Abstract
Transmissible gastroenteritis virus (TGEV) isolates that have been adapted to passage in cell culture maintain their infectivity in vitro but may lose their pathogenicity in vivo. To better understand the genomic mechanisms for viral attenuation, we sequenced the complete genomes of two virulent TGEV strains and their attenuated counterparts: virulent TGEV Miller M6 and attenuated TGEV Miller M60 and virulent TGEV Purdue and attenuated TGEV Purdue P115, together with the ISU-1 strain of porcine respiratory coronavirus (PRCV-ISU-1), a naturally occurring TGEV deletion mutant with an altered respiratory tropism and reduced virulence. Pairwise comparison at both the nucleotide (nt) and amino acid (aa) levels between virulent and attenuated TGEV strains identified a common change in nt 1753 of the spike gene, resulting in a serine to alanine mutation at aa position 585 of the spike proteins of the attenuated TGEV strains. Alanine was also present in this protein in PRCV-ISU-1. Particularly noteworthy, the serine to alanine mutation resides in the region of the major antigenic site A/B (aa 506–706) that elicits neutralizing antibodies and within the domain mediating the cell surface receptor aminopeptidase N binding (aa 522–744). Comparison of the predicted polypeptide products of ORF3b showed significant deletions in the naturally attenuated PRCV-ISU-1 and TGEV Miller M60; these deletions occurred at a common break point, suggesting a related mechanism of recombination that may affect viral virulence or tropism. Sequence comparisons at both genomic and protein levels indicated that PRCV-ISU-1 had a closer relationship with TGEV Miller strains than Purdue strains. Phylogenetic analyses showed that virulence is an evolutionarily labile trait in TGEV and that TGEV strains as a group share a common ancestor with PRCV.
Keywords: Transmissible gastroenteritis virus, Porcine respiratory coronavirus, Complete genomic sequence, Virulence, Attenuation, Tropism shift
Introduction
Transmissible gastroenteritis virus (TGEV) is a group 1 coronavirus. It was identified as an etiological agent of transmissible gastroenteritis in swine in 1946 in the United States (Doyle, 1951, Doyle LP, 1946) and in the following two decades after its discovery, it was reported in England, Belgium, Japan, China, Australia and Africa (Kemeny and Woods, 1977, Pritchard, 1987, Wood et al., 1981, Woods, 1976, Woods and Wesley, 1986). Transmissible gastroenteritis virus replicates in the cytoplasm of villous epithelial cells in the small intestine, leading to severe villous atrophy and malabsorptive diarrhea, resulting in almost 100% mortality in seronegative suckling pigs (Saif and Sestak, 2006). The virus is an enteropathogen, although TGEV also replicates in the upper respiratory tract (Underdahl et al., 1975) with transient nasal shedding in experimentally inoculated pigs (VanCott et al., 1993).
Transmissible gastroenteritis virus has a positive-sense, single-stranded RNA genome of ∼ 28.5 kb in size. The genomic sequence contains 9 open-reading frames (ORFs) that encode 4 structural proteins [spike (S), envelope (E), membrane (M) and nucleoprotein (N)] and 5 nonstructural proteins (replicase 1a and 1b, 3a, 3b, and protein 7) arranged on the genome in the order 5′-replicase (1a/1b)-S-3a-3b-E-M-N-7-3′. The S glycoprotein of TGEV forms large, petal-shaped spikes on the surface of the virion which are responsible for binding to specific receptors on the membranes of susceptible cells. Aminopeptidase N (APN) is the major cell surface receptor for TGEV (Delmas et al., 1992) and several co-receptors including sialoglycoproteins have been implicated in conferring enteric tropism to TGEV (Ballesteros et al., 1997, Delmas et al., 1993). Four major antigenic sites, A, B, C and D were characterized on the N terminus of the S protein using monoclonal antibodies (Correa et al., 1988, Delmas et al., 1986, Delmas et al., 1990, Simkins et al., 1992). Monoclonal antibodies against S can neutralize virus infectivity either by blocking virus attachment to cells or through interfering with virus endocytosis or membrane fusion (Sune et al., 1990). The adjacent sites A and B were mapped to a region of approximately 200 amino acids beginning from residue 506 as its N terminal boundary (Godet et al., 1994). Antigenic sites A/B also overlap with the domain of the S protein encompassing amino acids 522–744 that mediate aminopeptidase N (APN) receptor binding (Godet et al., 1994). Three antigenic sites were also defined in the N protein (Martin Alonso et al., 1992, Simkins et al., 1989).
