) cases with LOH | No. (%) samples with LOH | No. (%) cases with same LOH in normal and cancer | No. (%) samples with same LOH in normal and cancer | Extent of LOH |
|---|---|---|---|---|---|
| Ducts/TDLUs | 8/18 (44%) | 14/109 (13%) | 1/8 (13%) | 1/14 (7%) | Single locus |
| In situ cancer | 14/14 (100%) | 29/32 (91%) | NA | NA | Multiple ≫ single |
| Invasive cancer | 9/10 (90%) | 19/20 (95%) | NA | NA | Multiple ≫ single |
Histologically Normal Epithelium
Eight of 18 (44%) cases contained one or more ducts/TDLUs with LOH. Overall, 14 of 109 (13%) normal-appearing samples contained 14 LOH, consistent with proportions reported previously. 5 Because the 95% confidence interval for this 13% rate is 7 to 19%, but the maximal estimated rate of artifactual LOH is 4%, these abnormalities are unlikely to be because of chance. In addition, the magnitude of AI in these 14 ducts/TDLUs was greater than in the eight control reactions whose ratios outside the cutoff [mean allele ratio, 2.15 (53% imbalance) versus 1.52 (34% imbalance), P = 0.03].
Ten cases contained no abnormalities; four cases contained a single duct/TDLUs with LOH; in three cases, separate ducts/TDLUs contained distinct sites of LOH. In two cases, two normal-appearing samples had LOH of the same marker and allele (Table 3) ▶ . These were considered independent, but could have represented a single, convoluted duct. There was a trend toward examination of more ducts/TDLUs in cases with LOH than cases without LOH (7.5 versus 4.5, P = 0.10), suggesting that detection of abnormalities may be related to the number of ducts/TDLUs examined. Detection of LOH in normal-appearing epithelium was not influenced by the subject’s age at diagnosis (45 years in women with LOH versus 46 years in women without LOH). All 14 samples contained a single abnormality.
Table 3.
Heterogeneous LOH in Histologically Normal Epithelium (N) versus Co-Existing Cancers
| Case no. | LOH |
|---|---|
| 2004 | |
| 2008 | |
| 2012 | |
| 2014 | |
| 2028 | 1 N has LOH at 16q, not seen in CIS |
| 1 N has LOH at 1q, not seen in CIS | |
| 2031 | 1 N has LOH at 16q site not seen in CIS and Inv |
| 2032 | |
| 2034 | 1 N has LOH at 17q, not seen in CIS |
| 1 N (N7) has LOH at 11p, not seen in CIS | |
| 2 Ns have LOH at 11p (distinct from N7), not seen in CIS | |
| 2044 | 1 N has LOH of opposite 1q allele as Inv |
| 0038 | |
| 0039 | 2 Ns have LOH at 17q site, not seen in CIS |
| 0052 | |
| 0053 | 1 N has LOH at 7q which is lost in CISand Inv |
| 1 N has LOH of opposite 1q allele as CIS and Inv | |
| 0070 | |
| 0071R | 1 N has LOH of opposite 1q allele as CIS and Inv |
| 0071L | |
| 0072 | 1 N has LOH of opposite 16q allele as CIS |
| 0074 |
To confirm each case’s constitutional pattern at each marker, stromal or nodal tissue was examined in the seven cases in which the tissue was available. Although LOH has been reported in stroma, 11,12 we microdissected multiple samples to obtain each case’s normal somatic pattern. In all seven cases, the stromal or nodal pattern at each marker was the same as the predominant pattern in normal-appearing breast epithelium (see section below). In the 11 cases that lacked stroma or lymph nodes, an average of 5.7 (range, 4 to 11) normal ducts/TDLUs were available to determine each marker’s normal pattern. Therefore, LOH in a single, normal-appearing sample is less likely to represent an aberration occurring early in breast development and more likely to represent a later genetic event.
Cancers
All cancers contained genetic abnormalities, usually multiple (Table 2) ▶ . Overall, 29 of 32 (91%) CIS samples from 14 of 14 (100%) evaluable cases contained 124 LOH; and 19 of 20 (95%) Inv samples from 9 of 10 (90%) cases contained 98 LOH. The pattern of LOH among all CIS or all Inv samples within a case was usually identical, implying that they derived from a single clone (data not shown). When they differed, it was most commonly because of the presence of a normal pattern, suggesting contamination with normal cells.
