Abstract
t(11;18)(q21;q21) and aneuploidy are recurrent chromosomal aberrations in mucosa-associated lymphoid tissue (MALT) lymphomas. To investigate their relationship and clinical significance, we developed a two-color fluorescence in situ hybridization (FISH) technique to detect t(11;18) and aneuploidy in nuclei isolated from paraffin-embedded tissue. Thirty-seven MALT lymphomas (all previously evaluated for t(11;18) by reverse transcriptase-polymerase chain reaction), 1 large cell lymphoma (LCL) arising subsequent to MALT lymphoma, and 16 controls were tested by FISH using the t(11;18) probe set and multiple centromeric probes. t(11;18)(q21;q21) was present by FISH in 11 of 12 polymerase chain reaction-positive MALT lymphomas (92%). The LCL and its clonally identical antecedent MALT lymphoma both showed t(11;18). The LCL had trisomy 12, and a small subset of MALT lymphoma cells had trisomy 3 and/or 12. Only one other MALT lymphoma with t(11;18) showed aneuploidy (trisomy 3) in a small clone, whereas 15 of 25 t(11;18)-negative MALT lymphomas (60%) showed trisomy of chromosomes 18 (n = 12), 3 (n = 8), 7 (n = 2), and/or 11 (n = 1). t(11;18) and aneuploidy are primarily mutually exclusive events, suggesting different pathogenetic pathways in the development of MALT lymphomas. Both t(11;18) and aneuploidy were seen disproportionately in lung, and both were associated with recurrent disease.
Extranodal marginal zone B-cell lymphoma of mucosa-associated lymphoid tissue (MALT lymphoma), nodal marginal zone B-cell lymphoma (NMZL), and splenic marginal zone B-cell lymphoma (SMZL) are all considered low-grade B-cell malignant lymphomas. Because of the similarities in morphology and immunophenotype, it was initially suggested that these neoplasms were different manifestations of the same disease process, with the postulated cell of origin being the marginal zone B cell. The theory of a unified disease process was supported by cytogenetic data demonstrating that certain cytogenetic abnormalities, including trisomies of chromosomes 3, 7, 12, and 18, were recurrent findings in MALT lymphomas, NMZLs, and SMZLs, 1,2 whereas they were not commonly identified in other B-cell malignant lymphomas. 3 However, these findings were primarily based on fluorescence in situ hybridization (FISH) studies using centromeric probes that did not allow for detection of translocations or of anomalies involving chromosomes other than those specifically being tested.
Concurrently, the t(11;18)(q21;q21) was emerging as the most frequent structural chromosomal abnormality in MALT lymphomas, based on karyotypic data. 4-9 This translocation results in the fusion of two genes, API2 and MALT1 (also referred to as MLT), at the 11q21 and 18q21 breakpoints, respectively. 10 The API2 gene is a member of a five-gene family in humans that codes for proteins involved in apoptosis regulation, and the MALT1 gene codes for a paracaspase (caspase-like protein). 11 The API2-MALT1 fusion protein is thought to induce nuclear factor-κB, resulting in transcription activation. 12 We and others have recently shown that API2-MALT1 fusion is found in a subset of MALT lymphomas, but not in NMZLs or SMZLs, 13-17 suggesting that MALT lymphomas are pathophysiologically distinct from NMZLs and SMZLs.
However, many questions regarding the relationship between MALT lymphomas containing t(11;18) and those exhibiting aneuploidy remain unanswered. For example, it is not known whether t(11;18) and aneuploidy are seen in the same tumors or whether they are mutually exclusive cytogenetic events. Their presence in the same tumor would suggest karyotypic progression, whereas their presence in different tumors might indicate that different pathogenetic pathways exist in the development of MALT lymphomas. The identification of t(11;18) in gastric MALT lymphomas has been associated with resistance to Helicobacter pylori eradication therapy and advanced or disseminated disease, 18-20 but the clinical significance of t(11;18) in MALT lymphomas arising in other anatomical sites, and of aneuploidy in general, is unknown. Also, the 11;18 translocation is found in approximately 20 to 50% of MALT lymphomas, particularly those arising in pulmonary and gastrointestinal sites, 13-17,21 but the frequency of aneuploidy in different anatomical sites has not been examined.
These questions are difficult to answer using presently available techniques. Standard karyotyping is limited by low yield because of the low proliferative rate of the lymphoma cells, as well as by the difficulty in obtaining fresh MALT lymphoma specimens prospectively. The need for fresh or frozen tissue, in addition to its inability to detect aneuploidy, likewise limits the applicability of reverse transcriptase-polymerase chain reaction (RT-PCR). In response to these limitations, we developed a highly sensitive method using two-color FISH to detect t(11;18)(q21;q21) in nuclei isolated from paraffin-embedded tissue. Differently colored probes span API2 at 11q21 and MALT1 at 18q21, respectively, resulting in a dual fusion (D-FISH) pattern in cells with an 11;18 translocation. Because two fusion signals are produced, there is increased sensitivity for detection of abnormal cells. This method has the added advantage of being able to detect other abnormalities involving chromosomes 11 and 18, such as trisomy 11, trisomy 18, or translocations involving 11q21 or 18q21 and a partner chromosome other than 18q21 or 11q21, respectively. In addition, centromeric probes can be used to look for aneuploidy involving other chromosomes. Using FISH, paraffin-embedded tissue specimens from 37 MALT lymphomas from diverse anatomical sites and 16 controls were retrospectively evaluated for t(11;18)(q21;q21) and aneuploidy of chromosomes 3, 7, 11, 12, and 18. The findings were correlated with clinical data to address these questions.
Materials and Methods
Patient Samples
Fifty-three cases, including 37 MALT lymphomas (each previously analyzed for API2-MALT1 fusion transcripts using RT-PCR 13 ), 7 normal lymph nodes, 4 normal tonsils, 3 chronic lymphocytic leukemias/small lymphocytic lymphomas, and 2 SMZLs were selected from the files of the Division of Anatomical Pathology of the Mayo Clinic. The MALT lymphomas involved diverse extranodal sites including 11 lung, 6 parotid, 4 orbit, 3 stomach, 3 thyroid, 2 colon, 2 small bowel, 3 lacrimal gland, and 1 each from kidney, paravertebral region, and skin of scalp. Multiple specimens were examined from two patients: two gastric MALT lymphoma specimens dated 25 months apart from one individual, and two colon specimens [a MALT lymphoma and a diffuse large B-cell lymphoma (LCL) that occurred 12 months later] from a second patient. Clinical data were collected in all cases. All patients consented to research use of their tissue, and the study was approved by the Mayo Clinic Institutional Review Board. Each lymphoma met the morphological and phenotypic criteria (by frozen section immunohistochemistry or flow cytometry) of the World Health Organization Classification of Tumors of Hematopoietic and Lymphoid Tissues, 22 and in each case paraffin-embedded tissue was available for isolation of nuclei for FISH.