After continuous passage in cell culture, TGEV isolates gradually lose their virulence and viral tropisms may shift from enteric to respiratory (Furuuchi et al., 1978, Harada et al., 1969). Attenuation and tropism shift can also occur in nature, an example being the naturally occurring S gene deletion mutant, the porcine respiratory coronavirus (PRCV), which has both reduced pathogenicity and a predominantly respiratory tropism (Pensaert et al., 1986, Saif and Sestak, 2006, Wesley et al., 1990a, Wesley et al., 1990b). In contrast to TGEV, PRCV mainly infects the respiratory tract, causing mild to moderate pneumonia often with pronounced interstitial lung lesions, but with little or no replication in the intestine. Studies of PRCV have provided some insight into the determinants for the tropism and virulence changes in TGEV. At the genome level, PRCV and TGEV share high nucleotide sequence identity except that PRCV has a deletion in the 5′ end of the S gene and deletions in ORF3, the latter leading to lack of or a truncated protein expressed. The 5′ deletion in the S gene is widely believed to play a major role in the altered tissue tropism of PRCV (Wesley et al., 1990a, Wesley et al., 1990b, Wesley et al., 1991). Ballesteros et al. (1997) and Sanchez et al. (1999) identified two domains in the spike protein of attenuated TGEV Purdue strain PUR46-MAD responsible for binding APN and co-receptors and demonstrated that a respiratory Purdue Type TGEV (PTV) (formerly known as NEB72) lost the co-receptor binding site due to the S gene mutation. Point mutations in the spike gene leading to a shift from enteric to respiratory tropism were also found in high cell culture passaged TGEV strain TOY56 (Sanchez et al., 1992). Thus the spike protein has been recognized as not only a tropism but also a virulence determinant.
Although it is generally accepted that deletions in the spike and ORF3 genes contribute to tropism change and attenuation of PRCV, other genes may also be involved. For instance, amino acid mutations in the M protein affect the ability of attenuated Purdue TGEV P115 to induce IFN-alpha, implying a potential role for the M protein in altered host response and virulence (Laude et al., 1992). Other evidence has implicated a role for nonstructural proteins 3a and 3b in determining virulence of these swine enteric and respiratory coronaviruses (Paul et al., 1997). An infectious clone of TGEV Purdue strain PUR46-MAD with ORF3 gene deletions showed a slightly reduced pathogenicity in vivo, but normal replication in cell culture (Sola et al., 2003). Similar effects of ORF3 deletion were also observed for the group 3 coronavirus, infectious bronchitis virus (Hodgson and Cavanagh, 2006, Shen et al., 2003). However, in one study a TGEV strain 96–1933 with a deletion in ORF3a was demonstrated to maintain enteric virulence (McGoldrick et al., 1999), suggesting that ORF3a is not essential for virulence, although the virulence of this virus needs to be further confirmed because the virus isolated and sequenced was not plaque purified and was not tested in pigs after plaque purification.
To determine the molecular basis for TGEV attenuation, we analyzed the nucleotide and deduced amino acid sequences of structural and nonstructural proteins of two virulent/attenuated TGEV pairs as well as the PRCV strain ISU-1. Determining tropism and virulence factors at the genomic level for TGEV/PRCV strains should enhance our understanding of the evolution of coronaviruses including the newly emerged severe acute respiratory syndrome coronavirus (SARS-CoV) (Marra et al., 2003, Rota et al., 2003, Saif, 2004, Saif, 2005).
Results
Assembly and validation of TGEV genomic sequences
Sequencing reads were downloaded, trimmed to remove amplicon primer-linker sequences as well as low quality sequence and assembled using TIGR Assembler (www.tigr.org/software/assembler/). To close gaps between assembled contigs, strain-specific primers were designed, RT-PCR was performed and amplicons were sequenced as described in Materials and methods. Additional primer design, cDNA synthesis and sequencing were performed to ensure greater than 4× sequence coverage along the coronavirus genomes. Assemblies were manually edited using CloE (Closure Editor), a TIGR program for editing assemblies. All apparent polymorphisms were checked against reference data and ambiguities were analyzed by RT-PCR and cloning.
The final genome assemblies have been deposited in GenBank. The GenBank accession numbers are as follows: Virulent TGEV Miller M6:DQ811785; attenuated TGEV Miller M60: DQ811786; PRCV-ISU-1: DQ811787; attenuated TGEV Purdue P115: DQ811788; virulent TGEV Purdue: DQ811789. The genome lengths in nt for TGEV virulent Miller M6, attenuated Miller M60, virulent Purdue, attenuated Purdue P115 and PRCV-ISU-1 are 28542, 27915, 28577, 28571, and 27546, respectively.
Genetic characterization of the TGEV and PRCV genomes
Genome sequence alignment of the TGEV strains and PRCV with the reference genome sequence of TGEV strain PUR46-MAD from GenBank (accession number NC_002306) showed comparable genome size and identical gene order. All 5 genomes start with 5′ untranslated regions similar to that of the reference strain above and end with a polyA tail except that the sequence for M60 is 69 nt short of reaching the polyA end. The TGEV and PRCV strains share high genomic sequence identities, ranging from 96.2% to 99.9% (Table 1 ). PUR46-MAD and Purdue P115 showed 99.9% genomic sequence similarity, because TGEV PUR46-MAD strain was derived from TGEV Purdue P115 and similarly attenuated from the virulent TGEV Purdue strain after high passage in cell culture (Penzes et al., 2001, Sanchez et al., 1992).
Table 1.