Stroma and Lymph Node
No LOH was seen in nine samples from three patients with stroma examined. In one of four (25%) cases (case 0072), one of six pathologically uninvolved lymph nodes demonstrated one LOH. This may represent the baseline rate of mutation in lymphoid tissue, although an occult metastasis cannot be ruled out because the cancer contained the same abnormality.
LOH in Relation to Distance between Ducts/TDLUs and Cancer
We asked whether LOH in ducts/TDLUs was related to a sample’s distance from the cancer. Twenty-eight of 109 (26%) ducts/TDLUs examined were located on the same block as cancers. Four of these 28 (14%) (all from case 2034), had LOH, but none of these 4 LOH were present in adjacent cancers. LOH was also seen in 10 of 81 (12%) ducts/TDLUs located on blocks not containing cancer. As described above, in only one case was a precursor-product relationship possible. Thus, although duct structure is convoluted, it seems that ducts/TDLUs more distant from the cancer were equally likely to contain abnormalities as samples nearer to the cancer. In fact, normal-appearing ducts/TDLUs adjacent to cancers did not contain abnormalities present in the malignancy.
Extent of LOH: Single Versus Multiple Loci
As shown in Table 2 ▶ , LOH in normal-appearing ducts/TDLUs encompassed only single markers, ie, additional informative loci on the same chromosome arm showed no LOH. In contrast, LOH in cancers usually encompassed all informative markers on a chromosome arm. Overall, LOH at all informative loci on one or more arms was seen in 0 of 18 (0%) histologically normal specimens, 10 of 14 (71%) CIS, and 9 of 9 (100%) Inv cancers.
Chromosome Arms Demonstrating LOH
LOH in normal-appearing tissue could indicate chromosome regions harboring tumor suppressor genes important early in breast carcinogenesis. 13 To identify these regions, we determined the proportional LOH on each chromosome arm for normal and malignant epithelial samples. The mean proportional LOH (or FAL) was 0.01 for normal epithelium, 0.27 for in situ cancer, and 0.30 for invasive cancers; the values for cancers are consistent with previous reports. 14,15 Results are shown in Table 4 ▶ .
Table 4.
Proportional LOH in Chromosome Arms
| Arm | Proportional LOH | ||
|---|---|---|---|
| Normal | CIS | Inv | |
| 1p | 0 | 0 | 0 |
| 1q | .02 | .48 | .50 |
| 3p | 0 | .19 | 0 |
| 7q | .01 | .07 | .06 |
| 11p | .02 | .35 | .23 |
| 11q | 0 | .32 | .41 |
| 16q | .02 | .65 | .79 |
| 17p | 0 | .38 | .64 |
| 17q | .02 | .29 | .37 |
| Xq | 0 | 0 | 0 |
| Mean: | 0.01 | 0.27 | 0.30 |
Perhaps because the overall number of LOH was relatively small, no significant global differences in proportional LOH between arms were detected in normal tissues (P = 0.39). [Global differences were suggested in in situ (P = 0.09) and detected in invasive cancers (P = 0.02).] However, we observed that arms with frequent, or rare, LOH in malignant tissue often showed the same relative frequency of LOH in normal-appearing tissue. For instance, in both normal and malignant tissue frequent LOH was seen on 1q and 16q, and infrequent LOH on 1p, 3p, Xq, and probably 7q. However, two arms had inconsistencies: LOH at 11q and 17p were common in cancer but completely absent in normal tissue. More focused analyses, comparing specific arms using the same analytical strategy, indicated significantly less LOH in normal tissue at sites on 11q and 17p than at sites on 1q (P = 0.002), 16q (P = 0.01), and possibly 17q (P = 0.06).
