Abstract
The chromosomal region 17q23 has been shown to be commonly amplified in breast tumors, especially those with poor prognosis. In addition to breast cancer, studies by comparative genomic hybridization have implicated the involvement of 17q23 in other tumor types as well. Here we performed a large-scale survey on the distribution and frequency of the 17q23 copy number increases across different tumor types using fluorescence in situ hybridization on tissue microarrays containing 4788 specimens. A total of 4429 tumor samples representing 166 different tumor categories and 359 normal tissue samples from 40 different tissue categories were analyzed. Successful hybridizations were observed in 3520 of the 4788 specimens (74%). Increased 17q23 copy number was detected in 15% of the evaluable specimens with tumors originating from the lung, mammary gland, and soft tissue being most frequently affected. Interestingly, high-level amplification was detected only in 2% of the tumors and was generally restricted to mammary tumors. In addition, we observed an association between the frequency of increased 17q23 copy number and tumor progression in various tumor types. These results indicate that increased 17q23 copy number occurs frequently in several different tumor types suggesting that increased dosage of genes in this region might play a role in development and progression of many tumor types.
Development and progression of cancer is driven by a step-wise accumulation of genetic alterations. Activation of oncogenes and other growth-promoting genes is an important part of this process and gene amplification is one of the mechanisms that leads to the activation of such genes. Several different chromosomal regions have been shown to be frequently amplified in human tumors and, in some instances, subsequent studies have successfully identified genes from these amplified regions that have a critical role in cancer development. 1,2
Genome-wide copy number analysis by comparative genomic hybridization has illustrated that 17q23 is one of the most frequently amplified chromosomal regions in breast cancer. 3,4 Recently, the 17q23 amplification was shown to be associated with poor prognosis of breast cancer 5 indicating that an increased dosage of one or more genes in this region is involved in the progression of this disease. In addition to breast cancer, increased copy number of the 17q23 region has also been observed by comparative genomic hybridization in tumors of the adrenal gland, 6 brain, 7 esophagus, 8 lung, 9,10 ovary, 11 stomach, 12 urinary bladder, 13 and uterus 14-16 indicating that 17q23-specific genes may also be important in the development of other tumor types.
Although the 17q23 amplification has been observed by comparative genomic hybridization in several different tumor types, these analyses have not covered the entire tumor spectrum and usually only a limited number of samples from a given tumor type has been analyzed. In addition, detailed analysis of the 17q23 amplicon has only been performed in breast cancer where we recently performed a microarray-based characterization of the 17q23 amplicon in breast cancer cell lines and in a large collection of primary breast tumors. 17 This analysis illustrated that in most breast cancers the amplification extends over a very large region at 17q23 and leads to simultaneous activation of several genes. 17
The purpose of the present study was to survey the distribution and frequency of the 17q23 amplification across a large variety of tumor types. We used the recently developed tissue microarray (TMA) technology in which hundreds of individual tumor biopsies from archival paraffin-embedded tissue blocks are arrayed into a new paraffin block. 18 Sections from such a TMA can be used for parallel high-throughput analysis of hundreds of tumor samples in a single experiment. Here, we studied the 17q23 amplification in vivo in 3520 tissue specimens using fluorescence in situ hybridization (FISH) to TMAs.
Materials and Methods
TMA
A total of 4788 tissue samples, representing 3709 primary tumors (from 135 different tumor categories), 720 metastases (of 31 different tumor categories), and 359 normal tissues (of 40 different tissue categories), were used in this study. The tissue samples were derived from the following organs or anatomical sites: fetus and placenta, brain, salivary glands, oral cavity, esophagus, stomach, small intestine, colon, appendix, anus, gallbladder, pancreas, liver, larynx, lung, kidney, urinary bladder, prostate, testis, ovary, uterus, vagina and vulva, mammary gland, adrenal gland, thyroid gland, parathyroid gland, thymus, nodal and lymphatic tissue, skin, and soft tissue.
The tissue specimens were placed on 10 different TMA blocks, each containing ∼500 specimens. The TMAs were constructed as previously described. 18 Briefly, a tissue-arraying instrument (Beecher Instruments, Silver Springs, MD) was used to create holes in a recipient paraffin block and to acquire cylindrical core tissue biopsies with a diameter of 0.6 mm from histologically representative areas of the donor blocks. The tissue core biopsies were transferred to the recipient paraffin block at defined array positions. Each tumor was represented only once on the array. Five-μm sections were cut from the TMA block using a microtome and an adhesive-coated tape-sectioning system (Instrumedics, Inc., Hackensack, NJ).
Copy Number Analysis by FISH
Two overlapping bacterial artificial chromosome clones (hRPK.15_K_2/AC005901 and hRPK.332_H_18/AC005746) were selected for FISH analysis. These clones represent the previously identified core region of the 17q23 amplification in breast cancer 17 and include the TBX2 gene that has been previously implicated as one of the putative targets for the 17q23 amplification in breast cancer. 19-21 The clones were labeled with SpectrumOrange (Vysis Inc., Downers Grove, IL) using random priming. A SpectrumGreen-labeled chromosome 17 centromere probe (Vysis) was used as reference.