Bacterial Artificial Chromosome (BAC) Pool Screening and BAC Probe Preparation
BAC pools (catalog no. 96011; Research Genetics, Huntsville, AL) were screened by PCR to identify clones containing API2 or MALT1 coding sequences. The screening resulted in the identification of two API2-containing BACs (250B13 and 340O13) and one MALT1-containing BAC (441H21). BAC DNAs were harvested from bacterial cultures (Qiagen, Valencia, CA) and labeled in a 45-μl volume containing the following components: 1 μg DNA, 5 μl dNTP (minus dCTP), 12.5 nmol each Texas Red-5dUTP and dCTP (BACs 250B13 and 340O13) or 12.5 nmol each fluorescein-12-dUTP and -dCTP (BAC 441H21), and 1× bionic enzyme (Life Technologies, Inc., Rockville, MD). Mixtures were incubated at 15°C for 2 hours and the reactions terminated with 5 μl of stop buffer (Life Technologies, Inc.). Labeled probe was then precipitated with 10 μg of COT-1 DNA (Life Technologies, Inc.), 1/10th volume sodium acetate, and 1 ml of absolute EtOH. Precipitated probes were then suspended in 12 μl of water.
FISH Analysis
FISH was performed as previously described. 23 Briefly, 50-μm sections were cut from each paraffin block and deparaffinized with xylene. Nuclei were extracted using pepsin, filtered, pelleted, resuspended, and placed on a slide. The slides were sequentially treated with methanol:acetic acid, 2× standard saline citrate, hot citric acid, and pepsin. Probe was added to each slide, the DNA and probe were co-denatured and hybridized, and the slides were washed and counterstained. Cells were viewed with a fluorescent microscope. The API2 and MALT1 probes were conjugated with Texas red and fluorescein, respectively. Centromere-specific α-satellite (CEN) probes for chromosomes 3, 11, and 12 were directly labeled with SpectrumOrange, and those for chromosomes 7 and 18 were directly labeled with SpectrumGreen. DNA probes for BCL2 (telomeric to the MALT1 gene at 18q21) and IGH (at 14q32) were directly conjugated with SpectrumOrange and SpectrumGreen, respectively (Vysis, Inc., Downers Grove, IL). As the signals were observed with filters through which the SpectrumOrange and Texas red fluorophores appear red, and the SpectrumGreen and fluorescein fluorophores appear green, the colors will be referred to as red (R) and green (G) in this article. Yellow fusion signals will be referred to as F. The API2-MALT1 probe set was applied to all cases and controls. CEN 3 and CEN 7 were applied as a set to all cases but one and all controls but one. CEN 12 and CEN 18 were applied as a second set to all cases and all controls but one. The probes for BCL2-IGH and CEN 11 were applied separately to selected cases.
Statistical Studies
FISH specimens were studied in random order in a blinded manner by two microscopists (XYW and RGM). Each microscopist scored 100 consecutive qualifying interphase nuclei from different areas of the same slide. The normal range was calculated using lymph nodes and tonsils from 11 patients without a malignant hematological disorder. The upper bound of the normal range was determined using a one-sided 95% confidence interval for observing the maximum number of nuclei for each false-positive signal pattern seen in 200 scoreable nuclei using the binomial distribution.
Immunoglobulin Gene Rearrangement Studies and Gene Sequencing
Immunoglobulin gene rearrangement and gene sequencing studies were performed on paraffin-embedded tissue from case 6 (colonic MALT lymphoma and subsequent colonic LCL specimens). DNA from each specimen was isolated and amplified using consensus primers directed to framework 3 (FR3A: ACACGGC(C/T)(G/C)TGTATTACTGT) and the joining region (LJH: TGAGGAGACGGTGACC) of IgH. PCR was performed in a 25-μl reaction volume containing patient DNA, 1.5 μmol/L MgCl2, 200 mmol/L dNTPs, 10 μmol/L primers, and 1.5 U AmpliTaq gold (PE Applied Biosystems, Foster City, CA). The PCR reaction was performed at 95°C for 10 minutes for an initial denaturation step, then 94°C for 1 minute, 60°C for 1 minute, 72°C for 1 minute for a total of 35 cycles, and a final extension at 72°C for 10 minutes. The PCR products were run on an 8% (19:1) polyacrylamide gel at 200 V for 60 minutes. Under UV light, the clonal PCR bands were isolated with a scalpel, placed in Tris/ethylenediaminetetraacetic acid and centrifuged at 12,000 rpm. The supernatant was used as template for automated sequencing with 5 pmol of the LJH primer on an ABI Prism 377 sequencer (PE Applied Biosystems). The obtained sequences were analyzed using a BLAST online server in the National Center for Biotechnology Information at the National Library of Medicine (Bethesda, MD).
Results
FISH: Normal Range
Table 1 ▶ summarizes the statistics for analysis of 200 scoreable nuclei from each of 11 normal paraffin-embedded specimens for false-positive patterns that would result in erroneous determination of API2-MALT1 fusion. Based on this analysis, a specimen would be classified as abnormal consistent with API2-MALT1 fusion if the number of nuclei observed with 1R1G2F or 1R1G3F was >3 (1.5%), 2R1G1F was >10 (5%), 1R2G1F was >3 (1.5%), and 1R1G1F was >15 (7.5%). A specimen would be classified as abnormal consistent with an additional complete or partial chromosome 11 or a structural rearrangement of API2 without fusion with MALT1 if the number of nuclei observed with 3R2G was >9 (4.5%). A specimen would be classified as abnormal consistent with an additional complete or partial chromosome 18 or a structural rearrangement of MALT1 without fusion with API2 if the number of nuclei observed with 2R3G was >8 (4.0%).
Table 1.