LOH in Co-Existing Normal-Appearing and Malignant Epithelium
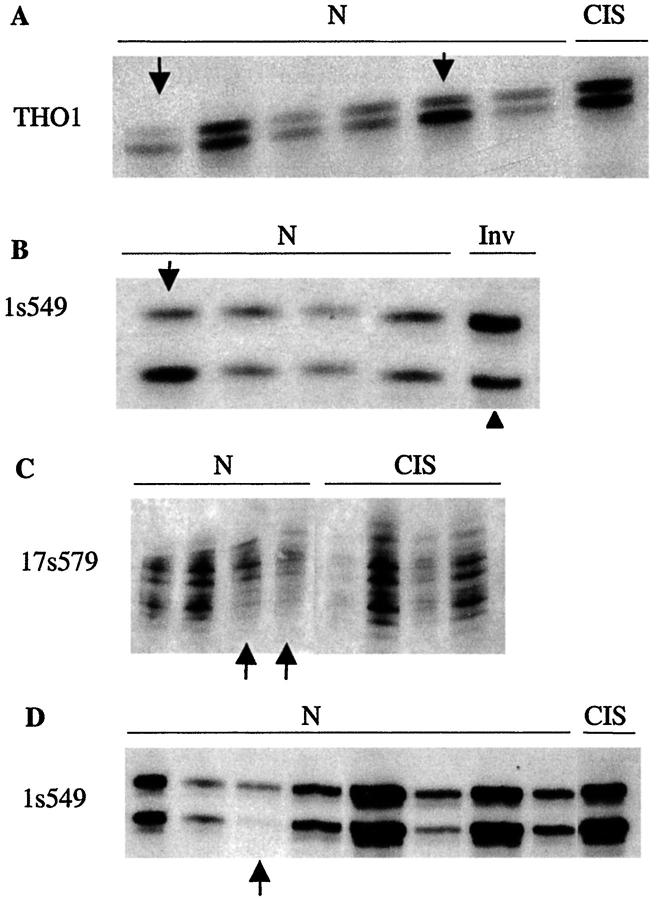
We evaluated whether identical abnormalities were present in morphologically normal ducts/TDLUs and co-existing cancers. If so, then the normal-appearing sample could represent a clonal precursor of the cancer. In contrast, LOH in normal but not malignant tissue suggests the presence of two independent clones. We found only 1 of 14 (7%), ducts/TDLUs with LOH, from 1 of 8 (13%) cases, that had the same LOH as the co-existing cancer (Table 2) ▶ . Representative examples are presented in Figure 3 ▶ and Table 3 ▶ summarizes the results. In the single case (case 0053) with LOH of the same site and allele in a normal-appearing ducts/TDLUs and the cancer, it is possible that the 7q LOH was coincidental, because this LOH was not seen in proliferative lesions that shared other LOH with the cancer (data not shown). We did find LOH of a marker’s opposite alleles in ducts/TDLUs compared with co-existing cancers (Figure 3B) ▶ . Thus, the great majority of ducts/TDLUs with LOH (13 of 14, or 93%) were clonally distinct from co-existing cancers.
Figure 3.
Morphologically normal ducts/TDLUs have LOH distinct from LOH in co-existing cancers. Examples from three cases (A and D, case 2034; B, case 2044; C, case 039) demonstrating that genetically abnormal ducts/TDLUs do not commonly share LOH with co-existing cancers. Arrows indicate lost alleles in duct/TDLUs, arrowhead indicates lost alleles in cancers, markers are listed at the left of each panel, and lesions are indicated by lettering across the top of each panel. N, normal; CIS, carcinoma in situ; INV, invasive tumor.
Discussion
This study reports the largest investigation to date of LOH or AI in histologically normal breast epithelium. The results demonstrate a consistent, low level of LOH [14 of 109 (13%) samples, overall proportional LOH = 0.01] in normal-appearing ducts/TDLUs from a large subset [8 of 18 (44%)] of breast cancer cases. The frequency and the magnitude of LOH/AI are greater than expected by chance. Ducts/TDLUs with LOH were found throughout the breast specimens. All LOH in normal-appearing epithelium involved single markers, whereas LOH in cancers commonly encompassed all informative markers on a chromosome arm. Remarkably, ducts/TDLUs with LOH were rarely implicated as cancer precursors, because their LOHs involved different markers, or different alleles of the same marker, as the co-existing cancer. Global differences in LOH per arm could not be demonstrated in normal-appearing tissue (perhaps because the overall number of LOHs is low), but sites of LOH did not appear to be completely random. Although the number of observations is small, we found less LOH at 11q and 17p than at 1q, 16q, and possibly 17q.