Dual-color FISH was performed as previously described. 22 In brief, TMA sections were treated according to the Paraffin Pretreatment Reagent kit protocol (Vysis), denaturated at 94°C for 5 minutes in Tth-buffer [10 mmol/L Tris-HCl, pH 8.9 (25°C), 0.1 mol/L KCl, 1.5 mmol/L MgCl, 50 μg/ml bovine serum albumin, 0.05% Tween 20 (v/v)], treated with Proteinase K (10 μg/ml in phosphate-buffered saline) at 37°C for 10 minutes, dehydrated, and air-dried. Hybridization was performed overnight. After hybridization the slides were washed in 0.4× standard saline citrate/0.3% Nonidet P-40 at 72°C for 3 minutes and then counterstained with 4′, 6-diamidino-2-phenylindole in anti-fade solution.
FISH signals were evaluated using a Zeiss fluorescence microscope. Specimens were classified into three groups (normal, gain, and high-level amplification) based on the ratio between the 17q23 and the 17 centromere signals in the tumor cells. A minimum of 50 tumor cells was evaluated per specimen. The samples with a ratio of 1:1 were considered normal. If more than 10% of the tumor cells had a ratio different from 1:1 the specimens were classified as either gain (1:1<ratio <3:1) or high-level amplification (ratio ≥3:1 or tight clusters of signals).
Statistical Analysis
The association between increased 17q23 copy number and tumor progression was analyzed using Fisher’s exact test. All P values are two-sided.
Results
17q23 Copy Number Analysis by FISH to TMAs
The 17q23 copy number was determined in situ in 4788 tissue specimens. The tissue samples included 3709 primary tumors (from 135 different tumor categories), 720 metastases (from 31 different tumor categories), and 359 normal tissues (from 40 different tissue categories). Successful hybridizations were observed in 3520 of the 4788 specimens (74%) in the TMAs (Figure 1) ▶ . The failures were because of missing or unrepresentative samples (9%), autofluorescence of the tissue (10%), or poor hybridization quality (7%). Tissues containing large amounts of fat, such as lipomas, were most frequently nonanalyzable, most likely because of the melting of the fat during the hybridization procedure. 22
Figure 1.
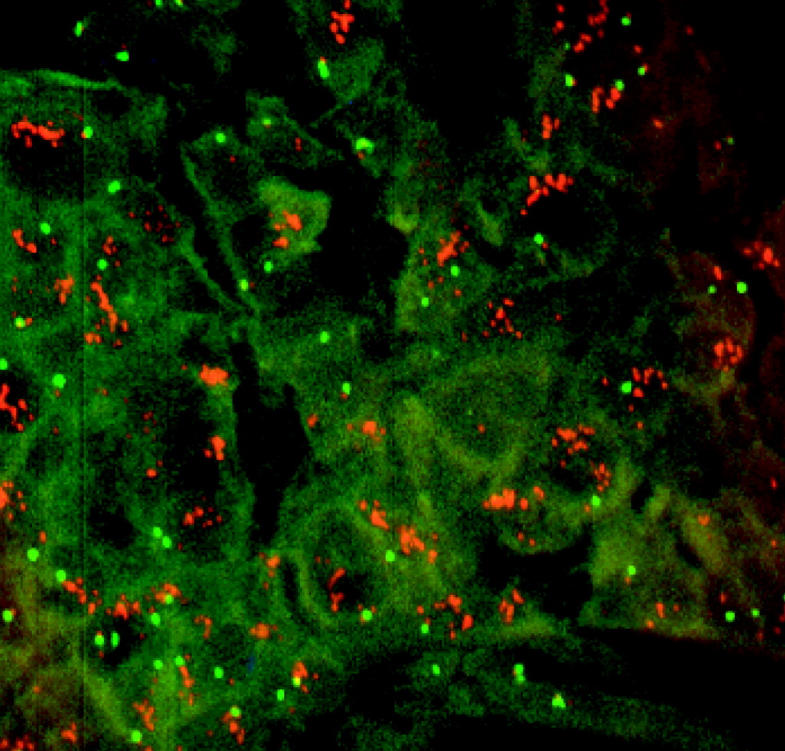
FISH analysis on TMA. The SpectrumOrange-labeled 17q23-specific probe (red signals) and SpectrumGreen-labeled chromosome 17 centromere probe (green signals) were hybridized to the TMAs. An example of a ductal carcinoma of the breast tumor sample with high-level amplification of 17q23 is shown.
Analysis of the normal tissue specimens demonstrated that all except 1 of the 245 evaluable samples showed the expected two signals for both 17q23 and centromere 17 probes. A single specimen of normal liver showed five signals for 17q23 and four signals for 17 centromere. A retrospective histological analysis using an adjacent section from the TMA did not reveal any abnormal cells in this particular sample. As normal liver cells can be polyploid 23 we expect that this particular sample represents such a case.