Normal Range for Lymph Nodes/Tonsils Based on Analysis of 200 Nuclei
| Pattern | Mean | Standard deviation | Maximum | Minimum | Upper bound of 95% confidence interval | Cutoff |
|---|---|---|---|---|---|---|
| 1R1G2F | 0 | 0 | 0 | 0 | 0 | >1.5% |
| 1R2G1F | 0 | 0 | 0 | 0 | 0 | >1.5% |
| 2R1G1F | 0.5 | 1.5 | 5 | 0 | 5 | >5.0% |
| 1R1G1F | 5.8 | 2.1 | 9 | 2 | 9 | >7.5% |
| 3R2G | 1.3 | 1.3 | 4 | 0 | 4 | >4.5% |
| 2R3G | 2.2 | 1.2 | 3 | 0 | 3 | >4.0% |
R, red (API2) signal; G, green (MALT1) signal; F, fusion (API2-MALT1) signal.
FISH Analysis
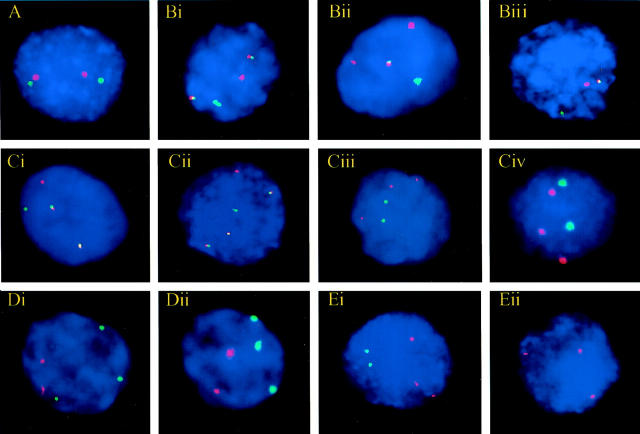
A t(11;18)(q21;q21) was identified by FISH in 11 of 12 MALT lymphomas in which API2-MALT1 fusion had been previously demonstrated by RT-PCR (Table 2) ▶ , including both the primary and recurrent gastric MALT lymphomas from the same patient. The mean percent and SD of nuclei per case with an abnormal FISH pattern indicative of t(11;18)(q21;q21) was 60.8 ± 13.3 with a range of 41 to 88. Of the 11 FISH-positive MALT lymphomas, 8 were 1R1G2F (Figure 1Bi) ▶ , 2 were 2R1G1F (Figure 1Bii) ▶ , and 1 was 1R1G1F (Figure 1Biii) ▶ . The 2R1G1F pattern most likely resulted from deletion of a portion of the MALT1 FISH target sequence telomeric to the 18q21 breakpoint and the 1R1G1F pattern presumably occurred because of deletions of portions of both 11q and 18q. Similar losses of hybridization sites have been associated with translocations involving BCR and ABL 24 and MLL. 25
Table 2.
Results of FISH for t(11;18)(q21;q21) and CEN 3, 7, 12, and 18 on MALT Lymphomas Previously Shown to Be Positive for API2-MALT1 Fusion by RT-PCR, Arranged in Descending Order by Percent Nuclei Abnormal for API2-MALT1 Fusion by FISH
| Case | Diagnosis | Site | t(11;18) FISH pattern | t(11;18) Abnormal nuclei (%) | CEN 3 3 signals (%) | CEN 7 3 signals (%) | CEN 12 3 signals (%) | CEN 18 3 signals (%) |
|---|---|---|---|---|---|---|---|---|
| 1 | MALT lymphoma | Small bowel | 1R1G2F | 73 | 1.5 | 0 | 0 | 0 |
| 2 | MALT lymphoma | Lung | 1R1G2F | 71 | 0 | 0 | 1.5 | 0 |
| 3 | MALT lymphoma | Lung | 1R1G2F | 69 | 0.5 | 0 | 1 | 1 |
| 4 | MALT lymphoma | Small bowel | 1R1G1F | 63 | 1 | 0 | 0 | 0 |
| 5 | MALT lymphoma | Lung | 1R1G2F | 62 | 6.5 | 0 | 0.5 | 0.5 |
| 6A | MALT lymphoma | Colon | 1R1G2F | 61 | 20 | 0 | 13 | 0 |
| 6B | LCL (transformation) | Colon | 1R1G3F | 88 | 1 | 0 | 76.5 | 0.5 |
| 7A | MALT lymphoma | Stomach | 2R1G1F | 55 | 2.5 | 1 | 0 | 0.5 |
| 7B | MALT lymphoma (recurrence) | Stomach | 2R1G1F | 53 | 1.5 | 0 | 0.5 | 0 |
| 8 | MALT lymphoma | Colon | 1R1G2F | 54 | 1 | 0.5 | 1 | 1 |
| 9 | MALT lymphoma | Lung | 1R1G2F | 49 | 0 | 0.5 | 0 | 0 |
| 10 | MALT lymphoma | Lung | 2R1G1F | 44 | 0 | 0 | 1 | 0.5 |
| 11 | MALT lymphoma | Lung | 1R1G2F | 41 | 0.5 | 0 | 0 | 0.5 |
| 12 | MALT lymphoma | Parotid | 2R2G | 0 | 0.5 | 0.5 | 0.5 | 0 |
R, red (API2) signal; G, green (MALT1) signal; F, fusion (API2-MALT1) signal.
Figure 1.
Representative FISH patterns in normal and abnormal interphase cells. A: Normal, 2R2G, using the AP12-MALT1 probe. B: Abnormal FISH patterns using the API2-MALT1 probe. Bi: t(11;18)(q21;q21), 1R1G2F. Dual fusion (D-FISH) pattern. Bii: t(11;18)(q21;q21), 2R1G1F. Extra signal-red (ES-R) pattern. Results from deletion of a portion of the MALT1 FISH target sequence telomeric to the 18q21 breakpoint. Biii: t(11;18)(q21;q21), 1R1G1F. Single fusion (S-FISH) pattern. Results from deletions of portions of both 11q and 18q. C: FISH patterns of a t(11;18)-positive MALT lymphoma that transformed to a t(11;18)-positive LCL. Ci: MALT lymphoma interphase nucleus with t(11;18)(q21;q21). 1R1G2F using the API2-MALT1 probe. Cii: LCL interphase nucleus with t(11;18)(q21;q21). 1R1G3F using the API2-MALT1 probe. Ciii: MALT lymphoma interphase nucleus with trisomy 12 (subset of neoplastic cells). 3R2G using CEN 12 (R) and CEN 18 (G) probes. Civ: LCL interphase nucleus with trisomy 12. 3R2G using CEN 12 (R) and CEN 18 (G) probes. D: FISH patterns of a MALT lymphoma with trisomy 18 without t(11;18). Di: 2R3G, using the API2-MALT1 probe. Dii: 2R3G using CEN 12 (R) and CEN 18 (G) probes. E: FISH patterns of a MALT lymphoma with trisomy 11 without t(11;18). Ei: 3R2G, using the API2-MALT1 probe. Eii: Three signals using the CEN 11 probe (R).