Pairwise distance of genomes of PRCV-ISU-1, the four TGEV strains and the reference TGEV PUR46-MAD strain (GenBank Accession number NC_002306)a
| Reference TGEVNC_002306 | Attenuated TGEV Miller M60 | Virulent TGEV Miller M6 | Virulent TGEV Purdue | Attenuated TGEV Purdue P115 | PRCV-ISU-1 | |
|---|---|---|---|---|---|---|
| Reference TGEV-NC_002306 | *** | 98.7 | 98.8 | 99.8 | 99.9 | 97.5 |
| Attenuated TGEV Miller M60 | 1.2 | *** | 99.9 | 98.8 | 98.7 | 96.2 |
| Virulent TGEV Miller M6 | 1.2 | 0.1 | *** | 98.9 | 98.7 | 98 |
| Virulent TGEV Purdue | 0.1 | 1.1 | 1.1 | *** | 99.8 | 97.6 |
| Attenuated TGEV Purdue P115 | 0.1 | 1.3 | 1.2 | 0.2 | *** | 97.5 |
| PRCV-ISU-1 | 2.5 | 2 | 2 | 2.4 | 2.6 | *** |
Pairwise distance was calculated with ClustalW program using DNASTAR software. Percent similarity in upper right portion of table (above asterisks) and percent divergency in lower portion of table.
The 5 genomes contained 8 to 9 open-reading frames (ORFs) typical of other TGEV and PRCV strains. The length in amino acids of the structural and nonstructural proteins of the four TGEV strains and PRCV-ISU-1 is summarized in Table 2 . No protein 3a is encoded in PRCV-ISU-1 due to a 184 nt deletion at the 5′ end of the 3a gene.
Table 2.
Length in amino acids of predicted structural and nonstructural proteins of the four TGEV isolates and PRCV-ISU-1
| Virulent TGEV Miller M6 | Attenuated TGEV Miller M60 | Virulent TGEV Purdue | Attenuated TGEV Purdue P115 | PRCV-ISU-1 | |
|---|---|---|---|---|---|
| Replicase 1a | 4017 | 4017 | 4017 | 4017 | 4014 |
| Replicase 1b | 2680 | 2680 | 2680 | 2680 | 2680 |
| S | 1449 | 1448 | 1449 | 1447 | 1222 |
| 3a | 72 | 72 | 71 | 71 | – |
| 3b | 244 | 67 | 244 | 244 | 205 |
| E | 82 | 82 | 82 | 82 | 82 |
| M | 262 | 264 | 262 | 262 | 261 |
| N | 382 | 382 | 382 | 382 | 382 |
| 7 | 78 | 78 | 78 | 78 | 78 |
Nucleotide and amino acid changes in structural and nonstructural proteins after attenuation
Each viral assembly was analyzed using Viral Genome ORF Reader (VIGOR), a program designed at TIGR to predict viral protein sequences. Using VIGOR, we checked segment length, alignments with reference sequences, fidelity of reading frames, correlated amino acid mutations with nucleotide polymorphisms and detected potential sequence errors. The open-reading frames of the structural and nonstructural proteins of the four TGEV strains were aligned using the ClustalW algorithm to identify amino changes after attenuation. Comparison of individual proteins from each of the TGEV isolates revealed a number of deletions, insertions and point mutations that are summarized in Table 3 . A total of 20 amino acid point mutations were found in Miller M60 and 32 point mutations were found in Purdue P115, when compared to their virulent counterparts (Table 3).
Table 3.
Amino acid mutations in the structural and nonstructural proteins of M60 and P115 relative to their virulent counterpartsa
| aa Positionb | M6c | M60c | Virulent Purduec | P115c | |||
|---|---|---|---|---|---|---|---|
| Replicase 1a | 572 | S | S | S | F | ||
| 724 | S | S | S | G | |||
| 799 | V | V | V | I | |||
| 984 | N | T | N | N | |||
| 1174 | E | A | E | E | |||
| 1541 | V | V | V | I | |||
| 1576 | S | S | S | G | |||
| 1579 | I | I | I | V | |||
| 1884 | Q | H | Q | Q | |||
| 2732 | T | T | T | I | |||
| 2816 | P | P | P | S | |||
| 2818 | D | D | D | A | |||
| 3366 | V | V | V | I | |||
| 3786 | A | A | A | S | |||
| 3883 | A | A | A | S | |||
| 4015 | P | Q | Q | Q | |||
| No. of mutations | 4 | 12 | |||||
| Replicase 1b | 120 | A | A | V | A | ||
| 1250 | N | D | N | N | |||
| 2520 | C | G | G | G | |||
| No. of mutations | 2 | 1 | |||||
| Spike | 32 | H | H | H | Q | ||