These results raise two primary points for consideration. First, what roles do ducts/TDLUs with LOH play? The ability to detect LOH indicates that a substantial proportion of the sample’s cells contain the identical genetic abnormality, ie, represent the progeny of a single cell. LOH at various sites might occur in individual cells, but it will be detected only if the cell subsequently undergoes clonal expansion. Clonal expansion can result from a growth advantage conferred on the cell by loss of a critical gene. (Although the function of the putative critical gene’s remaining allele is unknown, haploinsufficiency alone is capable of generating a phenotype.) Alternatively, clonal expansion could reflect normal mammary development because a genetic change occurring in the breast before puberty would probably be manifest as a detectable mutation in normal adult breast tissue. We favor the former explanation because LOH was found in scattered, single ducts/TDLUs, whereas the mammary gland’s stem-cell-derived monoclonal patches probably encompass larger areas and contain multiple TDLUs. 16-18
Also suggestive that at least some ducts/TDLUs with LOH result from loss of a critical gene, and may be meaningful clones, is that the sites of LOH were not entirely random. We noted LOH at 1q and 16q, and perhaps 17q, relatively frequently in both normal and malignant tissue, whereas LOH at 11q and 17p were noted only in malignancies. These results suggest that LOH at certain sites may be associated with clonal expansion, whereas LOH at other sites may be associated with later steps in tumor development. Based on a small number of LOH events, these results warrant further examination in a larger study. However, they are consistent with data suggesting that genes important early in breast tumorigenesis may be located on 1q and 16q, 14,19 whereas genes acting later may be on 17p (for instance, p53 20-22 ) or 11q. 22,23 We did not examine abnormalities of specific genes, but microsatellites were selected to be in the vicinity of several genes implicated in breast carcinogenesis, ie, p53 (on 17p13) CCND1 (cyclin D1 on 11q13), or ATM (on 11q23).
Thus, although the majority of normal-appearing clones may not be destined to evolve into invasive disease, we speculate that ducts/TDLUs with LOH, particularly at 1q or 16q, might constitute a reservoir from which more advanced lesions develop. This would be consistent with a previous report noting the same LOH in a cancer and adjacent TDLUs. 4 In the present study, ducts/TDLUs with the same LOH as the co-existing cancer may have been missed because of sampling (we did not enrich for ducts/TDLUs located near cancers), or because of obliteration of the original aberrant ducts/TDLUs by tumor growth. Their absence does not alter the conclusion that most ducts/TDLUs with LOH seem unrelated to co-existing cancers. Progression of more than one unrelated clone might contribute to the development of proliferative lesions, second malignancies or, uncommonly, synchronous independent tumors. Because it remains possible that an occult mutation is common to seemingly unrelated clones, progression of more than one clone could conceivably reflect clonal evolution and contribute to intratumoral heterogeneity. 24,25
An unanswered question that should help determine the roles played by ducts/TDLUs with LOH is whether the number of samples with LOH, or the fraction of cases with LOH in histologically normal epithelium, is increased in these cancer-containing breasts compared to presumed, but undefined, normal background rates. Additional studies will address this question more conclusively, but preliminary evidence has suggested that abnormalities increase in histologically normal epithelium as breast cancer risk increases. 5 This view would be consistent with breast cancer patients’ increased risk of cancer in the contralateral breast. 26,27 Because detection of LOH in the present study may have been related to the number of ducts/TDLUs examined (P = 0.10), if more samples were examined, an even larger proportion of cases might contain abnormalities. Similarly, because the location of ducts/TDLUs with LOH seemed unrelated to their distance from the cancer, if more samples were examined, more aberrant ducts/TDLUs might be found in each breast.