Excluding the MALT lymphoma that was associated with a subsequent LCL (discussed below), 10 of 11 t(11;18)-positive MALT lymphomas had a normal FISH pattern using centromeric probes to chromosomes 3, 7, 12, and 18. In the remaining case, 6.5% of the cells displayed three CEN 3 signals, compared to 62% of the cells harboring t(11;18). The normal lymph nodes, tonsils, chronic lymphocytic leukemias/small lymphocytic lymphomas, and SMZLs all had normal FISH patterns, except for one normal tonsil for which there were no centromeric probe data because of probe hybridization failure, and one chronic lymphocytic leukemia/small lymphocytic lymphoma that showed trisomy 12.
Twelve of the 25 t(11;18)-negative MALT lymphomas had a normal FISH pattern using the API2-MALT1 probe set (Figure 1A ▶ , Table 3 ▶ ). Twelve others had an abnormal 2R3G signal pattern (Figure 1Di) ▶ , and of these, 10 had three signals using CEN 18, confirming the presence of trisomy 18 (Figure 1Dii) ▶ . The remaining two had three BCL2 signals (located at 18q21), consistent with partial trisomy 18 (isochromosome 18q or a marker chromosome involving 18q). The last case had a 3R2G pattern using the API2-MALT1 probe set (Figure 1Ei) ▶ , and three signals using CEN 11, indicative of trisomy 11 (Figure 1Eii) ▶ . Finally, several cases had three signals using CEN 3 (n = 8) and/or CEN 7 (n = 2). CEN 3 and CEN 7 were not tested in one case because of lack of cells.
Table 3.
Results of FISH for t(11;18)(q21;q21), CEN 3, 7, 12, 18, and 18q21 (BCL2) on 25 MALT Lymphomas Previously Shown to Be Negative for API2-MALT1 Fusion by RT-PCR, Arranged in Descending Order by Percent Nuclei Abnormal for t(11;18) by FISH
| Case | Site | t(11;18) FISH pattern | t(11;18) Abnormal nuclei (%) | CEN 3 3 signals (%) | CEN 7 3 signals (%) | CEN 12 3 signals (%) | CEN 18 3 signals (%) | BCL2 (18q21) 3 signals (%) |
|---|---|---|---|---|---|---|---|---|
| 1 | Orbit | 2R3G | 56 | 56 | 3.5 | 0.5 | 0.5 | 37 |
| 2 | Parotid | 2R3G | 50 | 22 | 1 | 0 | 36 | ND |
| 3 | Lung | 2R3/4G | 41 | 0.5 | 0 | 0 | 34.5 | 25 |
| 4 | Parotid | 2R3G | 31.5 | 0 | 0 | 0 | 0 | 17 |
| 5 | Lacrimal gland | 2R3G | 30.0 | 0 | 0 | 0.5 | 15.5 | ND |
| 6 | Kidney | 2R3G | 29 | 36.5 | 29 | 0 | 17 | 16 |
| 7 | Stomach | 2R3G | 25 | 45 | 0 | 0 | 26.5 | 41 |
| 8 | Lacrimal gland | 2R3G | 25 | ND | ND | 0 | 17.5 | ND |
| 9 | Lung | 2R3/4G | 25 | 20.5 | 2.5 | 0 | 13 | ND |
| 10 | Lung | 2R3G | 24.5 | 36.5 | 1 | 1.5 | 34 | 29 |
| 11 | Lacrimal gland | 2R3G | 22.5 | 27.5 | 0 | 0 | 40.5 | 34 |
| 12 | Thyroid* | 3R2G | 21.5 | 2.5 | 0 | 0.5 | 0 | ND |
| 13 | Paravertebral region | 2R3G | 20.5 | 0.5 | 0 | 0 | 11.5 | 37 |
| 14 | Parotid | 2R2G | 0 | 0 | 0.5 | 1.5 | 1.5 | ND |
| 15 | Parotid | 2R2G | 0 | 0 | 2.5 | 1 | 1 | ND |
| 16 | Skin (scalp) | 2R2G | 0 | 0.5 | 10.5 | 0.5 | 0 | ND |
| 17 | Orbit | 2R2G | 0 | 0.5 | 0 | 0 | 0 | ND |
| 18 | Lung | 2R2G | 0 | 0.5 | 1 | 0 | 0.5 | ND |
| 19 | Thyroid | 2R2G | 0 | 0.5 | 1.5 | 0 | 0 | ND |
| 20 | Stomach | 2R2G | 0 | 2 | 2 | 0 | 0.5 | ND |
| 21 | Orbit | 2R2G | 0 | 0 | 0.5 | 0 | 0 | ND |
| 22 | Parotid | 2R2G | 0 | 0.5 | 0 | 0 | 1 | ND |
| 23 | Lung | 2R2G | 0 | 42.5 | 0.5 | 1.5 | 0 | ND |
| 24 | Orbit | 2R2G | 0 | 0.5 | 0.5 | 0 | 1 | ND |
| 25 | Thyroid | 2R2G | 0 | 0 | 0.5 | 0 | 0 | ND |
R, red (API2) signal; G, green (MALT1) signal; ND, not done.
*, 28% of nuclei had three signals using a CEN 11 FISH probe.
Transformation of t(11;18)-Positive MALT Lymphoma into t(11;18)-Positive LCL
A single case of a t(11;18)-positive MALT lymphoma that transformed into a t(11;18)-positive LCL was identified. The MALT lymphoma, which was initially diagnosed in the stomach and bone marrow in this 54-year-old female, was first treated with leukeran. Six years later, biopsy-proven MALT lymphoma was diagnosed in both the colon and the lung and the patient was treated with interleukin-4. Biopsy-proven recurrent colonic MALT lymphoma identified 9 months later was treated with rituximab, and 3 months later the patient underwent left hemicolectomy for a perforated colon associated with diffuse large B-cell lymphoma. She developed acute respiratory distress syndrome and passed away 3 days later.