| 53 | V | A | V | V | |||
| 86 | L | L | L | V | |||
| 97 | L | S | W | W | |||
| 99 | H | Y | H | H | |||
| 208 | L | L | L | F | |||
| 318 | L | F | F | F | |||
| 375 | N | N | Y | Deleted | |||
| 376 | D | D | D | Deleted | |||
| 403 | L | L | L | H | |||
| 417 | P | P | P | L | |||
| 418 | S | I | S | S | |||
| 496 | L | L | L | I | |||
| 536 | G | G | D | G | |||
| 562 | N | N | N | H | |||
| 564 | T | N | T | T | |||
| 585d | S | A | S | A | |||
| 675 | L | L | L | V | |||
| 796 | V | Deleted | V | V | |||
| 884 | L | Q | Q | Q | |||
| 942 | E | E | G | E | |||
| 1109 | T | T | T | I | |||
| 1234 | A | A | A | P | |||
| No. of mutations | 8 | 13 | |||||
| Protein 3a | 8 | N | N | D | Y | ||
| 29 | V | I | V | V | |||
| 72 | H | H | Deleted | Deleted | |||
| No. of mutations | 1 | 1 | |||||
| Protein 3b | 8 | N | N | N | S | ||
| 32 | Q | H | Q | Q | |||
| 33–209 | Deleted | ||||||
| No. of mutations | 1 | 1 | |||||
| E | 13 | N | T | N | N | ||
| 18 | S | S | S | N | |||
| 33 | L | S | S | S | |||
| No. of mutations | 2 | 1 | |||||
| M | 67–68 | Insertion | |||||
| 156 | N | N | N | K | |||
| No. of mutations | 0 | 4 | |||||
| N | 110 | D | D | E | D | ||
| 182 | S | S | F | S | |||
| 287 | I | V | I | I | |||
| No. of mutations | 1 | 2 | |||||
| Protein 7 | 71 | Y | F | Y | Y | ||
| No. of mutations | 1 | 0 |
Deduced protein sequences of all four TGEV viruses were aligned with ClustalW program in Lasergene software (DNASTAR Inc.).
Positions for amino acid changes are shown.
Amino acids are bold italics when mutations are present between virulent and attenuated TGEV strains.
The common mutation between the two virulent and attenuated pairs at aa 585 in spike protein is highlighted by underlining.
Spike gene
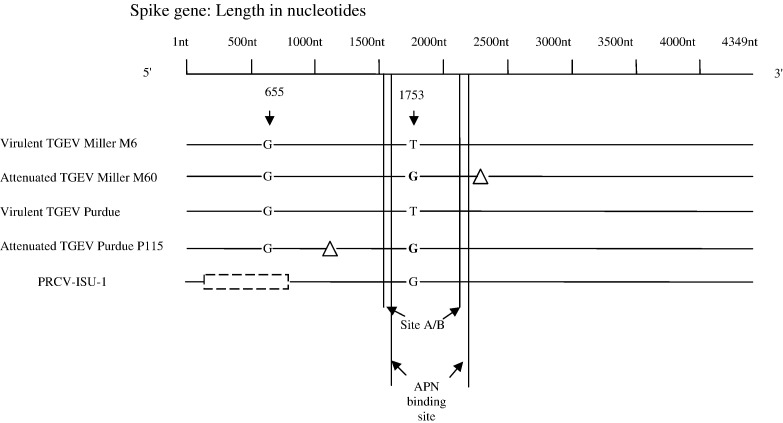
A schematic illustration of spike gene changes and deletions in TGEV isolates and PRCV is presented in Fig. 1 . The most striking variation between attenuated and virulent isolates was seen in the spike and ORF3a/3b genes. Large deletions were present in the spike gene of PRCV-ISU-1 and in the ORF3 gene of both PRCV-ISU-1 and M60 (Table 3, Table 4 ). A 3 nt deletion was present in the S gene of Miller M60 from 2385–2387, resulting in a spike protein of 1448 aa in length, 1 aa shorter compared to Miller M6. In Purdue P115 there was a 6 nt deletion in the S gene, leading to a spike protein 2 aa shorter than virulent Purdue strain (1447 vs. 1449 aa). Sequence analysis confirmed the previously reported 681 nt deletion in the 5′ end of the S gene in PRCV-ISU-1 (Bae et al., 1991, Wesley et al., 1991).
Fig. 1.
Schematic representation of the spike gene of the four TGEV strains and PRCV-ISU-1 showing deletions and mutations. △, 3 nt (nt 2385 to 2387) deletion in M60 spike gene and 6 nt (nt 1122 to 1127) deletion in P115 spike gene.  , 681 nt (nt 65 to 745) deletion in PRCV spike gene. The T to G (bold) mutation at nt 1753 in M60, P115 and PRCV leading to an S to A substitution at aa 585 in the spike protein is shown. The approximate antigenic region of site A/B (nt 1518–2118) and the aminopeptidase N (APN) binding site (nt 1566–2232) are indicated. The potential enteric tropism determining residue at nt position 655 identified by Ballesteros et. al. (1997) is shown.
, 681 nt (nt 65 to 745) deletion in PRCV spike gene. The T to G (bold) mutation at nt 1753 in M60, P115 and PRCV leading to an S to A substitution at aa 585 in the spike protein is shown. The approximate antigenic region of site A/B (nt 1518–2118) and the aminopeptidase N (APN) binding site (nt 1566–2232) are indicated. The potential enteric tropism determining residue at nt position 655 identified by Ballesteros et. al. (1997) is shown.