The second point for consideration is whether these results provide support for particular mechanisms implicated in human breast carcinogenesis. A majority [13 of 14 (93%)] of normal-appearing epithelial clones detected in the present study were distinct from, and thus were not precursors of, the co-existing cancer. The presence of multiple, distinct co-existing clones (ie, the cancer plus any ducts/TDLUs with LOH) is consistent with the hypothesis that some form of genetic instability, manifest in this study as LOH, may begin very early in breast tumorigenesis, while tissue is histologically normal. The geographic extent of any such area of instability is uncertain. Field abnormalities have been proposed previously in cancer-containing breasts. 28-30
The present results also suggest that the predominant mechanism(s) leading to LOH may change during tumor progression. LOH of limited chromosome regions, as we found in ducts/TDLUs, is consistent with mitotic recombination (although other mechanisms are plausible) 31-33 and might occur first. In contrast, LOH at all evaluable markers along an arm, as we found in cancers, would more likely result from loss of an entire chromosome (chromosome nondisjunction). 33 Chromosome number instabilities are characteristic of human cancers 34 and may be because of malfunction of the mitotic chromosome segregation apparatus. 35 Thus, mechanisms leading to LOH in limited chromosomal regions may contribute early to breast tumorigenesis, whereas mechanisms leading to whole chromosome abnormalities may contribute to late events. Consistent with this, a recent study posits aneuploidy as a late event in breast carcinogenesis, 36 and only rare cytogenetic abnormalities have been reported in normal-appearing tissue adjacent to cancers. 19,37
In summary, the current data indicate that clonal, genetically abnormal ducts/TDLUs are scattered throughout normal-appearing epithelium of cancerous breasts. These clones are distinct from the co-existing cancer, and could be a consequence either of normal development or of pathological events. It is possible that ducts/TDLUs with LOH form a reservoir from which cancers may develop if sufficient additional abnormalities accumulate. The presence of multiple clones (ie, the cancer plus any ducts/TDLUs with LOH) suggests that some type of genetic instability, affecting an undefined area of breast tissue, may contribute early to tumorigenesis.
Footnotes
Address reprint requests to Carol L. Rosenberg, M.D., Boston University Medical Center, 650 Albany St., EBRC-4, Boston, MA 02118. E-mail: crosenberg@medicine.bu.edu.
Supported by the Department of Defense Breast Cancer Research Program (grant DAMD 17-97-7191), the Massachusetts Department of Public Health Breast Cancer Research Program, and the National Institutes of Health (grant CA81078).
References
- 1.Kerangueven F, Noguchi T, Coulier F, Allione F, Wargniez V, Simony-Lafontaine J, Longy M, Jacquemier J, Sobol H, Eisinger F, Birnbaum D: Genome-wide search for loss of heterozygosity shows extensive genetic diversity of human breast carcinomas. Cancer Res 1997, 57:5469-5474 [PubMed] [Google Scholar]
- 2.Osborne RJ, Hamshere MG: A genome-wide map showing common regions of loss of heterozygosity/allelic imbalance in breast cancer. Cancer Res 2000, 60:3706-3712 [PubMed] [Google Scholar]
- 3.Lakhani SR: The transition from hyperplasia to invasive carcinoma of the breast. J Pathol 1999, 187:272-278 [DOI] [PubMed] [Google Scholar]
- 4.Deng G, Lu Y, Zlotnikov G, Thor AD, Smith HS: Loss of heterozygosity in normal tissue adjacent to breast carcinomas. Science 1996, 274:2057-2059 [DOI] [PubMed] [Google Scholar]