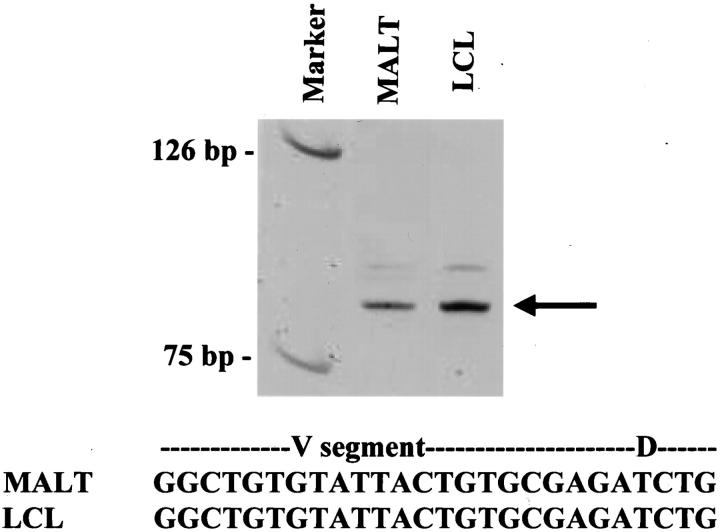
No blocks were available from the initial stomach and bone marrow specimens for FISH analysis. Immunoglobulin gene rearrangement studies and gene sequencing performed on the first colonic MALT lymphoma and the LCL identified products of identical molecular weight and sequence composition (Figure 2) ▶ , proving clonal identity between the two tumors. FISH studies (Table 2) ▶ showed t(11;18) in 61% of nuclei in the first colonic MALT lymphoma, and t(11;18) in 88% of nuclei in the LCL. The MALT lymphoma had a 1R1G2F pattern (Figure 1Ci) ▶ and the LCL had a 1R1G3F pattern (Figure 1Cii) ▶ . The MALT lymphoma had trisomy 3 and trisomy 12 in 20% and 13% of nuclei, respectively (Figure 1Ciii) ▶ , and the LCL lacked trisomy 3 but had trisomy 12 in 76.5% of nuclei (Figure 1Civ) ▶ .
Figure 2.
Demonstration of clonal identity between a MALT lymphoma and its subsequent LCL transformation by PCR and sequence analysis. Ethidium bromide-stained polyacrylamide gel (top) showed PCR products from the MALT lymphoma and the LCL were of identical size (arrow). Sequencing demonstrated identical nucleotides spanning the V-D junction (bottom), indicating clonal identity.
Clinical Data
This series of MALT lymphoma patients included 24 women and 13 men (F:M = 1.8:1), with a median age of 65 years (range, 43 to 83 years). Sixteen tumors (43%) recurred, seven (19%) involved the bone marrow, and three (8%) were associated with a subsequent LCL. At last follow-up (median, 56 months), 26 patients were alive and 10 were dead. One was lost to follow-up.
The seven women and five men whose tumors harbored t(11;18) had a median age of 67.5 years (range, 60 to 80 years). At last follow-up, six patients were alive and six were dead. The tumors involved the lung (n = 6), small bowel (n = 2), colon (n = 2), stomach (n = 1), and parotid (n = 1). Five tumors (42%) were recurrent (two small bowel, one colon, one parotid, and one lung) or subsequently recurred, and three (25%) showed bone marrow involvement at diagnosis. One tumor (described above) transformed to large cell lymphoma; a second tumor was associated with a subsequent LCL but tissue was not available from the LCL to investigate clonal identity between the two tumors or perform FISH studies. Both patients died of disease. Two patients died of other causes (metastatic rectal adenocarcinoma and postoperative complications), and in the remaining two patients the cause of death is unknown.
The 12 women and 3 men whose tumors showed aneuploidy but lacked t(11;18) had a median age of 61 years (range, 43 to 83 years). At last follow-up 12 patients were alive, 2 were dead, and 1 was lost to follow-up. These tumors involved diverse sites including lung (n = 4), lacrimal gland (n = 3), parotid (n = 2), and stomach, orbit, thyroid, kidney, skin of scalp, and paravertebral region (n = 1 each). Eight MALT lymphomas (53%) were recurrent (two lacrimal glands, one parotid, and one scalp specimen) or subsequently recurred, and four (27%) initially involved the bone marrow. One tumor was associated with a subsequent LCL; the patient had had bone marrow involvement and multiple recurrences of MALT lymphoma before developing LCL, and ultimately died of disease. No tissue was available from the LCL to investigate clonal identity or perform FISH analysis. The other deceased patient died of other causes (complications from aortic aneurysm repair).
The five women and five men whose tumors showed no cytogenetic abnormalities had a median age of 63 years (range, 47 to 80 years). The MALT lymphomas involved the orbit (n = 3), parotid (n = 3), thyroid (n = 2), stomach (n = 1), and lung (n = 1). Two (20%) recurred in other extranodal sites, and none involved the bone marrow or were associated with a subsequent LCL. At last follow-up eight patients were alive and two were dead. One patient died of a CD10-positive large cell lymphoma that had antedated the MALT lymphoma, and one died of other causes.
Discussion
The t(11;18)(q21;q21) and aneuploidy of chromosomes 3, 7, 12, and 18 have been described with increased frequency in MALT lymphomas, but the relationship between t(11;18)-positive and aneuploidy-positive MALT lymphomas and the clinical significance of these cytogenetic abnormalities is unclear. Because of the limitations of conventional cytogenetics and RT-PCR, we developed a highly sensitive two-color FISH technique to identify t(11;18)(q21;q21) and aneuploidy to address these issues. The FISH method described here works well on both fresh and paraffin-embedded tissue, but the potential to study the latter greatly increases the applicability of the test. By isolating individual nuclei from paraffin-embedded tissue, several problems common with thin-section direct FISH techniques, such as interpretation of signals within overlapping nuclei and loss of signals within truncated nuclei, were avoided. This method consistently produced FISH signals from individual nuclei that were planar, bright, and discrete, and it works equally well on B5-fixed and formalin-fixed tissue.