Frequent Increase in 17q23 Copy Number in Several Tumor Types
Evaluable hybridization results were obtained from a total of 3275 tumor samples and increased copy number of the 17q23 region was detected in 477 (15%) samples representing 86 (52%) of the 166 tumor categories examined (Table 1) ▶ . The frequency of increased copy number varied considerably from one tumor type to another with 65 tumor categories having at least 10% of the specimens affected (Table 1) ▶ . Several organs or anatomical sites, such as brain, lung, mammary gland, prostate, skin, soft tissue, and thyroid gland, showed increased copy number in multiple tumor categories (Table 1) ▶ . Increased copy number was particularly frequent in the lung, mammary gland, and soft tissue tumor categories (Figure 2) ▶ .
Table 1.
Increased 17q23 Copy Number by FISH Analysis on TMA
| Organ/anatomic site | Tumor category | No. of tumors* | Increased copy number (%) | High-level amplification (%) |
|---|---|---|---|---|
| Adrenal gland | Pheochromocytoma | 20 | 30 | 5 |
| Brain | Astrocytoma | 22 | 9 | 0 |
| Brain | Glioblastoma multiforme | 31 | 6 | 0 |
| Brain | Meningeoma | 28 | 14 | 0 |
| Brain | Oligodendroglioma | 15 | 47 | 0 |
| Colon | Adenocarcinoma | 276 | 0.4 | 0 |
| Colon | Adenocarcinoma (metastases) | 32 | 9 | 0 |
| Esophagus | Squamous cell carcinoma | 15 | 33 | 0 |
| Gallbladder | Adenoma | 13 | 23 | 0 |
| Hematology | Non-Hodgkin lymphoma | 84 | 6 | 0 |
| Larynx | Squamous cell carcinoma | 21 | 43 | 0 |
| Liver | Hepatocellular carcinoma | 47 | 17 | 0 |
| Lung | Adenocarcinoma | 28 | 32 | 4 |
| Lung | Carcinoid | 11 | 9 | 0 |
| Lung | Large cell carcinoma | 34 | 56 | 0 |
| Lung | Small cell carcinoma | 15 | 20 | 0 |
| Lung | Squamous cell carcinoma | 27 | 30 | 0 |
| Mammary gland | DCIS | 21 | 10 | 5 |
| Mammary gland | Invasive carcinoma, ductal | 41 | 22 | 15 |
| Mammary gland | Invasive carcinoma, ductal (metastases) | 342 | 40 | 11 |
| Mammary gland | Invasive carcinoma, lobular | 28 | 7 | 0 |
| Mammary gland | Invasive carcinoma, lobular (metastases) | 69 | 16 | 4 |
| Mammary gland | Invasive carcinoma, medullary | 32 | 13 | 0 |
| Mammary gland | Invasive carcinoma, mucinous | 11 | 27 | 9 |
| Metastases | Adenocarcinoma (unknown primary) | 16 | 44 | 0 |
| Oral cavity | Squamous cell carcinoma | 19 | 5 | 0 |
| Oral cavity | Squamous cell carcinoma (metastases) | 26 | 12 | 0 |
| Ovary | Adenocarcinoma, endometroid | 35 | 11 | 3 |
| Ovary | Adenocarcinoma, serous | 45 | 9 | 0 |
| Pancreas | Adenocarcinoma | 22 | 14 | 0 |
| Parathyroid | Adenoma | 17 | 18 | 0 |
| Prostate | Adenocarcinoma (metastases) | 13 | 31 | 0 |
| Prostate | Adenocarcinoma, hormone refractory | 13 | 46 | 0 |
| Prostate | Adenocarcinoma, untreated | 63 | 3 | 0 |
| Salivary glands | Adenolymphoma | 25 | 4 | 0 |
| Skin | Cylindroma | 17 | 12 | 0 |
| Skin | Malignant melanoma | 63 | 16 | 0 |
| Skin | Squamous cell carcinoma | 28 | 14 | 4 |