Table 4.
Amino acid differences in the structural and nonstructural proteins of TGEV Miller and Purdue strainsa
| aa Positionb | M6 | M60 | Virulent Purdue | P115 | PRCV-ISU-1c | |
|---|---|---|---|---|---|---|
| Replicase 1a | 102 | F | F | L | L | F |
| 286 | T | T | A | A | A | |
| 343 | I | I | V | V | I | |
| 355 | E | E | D | D | E | |
| 521 | A | A | V | V | A | |
| 590 | M | M | V | V | V | |
| 639 | L | L | F | F | L | |
| 803 | E | E | K | K | E | |
| 821 | A | A | S | S | A | |
| 888 | D | D | E | E | D | |
| 1008 | N | N | D | D | N | |
| 1023 | I | I | T | T | I | |
| 1029 | A | A | V | V | A | |
| 1035 | V | V | A | A | A | |
| 1079 | T | T | N | N | N | |
| 1299 | L | L | P | P | L | |
| 1708 | R | R | K | K | K | |
| 1800 | R | R | K | K | K | |
| 1894 | V | V | A | A | A | |
| 2178 | N | N | S | S | N | |
| 2267 | L | L | F | F | F | |
| 2420 | A | A | V | V | V | |
| 2741 | V | V | I | I | I | |
| 2763 | V | V | I | I | V | |
| 3232 | F | F | V | V | F | |
| 4000 | S | S | F | F | S | |
| Replicase 1b | 272 | H | H | Y | Y | Y |
| 459 | H | H | Q | Q | Q | |
| 480 | S | S | N | N | S | |
| 718 | V | V | I | I | V | |
| 1290 | V | V | A | A | V | |
| 1479 | S | S | A | A | A | |
| 1957 | T | T | M | M | M | |
| 2081 | R | R | K | K | R | |
| 2123 | N | N | K | K | N | |
| Spike | 48 | S | S | P | P | |
| 72 | N | N | D | D | ||
| 100 | K | K | R | R | AA 23–250 | |
| 184 | A | A | E | E | Deleted | |
| 218 | T | T | V | V | ||
| 384 | S | S | F | F | S | |
| 389 | M | M | I | I | I | |
| 487 | Y | Y | H | H | Y | |
| 590 | V | V | I | I | V | |
| 649 | D | D | E | E | D | |
| 815 | L | L | F | F | L | |
| 951 | D | D | H | H | D | |
| 967 | A | A | D | D | S | |
| 1239 | L | L | S | S | L | |
| Protein 3a | 50 | R | R | K | K | No protein 3A due to deletion in 3A gene |
| 67 | T | T | S | S | ||
| 68 | Q | Q | H | H | ||
| 69 | N | N | I | I | ||
| 70 | P | P | V | V | ||
| 71 | K | K | V | V | ||
| Protein 3b | 31 | K | K | Q | Q | Q |
| 239 | R | R | H | H | R | |
| E | 58 | V | V | A | A | V |
| 82 | V | V | A | A | V | |
| M | 24 | D | D | S | S | Deleted |
| 27 | G | G | D | D | G | |
| 64 | I | I | V | V | I | |
| 96 | I | I | V | V | I | |
| N | 28 | S | S | N | N | S |
| 262 | S | S | T | T | S | |
| 320 | E | E | G | G | E | |
| 355 | E | E | D | D | E | |
| 376 | M | M | I | I | M | |
| Protein 7 | 4 | L | L | F | F | L |
| 14 | T | T | I | I | T | |
| 39 | D | D | N | N | D | |
| 60 | L | L | V | V | L | |
| 76 | I | I | T | T | I |
Deduced protein sequences of all four TGEVs and PRCV-ISU-1 were aligned with ClustalW program in Lasergene software (DNASTAR Inc.).
Positions at which amino acid is different were listed.
The aa of PRCV-ISU-1 at the divergent positions is listed. Letters are in bold when PRCV-ISU-1 has identical aa as Miller strains at the divergent positions.
A common T (virulent TGEV strains) to G mutation for both M60 and Purdue P115 was located at nt 1753 of the spike genes. The PRCV-ISU-1 strain had a G at the corresponding nucleotide position. This mutation that resulted in a serine to alanine mutation at aa 585 of the spike protein, was the only common amino acid change found between both the Miller and Purdue strains after attenuation. Because the alanine residue was also observed at the corresponding position (585) in the spike protein of PRCV-ISU-1, it may represent a genetic marker of attenuation among TGEV and PRCV strains but further analysis using reverse genetics and experimental animal studies is needed to confirm this possibility.