The API2 and MALT1 probes in the present study were designed to produce a dual fusion pattern in positive cases by spanning both breakpoints. In most patients with a t(11;18), this method detects disease involving more than 1.5% of nuclei when 200 nuclei are examined. It is thus more sensitive than previously reported FISH techniques in which only the 11q21 breakpoint was spanned. 14-16 Using this technique, we achieved a sensitivity of 92% and a specificity of 100%. The single false-negative specimen was minimally involved by tumor, with too few lymphoma cells to exceed the 1.5% cutoff point. This probe set also detects other abnormalities involving chromosomes 11 and 18. Centromeric probes were used to confirm trisomy 11 and 18 and to detect trisomy 3, 7, and 12. The combination of the API2-MALT1, BCL2-IGH, and CEN 18 FISH probes permitted detection of abnormalities involving only the long arm of chromosome 18, as both MALT1 and BCL2 are located at 18q21 although the probes show no overlap.
The present study demonstrates that t(11;18) and aneuploidy are usually mutually exclusive events in MALT lymphoma. Two exceptions were identified: both were recurrent MALT lymphomas in which the majority of the cells showed API2-MALT1 fusion, whereas only a minority of cells showed aneuploidy. One tumor subsequently transformed to a clonally identical, t(11;18)-positive LCL involving the same site. It has been suggested that MALT lymphomas harboring t(11;18) are unlikely to transform to a higher grade lymphoma, 8,14,16 and this is the first reported case of a t(11;18)-positive LCL to date. The MALT lymphoma contained small subsets of trisomy 12 cells and trisomy 3 cells, whereas all of the LCL cells contained trisomy 12 as well as an extra API2-MALT1 fusion signal but lacked trisomy 3. The fact that both minor clones containing aneuploidy arose in recurrent tumors suggests that aneuploidy may occasionally be a secondary chromosomal abnormality in t(11;18)-positive MALT lymphomas, and may contribute to the tumor’s ability to recur or transform. Two MALT lymphomas have been previously described in which the major clone harbored t(11;18)(q21;q21) whereas a second, minor clone or subclone harbored trisomy 3. 5,26
Fifteen of 25 (60%) t(11;18)-negative MALT lymphomas harbored numerical chromosomal abnormalities, including trisomy 3, 7, 11, and/or 18. Additions of portions of chromosomes 3, 7, and 12, as well as trisomies of other chromosomes would not be detected using this probe set, so the frequency of numerical chromosomal abnormalities in MALT lymphomas may in fact be higher. Trisomy of chromosomes 3, 7, 12, and 18 were detected in 27%, 5%, 3%, and 27% of MALT lymphomas in the present study, respectively, and have been previously described in 12 to 85%, 2,3,27-29 3 to 15%, 1,2,27-29 3 to 38%, 1,2,27-29 and 7 to 36% 2,27-29 of low-grade MALT lymphomas, respectively. Trisomy 11, which was identified in one case, and an additional portion of trisomy 18, which was identified in two cases, have also been previously described. 3,28 Trisomy 3 has been considered the most common numeric chromosomal abnormality in MALT lymphomas, but in our study complete or partial trisomy 18 was most frequent, being seen in a third of MALT lymphomas and in nearly half of the API2-MALT1 fusion-negative cases. In addition, aneuploidy involving one (n = 8), two (n = 7), or three (n = 1) different chromosomes could be seen in the same tumor.
The observation that the t(11;18) and aneuploidy are usually mutually exclusive events indicates that at least two different pathways leading to the development of MALT lymphoma exist. In t(11;18)-positive cases the presence of the API2-MALT1 fusion protein is thought to result in induction of nuclear factor-κB, leading to enhanced transcription activation. 11,12 The pathogenetic role of aneuploidy in MALT lymphoma remains speculative at this point, although tumor growth may be induced as a result of a higher copy number of proliferation-related genes.
Patients with t(11;18)-positive or aneuploidy-positive MALT lymphomas had similar clinical behavior, with approximately half of the tumors recurring, and one-fourth of the tumors involving the bone marrow. In contrast, MALT lymphomas that lacked chromosomal abnormalities recurred in only 20% of cases and never involved the bone marrow. When only the 27 initial diagnostic specimens were considered, both API2-MALT1 fusion and aneuploidy were disproportionately detected in the lung. Indeed, 90% of the primary pulmonary MALT lymphomas had either t(11;18) or trisomy 3 and/or 18. Primary pulmonary MALT lymphomas may have a poorer prognosis than those arising in other sites. 30 These findings suggest that the presence of structural or numeric chromosomal abnormalities may confer a poorer prognosis in MALT lymphomas. API2-MALT1 fusion was also identified in primary gastrointestinal MALT lymphomas (one each in stomach and colon). Previous studies have detected API2-MALT1 fusion transcripts in 32% of stomach and 43% of lung MALT lymphomas, but it is not known whether these represented primary tumors. 14-16,20,21 In the present study, aneuploidy-positive primary MALT lymphomas involved a variety of anatomical sites in addition to lung (one each in stomach, parotid, thyroid, orbit, lacrimal gland, kidney, and paravertebral region), and MALT lymphomas without chromosomal abnormalities most frequently involved the orbit, parotid, and thyroid.
(Note: After submission of our manuscript, Starostik and colleagues, by performing microsatellite screening of 24 gastric MALT lymphomas, reported that t(11;18)-positive MALT lymphomas were primarily devoid of additional genetic abnormalities, whereas t(11;18)-negative MALT lymphomas often contained one or more allelic imbalances. 31 Barth and colleagues have also demonstrated by comparative genomic hybridization (CGH) and FISH that MALT lymphomas with t(11;18) lacked other abnormalities by CGH, whereas t(11;18)-negative MALT lymphomas showed frequent gains and losses of chromosomal material. 32 Their studies and our study all demonstrate that MALT lymphomas develop along two different pathogenetic pathways: those with t(11;18) and few if any additional genetic abnormalities, and those without t(11;18) but with one or more other genetic abnormalities. In the t(11;18)-negative group, similar regions showed genomic DNA amplification, chromosomal gains, and aneuploidy, including amplification of 3q26.2-27 (21%)/gain on chromosome 3 (31%)/trisomy 3 (22%), amplification of 11q23-24 (8%)/gain on 11q(6%)/trisomy 11 (3%), and amplification of 18q21 (8%)/gain on 18 (13%)/complete or partial trisomy 18 (32%). Additional studies may be helpful in determining the genetic mechanisms by which these DNA amplifications/chromosomal gains/aneuploidies are involved in lymphomagenesis. Because several of the allelic imbalances were identical to those seen in a series of previously studied gastric DLBCL, Starostik and colleagues suggested that a subset of t(11;18)-negative MALT lymphomas with allelic imbalances may eventually transform to DLBCL, whereas t(11;18)-positive MALT lymphomas are unlikely to do so. 33 However, we have demonstrated that t(11;18)-positive MALT lymphomas do indeed have the potential to transform into t(11;18)-positive LCL.)