| Soft tissue | Leiomyosarcoma | 46 | 41 | 0 |
| Soft tissue | Liposarcoma | 22 | 32 | 0 |
| Soft tissue | Malignant fibrous histiocytoma | 18 | 50 | 0 |
| Soft tissue | Malignant mesenchymoma | 10 | 10 | 0 |
| Soft tissue | Malignant mesothelioma | 10 | 50 | 0 |
| Soft tissue | Malignant schwannoma | 10 | 10 | 10 |
| Soft tissue | Neurofibroma | 23 | 9 | 0 |
| Soft tissue | PNET | 19 | 58 | 0 |
| Soft tissue | Rhabdomyosarcoma | 14 | 57 | 0 |
| Soft tissue | Schwannoma | 45 | 11 | 0 |
| Stomach | Adenocarcinoma | 113 | 15 | 2 |
| Thymus | Thymoma | 20 | 5 | 0 |
| Thyroid gland | Adenoma | 23 | 13 | 0 |
| Thyroid gland | Carcinoma, follicular | 29 | 3 | 0 |
| Thyroid gland | Carcinoma, medullary | 13 | 8 | 0 |
| Thyroid gland | Carcinoma, papillary | 26 | 4 | 0 |
| Urinary bladder | TCC, invasive | 46 | 17 | 0 |
| Urinary bladder | TCC, non invasive | 74 | 5 | 1 |
| Uterus | Adenocarcinoma, endometroid | 117 | 4 | 0.9 |
| Uterus, cervix | Carcinoma in situ | 13 | 8 | 0 |
| Uterus, cervix | Squamous cell carcinoma | 24 | 4 | 0 |
| Vagina and vulva | Squamous cell carcinoma, vulva | 42 | 14 | 0 |
| Brain | Medulloblastoma | 4 | 75 | 0 |
| Esophagus | Adenocarcinoma | 3 | 33 | 0 |
| Esophagus | Squamous cell carcinoma (metastases) | 2 | 50 | 0 |
| Larynx | Squamous cell carcinoma (metastases) | 3 | 33 | 0 |
| Lung | Adenocarcinoma (metastases) | 8 | 50 | 0 |
| Lung | Squamous cell carcinoma (metastases) | 6 | 17 | 0 |
| Mammary gland | Adenocarcinoma (metastases) | 7 | 43 | 0 |
| Mammary gland | Invasive carcinoma, cribriform | 6 | 17 | 17 |
| Mammary gland | Invasive carcinoma, medullary (metastases) | 1 | 100 | 100 |
| Mammary gland | Invasive carcinoma, mucinous (metastases) | 1 | 100 | 100 |
| Mammary gland | Invasive carcinoma, papillary | 7 | 43 | 0 |
| Metastases | Squamous cell carcinoma (unknown primary) | 7 | 14 | 0 |
| Oral Cavity | Adenocarcinoma (metastases) | 2 | 50 | 0 |
| Ovary | Adenocarcinoma (metastases) | 5 | 20 | 0 |
| Ovary | Carcinoma | 2 | 50 | 0 |
| Pancreas | Adenocarcinoma (metastases) | 3 | 67 | 0 |
| Papilla vateri | Adenocarcinoma (metastases) | 2 | 100 | 100 |
| Skin | Squamous cell carcinoma (metastases) | 3 | 33 | 0 |
| Soft tissue | Angiosarcoma | 1 | 100 | 0 |
| Soft tissue | Dermatofibrosarcoma | 4 | 50 | 25 |
| Soft tissue | Fibrosarcoma | 6 | 50 | 0 |
| Soft tissue | Haemangiopericytoma | 5 | 40 | 0 |
| Stomach | Adenocarcinoma (metastases) | 7 | 29 | 0 |
| Thyroid gland | Carcinoma, anaplastic | 6 | 33 | 33 |
| Uterus | Malignant muellerian mixed tumor | 4 | 50 | 0 |
| Uterus, cervix | Adenocarcinoma | 4 | 25 | 0 |
*Number of interpretable tumors is shown. Tumor categories with less than 10 analyzable samples are shown at the bottom of the table.
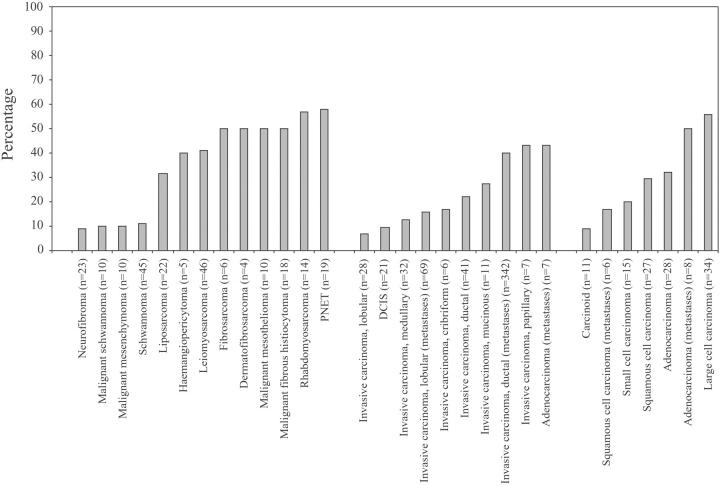
Figure 2.
The frequency of increased 17q23 copy number in different tumor categories originating from lung, mammary gland, and soft tissue.