ORF3a/3b gene
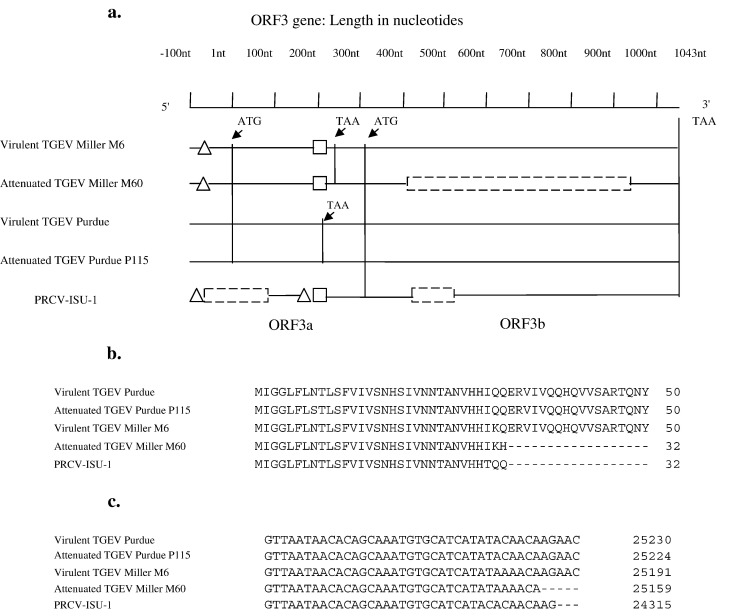
The ORF3a/3b deletions are represented schematically in Fig. 2a. PRCV-ISU-1 showed a 184 nt deletion in the ORF3a gene, disrupting the predicted open-reading frame of nonstructural protein 3a. A 3 nt deletion upstream and a 5 nt deletion downstream of the 184 nt deletion were also found. Furthermore, PRCV-ISU-1 contained a 117 nt in frame deletion in ORF3b gene, leading to a shorter nonstructural protein 3b relative to TGEV strains. Sequence comparison of ORF3 genes between the Miller and Purdue strains revealed 2 large deletions (16 and 29 nt respectively) in the ORF3a gene of the Miller strains. A previously undescribed 29 nt deletion was also present in PRCV-ISU-1 at exactly the same position as Miller strains when compared to Purdue strains. Miller M60 had a 531 nt in frame deletion in ORF3b gene resulting in a truncated 3b protein of 67 aa long. The deletion in M60 ORF3b gene was noted previously by our laboratory (Kwon et al., 1998) and was independently confirmed in this study by RT-PCR and sequence analysis (data not shown). Interestingly, these deletions occur at the same amino acid position (aa 33) in the predicted polypeptides of M60 and PRCV-ISU-1 (Fig. 2b); the apparent recombination point in the nucleotide sequence occurs within 2 nucleotides of each other as determined by ClustalW alignment (Fig. 2c).
Fig. 2.
(a) Schematic representation of ORF3a/3b gene (nt 1 to 1043) and the 100 nt preceding the gene (nt − 100 to 0): Gene start codon ATG for nonstructural protein 3a starts from nt 1 and ends at nt 247 for M6 and M60 and nt 215 for virulent Purdue and Purdue P115. No nonstructural protein 3a is encoded for PRCV-ISU-1 due to the 184 nt deletion that includes the ATG start codon. Nonstructural protein 3b starts from nt 310 and ends at nt 1043. △, 3 nt (nt − 100 to − 98) deletion at 5′ end and 5 nt (nt 163 to 167) deletion downstream in ORF3a of PRCV-ISU-1. In TGEV M6 and M60, △ represents 16 nt (nt − 76 to − 61) deletions. □, 29 nt (nt 195 to 223) deletion in PRCV, M6 and M60.  , 531 nt (nt 405 to 935) deletion in M60 3b and 184 nt (nt − 87 to 97) and 117 nt (nt 407 to 523) deletions in PRCV 3b respectively. (b) ClustalW alignment of ORF3b predicted polypeptides of TGEV strains and PRCV-ISU-1 showing the deletion start point in Miller M60 and PRCV. (c) ClustalW alignment of the nucleotide sequence of TGEV strains and PRCV-ISU-1 in the region of ORF3b deletion start point.
, 531 nt (nt 405 to 935) deletion in M60 3b and 184 nt (nt − 87 to 97) and 117 nt (nt 407 to 523) deletions in PRCV 3b respectively. (b) ClustalW alignment of ORF3b predicted polypeptides of TGEV strains and PRCV-ISU-1 showing the deletion start point in Miller M60 and PRCV. (c) ClustalW alignment of the nucleotide sequence of TGEV strains and PRCV-ISU-1 in the region of ORF3b deletion start point.
3′ end genes
Analysis of the 3′ end of the viral genomes revealed four genes encoding E, M, N, and protein 7. No deletions or insertions were present in E, N, and protein 7 genes. The deduced E and N proteins and protein 7 among the 5 viruses were 82 aa, 382 aa and 78 aa, respectively. There was a small variation in M protein size with M60 having a 6 nt insertion in the M gene when compared to the other 3 TGEV strains, leading to a membrane protein 2 aa longer than those of M6 and Purdue strains (264 aa and 262 aa respectively). The PRCV-ISU-1 had a 3 nt deletion in the M gene, resulting in an M protein of 261 aa.