Acknowledgments
We thank Dr. John Lust and Mr. Ming Mai for their assistance in performing immunoglobulin gene rearrangement and gene sequencing studies, Ms. Stephanie Brockman and Ms. Sarah Paternoster for their technical assistance in the fluorescence in situ hybridization laboratory, and Ms. Jane Garlitch and Ms. Kristin Rieger for their expert secretarial assistance.
Footnotes
Address reprint requests to Ellen D. Remstein, M.D., Hilton Building, Room 1166, Mayo Clinic, 200 First St. SW, Rochester, MN 55905. E-mail: remstein.ellen@mayo.edu.
References
- 1.Brynes RK, Almaguer PD, Leathery KE, McCourty A, Arber DA, Medeiros LJ, Nathwani BN: Numerical cytogenetic abnormalities of chromosomes 3, 7, and 12 in marginal zone B-cell lymphomas. Mod Pathol 1996, 9:995-1000 [PubMed] [Google Scholar]
- 2.Wotherspoon AC, Finn TM, Isaacson PG: Trisomy 3 in low-grade B-cell lymphomas of mucosa-associated lymphoid tissue. Blood 1995, 85:2000-2004 [PubMed] [Google Scholar]
- 3.Dierlamm J, Pittaluga S, Wlodarska I, Stul M, Thomas J, Boogaerts M, Michaux L, Driessen A, Mecucci C, Cassiman J-J, DeWolf-Peeters C, Van den Berghe H: Marginal zone B-cell lymphomas of different sites share similar cytogenetic and morphologic features. Blood 1996, 87:1299-1307 [PubMed] [Google Scholar]
- 4.Levine EG, Arthur DC, Machnicki J, Frizzera G, Hurd D, Peterson B, Gajl-Peczalska KJ, Bloomfield CD: Four new recurring translocations in non-Hodgkin lymphoma. Blood 1989, 74:1796-1800 [PubMed] [Google Scholar]
- 5.Griffin CA, Zehnbauer BA, Beschorner WE, Ambinder R, Mann R: t(11;18)(q21;q21) is a recurrent chromosome abnormality in small lymphocytic lymphoma. Genes Chromosom Cancer 1992, 4:153-157 [DOI] [PubMed] [Google Scholar]
- 6.Horsman D, Gascoyne R, Klasa R, Coupland R: t(11;18)(q21;t21): a recurring translocation in lymphomas of mucosa-associated lymphoid tissue (MALT)? Genes Chromosom Cancer 1992, 4:183-187 [DOI] [PubMed] [Google Scholar]
- 7.Leroux D, Seité P, Hillion J, Le Marc’hadour F, Pégourié-Bandelier B, Jacob M-C, Larsen C-J, Sotto J-J: t(11;18)(q21;q21) may delineate a spectrum of diffuse small B-cell lymphoma with extranodal involvement. Genes Chromosom Cancer 1993, 7:54-56 [DOI] [PubMed] [Google Scholar]
- 8.Ott G, Katzenberger T, Greiner A, Kalla J, Rosenwald A, Heinrich U, Ott MM, Müller-Hermelink HK: The t(11;18)(q21;q21) chromosome translocation is a frequent and specific aberration in low-grade but not high-grade malignant non-Hodgkin’s lymphomas of the mucosa-associated lymphoid tissue (MALT)-type. Cancer Res 1997, 57:3944-3948 [PubMed] [Google Scholar]
- 9.Auer IA, Gascoyne RD, Connors JM, Cotter FE, Greiner TC, Sanger WG, Horsman DE: t(11;18)(q21;q21) is the most common translocation in MALT lymphomas. Ann Oncol 1997, 8:979-985 [DOI] [PubMed] [Google Scholar]
- 10.Dierlamm J, Baens M, Wlodarska I, Stefanova-Ouzounova M, Hernandez JM, Hossfeld DK, De Wolf-Peeters C, Hagemeijer A, Van den Berghe H, Marynen P: The apoptosis inhibitor gene API2 and a novel 18q gene, MLT, are recurrently rearranged in the t(11;18)(q21;q21) associated with mucosa-associated lymphoid tissue lymphomas. Blood 1999, 93:3601-3609 [PubMed] [Google Scholar]
- 11.Uren AG, O’Rourke K, Aravind LA, Pisabarro MT, Seshagiri S, Koonin EV, Dixit VM: Identification of paracaspases and metacaspases: two ancient families of caspase-like proteins, one which plays a key role in MALT lymphoma. Mol Cell 2000, 6:961-970 [DOI] [PubMed] [Google Scholar]
- 12.Lucas PC, Yonezumi M, Inohara N, McAllister-Lucas LM, Abazeed ME, Chen FF, Yamaoka S, Seto M, Nüñez G: Bcl10 and MALT1, independent targets of chromosomal translocation in MALT lymphoma, cooperate in a novel NF-κB signaling pathway. J Biol Chem 2001, 276:19012-10919 [DOI] [PubMed] [Google Scholar]
- 13.Remstein ED, James CD, Kurtin PJ: Incidence and subtype specificity of API2-MALT1 fusion translocations in extranodal, nodal, and splenic marginal zone lymphomas. Am J Pathol 2000, 156:1183-1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosenwald A, Ott G, Stilgenbauer S, Kalla J, Bredt M, Katzenberger T, Greiner A, Ott MM, Gawin B, Döhner H, Müller-Hermelink HK: Exclusive detection of the t(11;18)(q21;q21) in extranodal marginal zone B cell lymphomas (MZBL) of MALT type in contrast to other MZBL and extranodal large B cell lymphomas. Am J Pathol 1999, 155:1817-1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baens M, Maes B, Steyls A, Geboes K, Marynen P, De Wolf-Peeter C: The product of the t(11;18), an API2-MLT fusion, marks nearly half of gastric MALT type lymphomas without large cell proliferation. Am J Pathol 2000, 156:1433-1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dierlamm J, Baens M, Stefanova-Ouzounova M, Hinz K, Wlodarska I, Maes B, Steyls A, Driessen A, Verhoef G, Gaulard P, Hagemeijer A, Hossfeld DK, De Wolf-Peeters C, Marynen P: Detection of t(11;18)(q21;q21) by interphase fluorescence in situ hybridization using API2 and MLT specific probes. Blood 2000, 96:2215-2218 [PubMed] [Google Scholar]