- 5.Larson PS, de las Morenas A, Cupples LA, Huang K, Rosenberg CL: Genetically abnormal clones in histologically normal breast tissue. Am J Pathol 1998, 152:1591-1598 [PMC free article] [PubMed] [Google Scholar]
- 6.Lakhani SR, Chaggar R, Davies S, Jones C, Collins N, Odel C, Stratton MR, O’Hare MJ: Genetic alterations in ‘normal’ luminal and myoepithelial cells of the breast. J Pathol 1999, 189:496-503 [DOI] [PubMed] [Google Scholar]
- 7.Washington C, Dalbegue F, Abreo F, Taubenberger JK, Lichy JH: Loss of heterozygosity in fibrocystic change of the breast: genetic relationship between benign proliferative lesions and associated carcinomas. Am J Pathol 2000, 157:323-329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zhuang Z, Goldstein SR, Weiss RA, Liotta L: Laser capture microdissection. Science 1996, 274:998-1001 [DOI] [PubMed] [Google Scholar]
- 9.Rosenberg CL, de las Morenas A, Huang K, Cupples LA, Faller DV, Larson PS: Detection of monoclonal microsatellite alterations in atypical breast hyperplasia. J Clin Invest 1996, 98:1095-1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Littell RC, Milliken GA, Stroup WW, Wolfinger RD: SAS System for Mixed Models. 1996. SAS Institute, Inc., Cary
- 11.Moinfar F, Man YG, Arnould L, Bratthauer GL, Ratschek M, Tavassoli FA: Concurrent and independent genetic alterations in the stromal and epithelial cells of mammary carcinoma: implications for tumorigenesis. Cancer Res 2000, 60:2562-2566 [PubMed] [Google Scholar]
- 12.Kurose K, Hoshaw-Woodard S, Adeyinka A, Lemeshow S, Watson PH, Eng C: Genetic model of multi-step breast carcinogenesis involving the epithelium and stroma: clues to tumour-microenvironment interactions. Hum Mol Genet 2001, 10:1907-1913 [DOI] [PubMed] [Google Scholar]
- 13.Chen LC, Kurisu W, Ljung BM, Goldman ES, Moore D, II, Smith HS: Heterogeneity for allelic loss in human breast cancer. J Natl Cancer Inst 1992, 84:506-510 [DOI] [PubMed] [Google Scholar]
- 14.Cornelisse CJ, Kuipers-Dijkshoorn N, van Vliet M, Hermans J, Devilee P: Fractional allelic imbalance in human breast cancer increases with tetraploidization and chromosome loss. Int J Cancer 1992, 50:544-548 [DOI] [PubMed] [Google Scholar]
- 15.Wistuba II, Tomlinson GE, Behrens C, Virmani A, Geradts J, Blum JL, Minna JD, Gazdar AF: Two identical triplet sisters carrying a germline brca1 gene mutation acquire very similar breast cancer somatic mutations at multiple other sites throughout the genome. Genes Chromosom Cancer 2000, 28:359-369 [DOI] [PubMed] [Google Scholar]
- 16.Tsai Y, Lu Y, Nichols PW, Zlotnikov G, Jones PA, Smith HS: Contiguous patches of normal human mammary epithelium derived from a single stem cell: implications for breast carcinogenesis. Cancer Res 1996, 56:402-404 [PubMed] [Google Scholar]
- 17.Diallo R, Schaefer KL, Poremba C, Shivazi N, Willmann V, Buerger H, Dockhorn-Dworniczak B, Boecker W: Monoclonality in normal epithelium and in hyperplastic and neoplastic lesions of the breast. J Pathol 2001, 193:27-32 [DOI] [PubMed] [Google Scholar]
- 18.Smith GH, Chepko G: Mammary epithelial stem cells. Microsc Res Tech 2001, 52:190-203 [DOI] [PubMed] [Google Scholar]
- 19.Cummings MC, Aubele M, Mattis A, Purdie D, Hutzler P, Hofler H, Werner M: Increasing chromosome 1 copy number parallels histological progression in breast carcinogenesis. Br J Cancer 2000, 82:1204-1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levine AJ: P53, the cellular gatekeeper for growth and division. Cell 1997, 88:323-331 [DOI] [PubMed] [Google Scholar]