For example, a total of 45 (35%) of the 129 interpretable lung tumor specimens showed increased copy number with large cell carcinomas (56% of the cases) and metastases of lung adenocarcinomas (50%) being most frequently affected. In addition, increased 17q23 copy number was also observed in adenocarcinomas (32%), squamous cell carcinomas (30%), small cell carcinomas (20%), squamous cell carcinoma metastases (17%), and carcinoid tumors (9%) of the lung (Table 1 ▶ , Figure 2 ▶ ). Similarly for tumors of the soft tissue, 76 (21%) of the 355 analyzable samples showed increased copy number with tumor categories, such as neuroepithelioma (PNET), rhabdomyosarcoma, malignant fibrous histiocytoma, and malignant mesothelioma, demonstrating increased copy number in more than half of the samples (Table 1 ▶ , Figure 2 ▶ ). Other very frequently affected tumor categories included oligodendroglioma, hormone refractory prostate cancer, squamous cell carcinoma of the larynx, leiomyosarcoma, and metastasis of ductal invasive breast cancer (Table 1) ▶ . Increased copy number of 17q23 was also observed in several tumor types that were represented by a small number of specimens (less than 10 analyzable samples) in the TMAs (Table 1) ▶ . For example, gain of 17q23 was detected in a single sample of angiosarcoma, three of four medulloblastomas, and in two of three metastases of pancreatic adenocarcinoma.
As expected, increased copy number of 17q23 was frequently observed in the tumor categories originating from the mammary gland. Out of the 589 mammary tumor samples analyzed 176 (30%) showed increased 17q23 copy number (Table 1) ▶ . The frequency of increased copy number varied among the different subtypes of mammary tumors. The most frequently affected tumor categories were metastases of ductal invasive carcinomas (40% of the cases), mucinous carcinomas (27%), and ductal carcinomas (22%).
High-Level Amplification of 17q23
Increased copy number in the form of high-level amplification of 17q23 was observed in 65 specimens (2%) distributed throughout 19 tumor categories representing adrenal gland, lung, mammary gland, ovary, skin, soft tissue, stomach, thyroid gland, urinary bladder, and uterus (Table 1) ▶ . Even though high-level amplification was observed in tumors from several organs the majority of affected specimens (78%) were derived from the mammary gland (Table 1 ▶ , Figure 2 ▶ ). The frequency of high-level amplification varied from 1 to 15% among the different malignancies. Invasive ductal breast cancer was the most common cancer subtype exhibiting 17q23 amplification (Table 1) ▶ . Interestingly, there was also evidence for frequent high-level amplification in some rare tumor types of which only a few samples could be analyzed, such as anaplastic thyroid carcinomas (amplification in two of the six samples analyzed), metastases of adenocarcinoma of papilla vateri (two of two), and dermatofibrosarcoma (one of four).
Analysis of Primary Tumors and Metastases
Increased copy number of 17q23 was observed in an average of 11% of primary tumors and 33% of metastases. For example, the average frequency of increased copy number was 14% in primary mammary tumors and 36% in breast cancer metastases. Similarly, increased 17q23 copy number was rare in untreated primary prostate tumors (3%) whereas it was frequent in late-stage prostate tumors (46% of hormone refractory adenocarcinomas and 31% of metastases). Furthermore, only 5% of the noninvasive transitional cell carcinomas of the urinary bladder were affected whereas 17% of the invasive tumors showed increased copy number. Statistical analysis in tumor categories in which both primary tumors and metastases were available showed a clear association between increased copy number at 17q23 and tumor progression (Table 2) ▶ . This association was statistically significant for colon adenocarcinomas, mammary ductal carcinomas, and prostate adenocarcinomas (Table 2) ▶ as well as when all primaries and metastases were combined (P < 0.0001).
Table 2.
Comparison on the Frequency of Increased 17q23 Copy Number Between Primary Tumors and Metastases from the Same Tumor Category*
| Organ/anatomic site | Tumor category | Frequency of increased copy number (n†) | Statistical significance | |
|---|---|---|---|---|
| Primary tumors | Metastases | |||
| Colon | Adenocarcinoma | 0.4% (276) | 9% (32) | P = 0.004 |
| Mammary gland | Carcinoma, ductal | 22% (41) | 40% (342) | P = 0.03 |
| Mammary gland | Carcinoma, lobular | 7% (28) | 16% (69) | n.s.‡ |
| Oral cavity | Squamous cell carcinoma | 5% (19) | 12% (26) | n.s.‡ |
| Prostate | Adenocarcinoma | 3% (63) | 31% (13) | P = 0.007 |
*For tumor categories with 10 or more evaluable specimens.
†Total number of tumors evaluated.
‡Not significant.
Discussion
This study represents the first large-scale analysis of copy number changes in thousands of tissue samples across all different tumor types using FISH to TMAs and thus illustrates the utility of TMA technology for large-scale surveys to characterize the disease specificity of genomic alterations at the population level. Here we used TMAs to evaluate the frequency of increased copy number at 17q23 in 4788 archival tissue specimens representing 206 different tissue categories. FISH on archival tissue sections can be difficult, and it is likely that for optimal performance case-by-case adjustments to hybridization protocols are required. However in this study, our overall success rate for FISH on TMAs constructed from archival specimens was 74% across a wide variety of tumor types. This result demonstrates that the concept of using FISH on multitumor TMAs is robust enough to be used as a screening tool for finding disease associations for molecular alterations.