Replicase 1a and 1b
About two thirds of the TGEV genome encodes the replicase genes 1a and 1b. The replicase genes are relatively conserved and no major deletions and insertions were present. In both the Miller and Purdue isolates, the replicase 1a gene was predicted to encode a protein of 4017 aa and the replicase 1b gene was predicted to encode a protein of 2680 aa. Comparison of the predicted polypeptide sequences indicated 4 aa changes in replicase 1a and 2 aa changes in replicase 1b for the M6/M60 pair. Twelve aa changes in replicase 1a and 1 aa change in replicase 1b were identified for virulent Purdue/P115 pair (Table 3). However, no common changes were found between the two pairs after attenuation.
Sequence differences between TGEV Miller and Purdue strains and their relationship to PRCV-ISU-1
Two short deletions were seen in the replicase 1a gene of PRCV-ISU-1, 6 nt from nt 3252 to 3257 and 3 nt from nt 3331 to 3333, respectively when compared to Miller and Purdue strains. More sequence differences were found between PRCV-ISU-1 and TGEV strains than between TGEV Miller and Purdue strains. TGEV virulent Miller M6 shared 98.9% genomic sequence identity with virulent Purdue strain. PRCV-ISU-1 had 98.0% and 97.6% genomic sequence identities with M6 and virulent Purdue, respectively. The high sequence homology among TGEV strains and PRCV-ISU-1 suggests these viruses are closely related, but PRCV-ISU-1 has a closer relationship with TGEV Miller strains than Purdue strains. To identify differences at the aa level, individual proteins of Miller and Purdue strains were compared. Amino acids of PRCV-ISU-1 at divergent positions were also listed (Table 4). Proteins of PRCV-ISU-1 were more biased toward Miller strains than Purdue strains, including S, protein 3a/3b, E, M, N and protein 7.
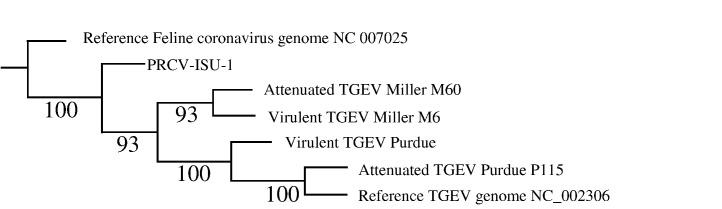
Phylogenetic analysis of TGEV and PRCV-ISU-1 genomic sequences
Sequence comparisons at both genomic and protein levels indicated that PRCV-ISU-1 had a closer relationship with TGEV Miller strains than Purdue strains. To further define the ancestry relatedness of PRCV-ISU-1 to TGEV stains, phylogenetic analysis of PRCV-ISU-1 was performed against the 4 TGEV strains and PUR46-MAD genomic sequence available in GenBank (accession number NC_002306). The phylogenetic tree was rooted with a feline coronavirus (accession number NC 007025) as an outgroup. The tree showed that Miller M6 and Miller M60 were most closely related with each other, consistent with the fact that both M6 and M60 are derived from Miller strain (Fig. 3 ). Purdue P115 clustered most closely with PUR46-MAD and together these two Purdue strains shared the closest relationship with virulent Purdue. PUR46-MAD strain is a derivative of the TGEV Purdue-P115 strain and both strains were derived from the virulent Purdue strain (Penzes et al., 2001, Sanchez et al., 1999). The tree also revealed that all TGEV strains were clustered into one clade, indicating TGEV strains share a common ancestor and as a group TGEV strains share a common ancestor with PRCV-ISU-1 (Fig. 3).
Fig. 3.
Phylogenetic analysis of genomic nucleotide sequences of the four TGEV strains and PRCV-ISU-1 with the reference TGEV genomes available in GenBank, and a Feline Coronavirus outgroup. The tree is based on muscle alignment of whole genomes. The tree search was conduced with TNT (Goloboff et al., 2005) with equally weighted parsimony counting gaps as a fifth state. Tree search was conducted using new technology parameters until a stable consensus was discovered. This search procedure produced a single tree of 7360 steps. Bootstrap values for 1000 resampling replicates are shown at nodes.
Discussion
We completed the entire genomic sequences of two virulent/attenuated TGEV pairs and the ISU-1 strain of PRCV. Although partial sequences of these strains were available in GenBank, to our knowledge this is the first report of the full genomic sequence of these pairs of viruses in comparison to one another and to a PRCV strain. The detailed comparison of the sequences of the 2 virulent and attenuated TGEV pairs aids in the identification of the genetic basis of coronavirus attenuation, which has not yet been clearly established and provides targets for verification of the role of such changes using infectious clones of TGEV. Sequences for TGEV genomes in public databases are lacking; before the present study there were only three complete genomic sequences of any TGEV strains in GenBank (accession number NC_002306, DQ443743 and DQ201447). Addition of the full genomic sequences for these 5 viruses will aid in understanding animal coronaviruses including their genetic structure, diversity and evolution.