- 21.Done SJ, Arneson NC, Ozcelik H, Redston M, Andrulis IL: P53 mutations in mammary ductal carcinoma in situ but not in epithelial hyperplasias. Cancer Res 1998, 58:785-789 [PubMed] [Google Scholar]
- 22.Tirkkonen M, Tanner M, Karhu R, Kallioniemi A, Isola J, Kallioniemi OP: Molecular cytogenetics of primary breast cancer by cgh. Genes Chromosom Cancer 1998, 21:177-184 [PubMed] [Google Scholar]
- 23.Aubele MM, Cummings MC, Mattis AE, Zitzelsberger HF, Walch AK, Kremer M, Hofler H, Werner M: Accumulation of chromosomal imbalances from intraductal proliferative lesions to adjacent in situ and invasive ductal breast cancer. Diagn Mol Pathol 2000, 9:14-19 [DOI] [PubMed] [Google Scholar]
- 24.Aubele M, Mattis A, Zitzelsberger H, Walch A, Kremer M, Hutzler P, Hofler H, Werner M: Intratumoral heterogeneity in breast carcinoma revealed by laser-microdissection and comparative genomic hybridization. Cancer Genet Cytogenet 1999, 110:94-102 [DOI] [PubMed] [Google Scholar]
- 25.Going JJ, Abd El-Monem HM, Craft JA: Clonal origins of human breast cancer. J Pathol 2001, 194:406-412 [DOI] [PubMed] [Google Scholar]
- 26.Broet P, de la Rochefordiere A, Scholl SM, Fourquet A, Mosseri V, Durand JC, Pouillart P, Asselain B: Contralateral breast cancer: annual incidence and risk parameters. J Clin Oncol 1995, 13:1578-1583 [DOI] [PubMed] [Google Scholar]
- 27.Rosen PP, Groshen S, Kinne DW, Norton L: Factors influencing prognosis in node-negative breast carcinoma: analysis of 767 t1n0m0/t2n0m0 patients with long-term follow-up. J Clin Oncol 1993, 11:2090-2100 [DOI] [PubMed] [Google Scholar]
- 28.Hassan HI, Walker RA: Decreased apoptosis in non-involved tissue from cancer-containing breasts. J Pathol 1998, 184:258-264 [DOI] [PubMed] [Google Scholar]
- 29.O’Connell JT, Shao ZM, Drori E, Basbaum CB, Barsky SH: Altered mucin expression is a field change that accompanies mucinous (colloid) breast carcinoma histogenesis. Hum Pathol 1998, 29:1517-1523 [DOI] [PubMed] [Google Scholar]
- 30.Umbricht CB, Evron E, Gabrielson E, Ferguson A, Marks J, Sukumar S: Hypermethylation of 14-3-3 sigma (stratifin) is an early event in breast cancer. Oncogene 2001, 20:3348-3353 [DOI] [PubMed] [Google Scholar]
- 31.Cavenee WK, Dryja TP, Phillips RA, Benedict WF, Godbout R, Gallie BL, Murphree AL, Strong LC, White RL: Expression of recessive alleles by chromosomal mechanisms in retinoblastoma. Nature 1983, 305:779-784 [DOI] [PubMed] [Google Scholar]
- 32.Luo G, Santoro IM, McDaniel LD, Nishijima I, Mills M, Youssoufian H, Vogel H, Schultz RA, Bradley A: Cancer predisposition caused by elevated mitotic recombination in bloom mice. Nat Genet 2000, 26:424-429 [DOI] [PubMed] [Google Scholar]
- 33.Thiagalingam S, Laken S, Willson JK, Markowitz SD, Kinzler KW, Vogelstein B, Lengauer C: Mechanisms underlying losses of heterozygosity in human colorectal cancers. Proc Natl Acad Sci USA 2001, 98:2698-2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitelman F, Johansson B, Mertens F: Catalogue of Chromosome Alterations in Cancer. 1994. Wiley, New York
- 35.Lengauer C, Kinzler KW, Vogelstein B: Genetic instabilities in human cancers. Nature 1998, 396:643-649 [DOI] [PubMed] [Google Scholar]
- 36.Rennstam K, Baldetorp B, Kytola S, Tanner M, Isola J: Chromosomal rearrangements and oncogene amplification precede aneuploidization in the genetic evolution of breast cancer. Cancer Res 2001, 61:1214-1219 [PubMed] [Google Scholar]
- 37.Botti C, Pescatore B, Mottolese M, Sciarretta F, Greco C, Di Filippo F, Gandolfo GM, Cavaliere F, Bovani R, Varanese A, Cianciulli AM: Incidence of chromosomes 1 and 17 aneusomy in breast cancer and adjacent tissue: an interphase cytogenetic study. J Am Coll Surg 2000, 190:530-539 [DOI] [PubMed] [Google Scholar]