- 17.Kalla J, Stilgenbauer S, Schaffner C, Wolf S, Ott G, Greiner A, Rosenwald A, Döhner H, Müller-Hermelink HK: Heterogeneity of the API2-MALT1 gene rearrangement in MALT-type lymphoma. Leukemia 2000, 14:1967-1974 [DOI] [PubMed] [Google Scholar]
- 18.Alpen B, Neubauer A, Dierlamm J, Marynen P, Thiede C, Bayerdorfer E, Stolte M: Translocation t(11;18) absent in early gastric marginal zone B-cell lymphoma of MALT type responding to eradication of Helicobacter pylori infection. Blood 2000, 95:4014-4015 [PubMed] [Google Scholar]
- 19.Liu H, Ruskon-Fourmestraux A, Lavergne-Slove A, Ye H, Molina T, Bouhnik Y, Hamoudi RA, Diss TC, Dogan A, Megraud F, Rambaud JC, Du MQ, Isaacson PG: Resistance of t(11;18) positive gastric mucosa-associated lymphoid tissue lymphoma to Helicobacter pylori eradication therapy. Lancet 2001, 357:39-40 [DOI] [PubMed] [Google Scholar]
- 20.Liu H, Ye H, Dogan A, Ranaldi R, Hamoudi RA, Bearzi I, Isaacson PG, Du M-Q: t(11;18)(q21;q21) is associated with advanced mucosa-associated lymphoid tissue lymphoma that expresses nuclear BLC10. Blood 2001, 98:1182-1187 [DOI] [PubMed] [Google Scholar]
- 21.Ye H, Dogan A, Liu H, Wotherspoon AC, Nicholson AG, Charlotte F, Leblond V, Speight P, Isaacson PG, Du MQ: Markedly variable frequencies of t(11;18)(q21;q21) in different MALT lymphomas. Blood 2001, 28:767A. [DOI] [PubMed] [Google Scholar]
- 22.Jaffe ES, Harris NL, Stein H, Vardiman JW: Pathology and genetics of tumours of haematopoietic and lymphoid tissues. World Health Organization Classification of Tumours. 2001, :pp 157-160 IARC Press, [Google Scholar]
- 23.Remstein ED, Kurtin PJ, Buño I, Bailey RJ, Proffitt J, Wyatt WA, Hanson CA, Dewald GW: Diagnostic utility of fluorescence in situ hybridization in mantle-cell lymphoma. Br J Haematol 2000, 110:856-862 [DOI] [PubMed] [Google Scholar]
- 24.Dewald GW, Wyatt WA, Silver RT: Atypical BCR and ABL D-FISH patterns in chronic myeloid leukemia and their possible role in therapy. Leukemia Lymphoma 1999, 34:481-491 [DOI] [PubMed] [Google Scholar]
- 25.Thirman MJ, Gill HJ, Burnett RC, Mbangkollo D, McCabe NR, Kobayashi H, Ziemin van-der-poel S, Kaneko Y, Morgan R, Sandberg AA, Chaganti RSK, Larson RA, Le Beau MM, Diaz MO, Rowley JD: A cDNA probe detects all rearrangements of the MLL gene in leukemias with common and rare 11q23 translocations. N Engl J Med 1993, 329:909-914 [DOI] [PubMed] [Google Scholar]
- 26.Hosaka S, Akamatsu T, Nakamura S, Kaneko T, Kitano K, Kiyosawa K, Ota H, Hosaka N, Miyabayashi H, Katsuyama T: Mucosa-associated lymphoid tissue (MALT) lymphoma of the rectum with chromosomal translocation of the t(11;18)(q21;q21) and an additional aberration of trisomy 3. Am J Gastroenterol 1999, 94:1951-1954 [DOI] [PubMed] [Google Scholar]
- 27.Ihrler S, Baretton GB, Menauer F, Blasenbreu-Vogt S, Löhrs U: Sjögren’s syndrome and MALT lymphomas of salivary glands: a DNA-cytometric and interphase-cytogenetic study. Mod Pathol 2000, 13:4-12 [DOI] [PubMed] [Google Scholar]
- 28.Hoeve MA, Gisbertz IAM, Schouten HC, Schuuring E, Bot FJ, Hermans J, Hopman A, Kluin PHM, Arends J-W, van Krieken JHJM: Gastric low-grade MALT lymphoma, high-grade MALT lymphoma and diffuse large B cell lymphoma show different frequencies of trisomy. Leukemia 1999, 13:799-807 [DOI] [PubMed] [Google Scholar]
- 29.Ott G, Kalla J, Steinhoff A, Rosenwald A, Katzenberger T, Roblick U, Ott MM, Müller-Hermelink HK: Trisomy 3 is not a common feature in malignant lymphomas of mucosa-associated lymphoma tissue type. Am J Pathol 1998, 153:689-694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kurtin PJ, Myers JL, Adlakha H, Strickler JG, Lohse C, Pankratz VS, Inwards DJ: Pathologic and clinical features of primary pulmonary extranodal marginal zone B-cell lymphoma of MALT type. Am J Surg Pathol 2001, 25:997-1008 [DOI] [PubMed] [Google Scholar]
- 31.Starostik P, Patzner J, Greiner A, Schwarz S, Kalla J, Ott G, Müller-Hermelink HK: Gastric marginal zone B-cell lymphomas of MALT type develop along Z distinct pathogenetic pathways. Blood 2002, 99:3-9 [DOI] [PubMed] [Google Scholar]
- 32.Barth TFE, Bentz M, Leithaüser F, Stilgenbauer S, Siebert R, Schlotter M, Schlenk RF, Dohner H, Moller P: Molecular-cytogenetic comparison of mucosa-associated marginal zone B-cell lymphoma and large B-cell lymphoma arising in the gastro-intestinal tract. Genes Chromosomes Cancer 2001, 31:316-325 [DOI] [PubMed] [Google Scholar]
- 33.Starostik P, Greiner A, Schultz A, Zettl A, Peters K, Rosenwald A, Kolve M, Müller-Hermelink HK: Genetic aberrations common in gastric high-grade large B-cell lymphoma. Blood 2000, 95:1180-1187 [PubMed] [Google Scholar]