The present study showed increased 17q23 copy number in 15% of the tumors representing 86 of the 166 different tumor categories analyzed. Increased copy number was observed frequently (in at least 10% of the specimens) in 65 of these categories, although there was considerable variation in frequency from one tumor type to another. The 166 different tumor categories evaluated in this study cover malignancies arising from almost every organ of the body. Interestingly, cancers emerging from only a few of these organs, such as small intestine, kidney, and testis, never exhibited abnormal 17q23 copy number. Based on our results, 17q23 copy number increases are common in human malignancies and may contribute to the development and progression of cancer in a variety of cellular environments.
The increased 17q23 copy number was observed most commonly in tumors derived from brain, lung, mammary gland, ovary, soft tissue, and urinary bladder. These results are in concordance with previous data from comparative genomic hybridization studies showing recurrent 17q23 gains in these tumor types. 3,4,7,9-11,13 We also observed increased 17q23 copy number in hormone refractory prostate cancer and prostate cancer metastases confirming a recent report on 17q23-q24 amplification in advanced prostate cancer. 24
As expected, increased 17q23 copy number was most common in the mammary tumors. The frequency of increased copy number varied among different subtypes of mammary tumors with an average of 14% in primary tumors. These results confirm the previous data on the high frequency of the 17q23 amplification in primary breast cancer. 5,17,20,21 The data also show that increased 17q23 copy number is not restricted to certain histological subtypes of breast cancer. The only breast tumor subtypes in which 17q23 copy number increases were not observed were phylloides tumor, tubular and apocrine carcinomas, as well as metastases of papillary adenocarcinoma. However, only a relatively small number of samples representing these tumor categories were examined in this study.
High-level amplification of 17q23 was observed in only 2% of the tumor samples. Although these tumors belong to 19 different tumor categories, most of them originate from the mammary gland indicating that high-level amplification of 17q23 is rather specific to mammary tumors. Several other chromosomal regions that are frequently amplified in mammary tumors do not show such tissue specificity. For example, the 20q13 amplification that was initially identified in breast cancer 25 is commonly observed in several other tumor types, such as bladder, colorectal, gastric, and ovarian cancers. 26-29 These results suggest that mammary tumors might gain a special advantage of a very high 17q23 gene dosage, or that the genes affected by this amplification have a special function in mammary tumorigenesis. It has to be noted, however, that this FISH study was performed using a custom-made 17q23-specific probe set that corresponds to the previously identified core region of the 17q23 amplification in breast cancer. 17 Although our previous data indicates that in breast tumors the region affected by increased copy number at 17q23 is rather large, 17 it is possible that smaller amplicons exist in other tumor types and that the location of the core of the amplicon might be different. Therefore, these amplicons could have been missed by the probe set used in this study.
Comparison of the frequency of increased 17q23 copy number between unmatched primary tumors and metastases illustrated a clear association between increased copy number and advanced tumor stage. This association was observed for all tumor categories in which primaries and metastases were available for analysis. In addition, a marked increase in the frequency of the 17q23 alterations was observed in transition from noninvasive to invasive bladder tumors and from untreated to hormone refractory prostate cancer. Together with the previously reported association between 17q23 amplification and poor prognosis in breast cancer, 5 these data strongly argue for an important role of one or several 17q23 genes for tumor progression.
Traditionally, discoveries of new amplification regions or putative amplification target genes originate from the analysis of a single tumor type. Subsequent studies are then required to evaluate the possible involvement of these newly discovered genes or loci in other tumor types. This is usually a laborious and slow process and therefore, leads to a considerable delay from the discovery of a potentially important genetic alteration to the definition of the tumor types in which it may play a role. In a pilot study using a mini multitumor TMA containing samples from 17 different tumor types, we have previously shown that representative frequencies of gene amplification can be obtained on TMAs. 30 Here we illustrate that using the TMA technology it is now possible to define virtually all tumor types that are influenced by a specific genetic alteration in one single high-throughput study of thousands of tumor specimens representing hundreds of different tumor types. The present study is the first in which almost all tumor types have been surveyed for the presence of a specific gene amplification. We predict that the use of the TMA technology for such surveys will facilitate rapid translation of gene discoveries to therapeutic and diagnostic applications in the future.
Acknowledgments
We thank Dr. Eigil Kjeldsen for his assistance in the statistical analysis.
Footnotes
Address reprint requests to Anne Kallioniemi, Laboratory of Cancer Genetics, Institute of Medical Technology, University of Tampere, P.O. Box 607, FIN-33014, University of Tampere, Finland. E-mail: anne.kallioniemi@uta.fi.
Supported in part by grants from the Danish Research Agency (52-00-0486 to C. L. A.), the Aarhus University Hospital Research Initiative (to C. L. A.), the Danish Cancer Society (98 216 54 to C. L. A.), and the Medical Research Fund of Tampere University Hospital (to M. B.).
References
- 1.Forozan F, Karhu R, Kononen J, Kallioniemi A, Kallioniemi OP: Genome screening by comparative genomic hybridization. Trends Genet 1997, 13:405-409 [DOI] [PubMed] [Google Scholar]
- 2.Gray JW, Collins C: Genome changes and gene expression in human solid tumors. Carcinogenesis 2000, 21:443-452 [DOI] [PubMed] [Google Scholar]
- 3.Courjal F, Theillet C: Comparative genomic hybridization analysis of breast tumors with predetermined profiles of DNA amplification. Cancer Res 1997, 57:4368-4377 [PubMed] [Google Scholar]
- 4.Tirkkonen M, Tanner M, Karhu R, Kallioniemi A, Isola J, Kallioniemi OP: Molecular cytogenetics of primary breast cancer by CGH. Genes Chromosom Cancer 1998, 21:177-184 [PubMed] [Google Scholar]
- 5.Barlund M, Forozan F, Kononen J, Bubendorf L, Chen Y, Bittner ML, Torhorst J, Haas P, Bucher C, Sauter G, Kallioniemi OP, Kallioniemi A: Detecting activation of ribosomal protein S6 kinase by complementary DNA and tissue microarray analysis. J Natl Cancer Inst 2000, 92:1252-1259 [DOI] [PubMed] [Google Scholar]
- 6.Dannenberg H, Speel EJ, Zhao J, Saremaslani P, van Der Harst E, Roth J, Heitz PU, Bonjer HJ, Dinjens WN, Mooi WJ, Komminoth P, de Krijger RR: Losses of chromosomes 1p and 3q are early genetic events in the development of sporadic pheochromocytomas. Am J Pathol 2000, 157:353-359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khan J, Parsa NZ, Harada T, Meltzer PS, Carter NP: Detection of gains and losses in 18 meningiomas by comparative genomic hybridization. Cancer Genet Cytogenet 1998, 103:95-100 [DOI] [PubMed] [Google Scholar]
- 8.Pack SD, Karkera JD, Zhuang Z, Pak ED, Balan KV, Hwu P, Park WS, Pham T, Ault DO, Glaser M, Liotta L, Detera-Wadleigh SD, Wadleigh RG: Molecular cytogenetic fingerprinting of esophageal squamous cell carcinoma by comparative genomic hybridization reveals a consistent pattern of chromosomal alterations. Genes Chromosom Cancer 1999, 25:160-168 [DOI] [PubMed] [Google Scholar]
- 9.Petersen I, Bujard M, Petersen S, Wolf G, Goeze A, Schwendel A, Langreck H, Gellert K, Reichel M, Just K, du Manoir S, Cremer T, Dietel M, Ried T: Patterns of chromosomal imbalances in adenocarcinoma and squamous cell carcinoma of the lung. Cancer Res 1997, 57:2331-2335 [PubMed] [Google Scholar]
- 10.Ried T, Petersen I, Holtgreve-Grez H, Speicher MR, Schrock E, du Manoir S, Cremer T: Mapping of multiple DNA gains and losses in primary small cell lung carcinomas by comparative genomic hybridization. Cancer Res 1994, 54:1801-1806 [PubMed] [Google Scholar]
- 11.Sonoda G, Palazzo J, du Manoir S, Godwin AK, Feder M, Yakushiji M, Testa JR: Comparative genomic hybridization detects frequent overrepresentation of chromosomal material from 3q26, 8q24, and 20q13 in human ovarian carcinomas. Genes Chromosom Cancer 1997, 20:320-328 [PubMed] [Google Scholar]
- 12.Koo SH, Kwon KC, Shin SY, Jeon YM, Park JW, Kim SH, Noh SM: Genetic alterations of gastric cancer: comparative genomic hybridization and fluorescence in situ hybridization studies. Cancer Genet Cytogenet 2000, 117:97-103 [DOI] [PubMed] [Google Scholar]
- 13.Voorter C, Joos S, Bringuier PP, Vallinga M, Poddighe P, Schalken J, du Manoir S, Ramaekers F, Lichter P, Hopman A: Detection of chromosomal imbalances in transitional cell carcinoma of the bladder by comparative genomic hybridization. Am J Pathol 1995, 146:1341-1354 [PMC free article] [PubMed] [Google Scholar]
- 14.Suehiro Y, Umayahara K, Ogata H, Numa F, Yamashita Y, Oga A, Morioka H, Ito T, Kato H, Sasaki K: Genetic aberrations detected by comparative genomic hybridization predict outcome in patients with endometrioid carcinoma. Genes Chromosom Cancer 2000, 29:75-82 [DOI] [PubMed] [Google Scholar]
- 15.Sonoda G, du Manoir S, Godwin AK, Bell DW, Liu Z, Hogan M, Yakushiji M, Testa JR: Detection of DNA gains and losses in primary endometrial carcinomas by comparative genomic hybridization. Genes Chromosom Cancer 1997, 18:115-125 [DOI] [PubMed] [Google Scholar]
- 16.Kiechle M, Hinrichs M, Jacobsen A, Luttges J, Pfisterer J, Kommoss F, Arnold N: Genetic imbalances in precursor lesions of endometrial cancer detected by comparative genomic hybridization. Am J Pathol 2000, 156:1827-1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Monni O, Barlund M, Mousses S, Kononen J, Sauter G, Heiskanen M, Paavola P, Avela K, Chen Y, Bittner ML, Kallioniemi A: Comprehensive copy number and gene expression profiling of the 17q23 amplicon in human breast cancer. Proc Natl Acad Sci USA 2001, 98:5711-5716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kononen J, Bubendorf L, Kallioniemi A, Barlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G, Kallioniemi OP: Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med 1998, 4:844-847 [DOI] [PubMed] [Google Scholar]
- 19.Bärlund M, Monni O, Kononen J, Torhorst J, Sauter G, Kallioniemi O-P, Kallioniemi A: Multiple genes at 17q23 undergo amplification and overexpression in breast cancer. Cancer Res 2000, 60:5340-5344 [PubMed] [Google Scholar]
- 20.Wu GJ, Sinclair CS, Paape J, Ingle JN, Roche PC, James CD, Couch FJ: 17q23 amplifications in breast cancer involve the PAT1, RAD51C, PS6K, and SIGma1B genes. Cancer Res 2000, 60:5371-5375 [PubMed] [Google Scholar]
- 21.Jacobs JJ, Keblusek P, Robanus-Maandag E, Kristel P, Lingbeek M, Nederlof PM, van Welsem T, van de Vijver MJ, Koh EY, Daley GQ, van Lohuizen M: Senescence bypass screen identifies TBX2, which represses Cdkn2a (p19(ARF)) and is amplified in a subset of human breast cancers. Nat Genet 2000, 26:291-299 [DOI] [PubMed] [Google Scholar]
- 22.Andersen CL, Hostetter G, Grigoryan A, Sauter G, Kallioniemi A: Improved procedure for fluorescence in situ hybridization on tissue microarrays. Cytometry 2001, 45:83-86 [DOI] [PubMed] [Google Scholar]
- 23.Gupta S: Hepatic polyploidy and liver growth control. Semin Cancer Biol 2000, 10:161-171 [DOI] [PubMed] [Google Scholar]
- 24.El Gedaily A, Bubendorf L, Willi N, Fu W, Richter J, Moch H, Mihatsch MJ, Sauter G, Gasser TC: Discovery of new DNA amplification loci in prostate cancer by comparative genomic hybridization. Prostate 2001, 46:184-190 [DOI] [PubMed] [Google Scholar]
- 25.Kallioniemi A, Kallioniemi OP, Piper J, Tanner M, Stokke T, Chen L, Smith HS, Pinkel D, Gray JW, Waldman FM: Detection and mapping of amplified DNA sequences in breast cancer by comparative genomic hybridization. Proc Natl Acad Sci USA 1994, 91:2156-2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Savelieva E, Belair CD, Newton MA, DeVries S, Gray JW, Waldman F, Reznikoff CA: 20q gain associates with immortalization: 20q13.2 amplification correlates with genome instability in human papillomavirus 16 E7 transformed human uroepithelial cells. Oncogene 1997, 14:551-560 [DOI] [PubMed] [Google Scholar]
- 27.Hidaka S, Yasutake T, Takeshita H, Kondo M, Tsuji T, Nanashima A, Sawai T, Yamaguchi H, Nakagoe T, Ayabe H, Tagawa Y: Differences in 20q13.2 copy number between colorectal cancers with and without liver metastasis. Clin Cancer Res 2000, 6:2712-2717 [PubMed] [Google Scholar]
- 28.Sakakura C, Hagiwara A, Yasuoka R, Fujita Y, Nakanishi M, Masuda K, Shimomura K, Nakamura Y, Inazawa J, Abe T, Yamagishi H: Tumour-amplified kinase BTAK is amplified and overexpressed in gastric cancers with possible involvement in aneuploid formation. Br J Cancer 2001, 84:824-831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanner MM, Grenman S, Koul A, Johannsson O, Meltzer P, Pejovic T, Borg A, Isola JJ: Frequent amplification of chromosomal region 20q12–q13 in ovarian cancer. Clin Cancer Res 2000, 6:1833-1839 [PubMed] [Google Scholar]
- 30.Schraml P, Kononen J, Bubendorf L, Moch H, Bissig H, Nocito A, Mihatsch MJ, Kallioniemi OP, Sauter G: Tissue microarrays for gene amplification surveys in many different tumor types. Clin Cancer Res 1999, 5:1966-1975 [PubMed] [Google Scholar]