The sequence analyses identified deletions that were reported previously including the 6 nt deletion in the S gene of the Purdue P115 compared to the virulent Purdue strain (Rasschaert and Laude, 1987) and 2 large deletions (16 and 29 nt respectively) in the ORF3a gene of the Miller strains when compared to the Purdue strains (Rasschaert et al., 1987, Wesley et al., 1989). Alignment of the genome sequences of the two virulent/attenuated pairs of TGEV strains revealed a common change at nt 1753 of the spike gene, resulting in a serine to alanine mutation in aa position 585 of the spike protein of the attenuated strains. This was the only common change at the protein level found in the attenuated viruses in comparison to their virulent counterparts. Interestingly, the naturally attenuated PRCV-ISU-1 also has an alanine residue at the corresponding position. This suggests that the alanine in place of serine may be a genetic marker for attenuation of TGEV strains and for PRCV which is also attenuated for pigs. A study by Sanchez et al. (1992) revealed that all TGEV strains analyzed had an alanine at aa 585 of the spike protein except for Mil65-AME. The MIL65-AME strain so designated by Enjuanes and colleagues is a derivative of our virulent Miller strain as demonstrated (Penzes et al., 2001). It was the only virulent enteric TGEV strain analyzed in their study and the other strains were either respiratory or attenuated by high passage through tissue culture.
A phenotypic change in TGEV resulting from a similar single nucleotide change at a different site in the spike protein has been reported previously. In a prior study, Ballesteros et al. (1997) and Sanchez et al. (1999) identified a G residue at nt position 655 of the spike protein that was essential to maintain enteric tropism of TGEV strain PUR46-MAD. Mutations at this nucleotide caused a shift from enteric to respiratory tropism of this virus. The 6 nt (nt 1122 to 1127, aa 375–376) deletion in the attenuated TGEV Purdue P115 spike gene, but not in the virulent Purdue strain, that we observed may also play a role in its attenuation. Penzes et al. (2001) observed the same deletion in the spike gene of attenuated Purdue strains PUR46-C8 and PUR46-MAD, but not in the in vivo maintained virulent strain PUR46-C11.
The serine to alanine mutation at aa 585 is located in the major antigenic sites A/B of TGEV spike proteins and within the binding site for receptor APN (Delmas et al., 1986, Delmas et al., 1990), suggesting that this change may have a significant influence in the receptor binding or neutralizing antibody interactions. Single amino acid changes in the spike protein of coronaviruses can have significant effects on antigenicity. For example, a single amino acid change in the antigenic domain II of the spike protein confers resistance to neutralization of a bovine coronavirus (Yoo and Deregt, 2001). The alanine residue is also present at the corresponding position in the spike protein of PRCV-ISU-1. Antigenic site A/B has been mapped from aa 506 to 706 of the spike protein (Godet et al., 1994); within this region there were 2 aa changes for M60 as compared to M6 and 4 aa changes for Purdue P115 as compared to virulent Purdue. However, the impact of the serine to alanine mutation on pig pathogenicity awaits generation of recombinant viruses by reverse genetics followed by their testing in vivo.
Although researchers have indicated that deletions in ORF3 may be involved in the virulence of TGEV (Paul et al., 1997) and deletion of ORF3 gene in a recombinant TGEV virus showed a limited effect on the viral virulence in vivo (Sola et al., 2003), only one mutation, but no deletions and insertions was found in the ORF3a and ORF3b proteins of the attenuated Purdue TGEV P115 when compared to the virulent Purdue TGEV and no mutations in ORF3 proteins were shared in the two virulent/attenuated pairs. It is noteworthy that we predicted the existence of protein 3b of virulent and attenuated Purdue TGEV viruses using the VIGOR program. Others suggested that the mRNA encoding protein 3b was not observed in TGEV Purdue, but only in Miller strains using infected cell lysates and Northern blot analysis (Izeta et al., 1999, Penzes et al., 2001) and therefore described it as a pseudogene in TGEV Purdue strains. A mutation was found in the highly conserved core sequence (CS, previously known as intergenic sequence IS) within coronavirus transcription regulatory motif, 5′-CUAAAC-3′, preceding ORF3b genes of the TGEV Purdue strains. This mutation replaced the last nucleotide C in CS with U. Because the CS represents signals for coronavirus transcription of subgenomic mRNAs (Lai and Cavanagh, 1997, Sawicki and Sawicki, 1990), it is speculated that the mutation renders the mRNA encoding the 3b protein undetectable by Northern blot for TGEV Purdue strains. However, the single mutation in the CS preceding the ORF3b gene of TGEV Purdue viruses may not completely abolish its transcription. It has been demonstrated that a core sequence differing at the last nucleotide from the canonical one (5′-CUAAAC-3′) maintains efficient transcription for the downstream gene (Sola et al., 2003). Because we observed this mutation in the CS of ORF3b gene for both virulent Purdue and attenuated Purdue P115 strains, protein 3b is unlikely to play a role in the attenuation of TGEV Purdue strains. Similarly, the absence of mutations or deletions in the nonstructural protein 7 of the virulent/attenuated Purdue TGEV pair indicates that changes in this protein alone are unlikely to be involved in the attenuation process, although evidence suggests that deletion of the entire ORF7 gene resulted in altered in vivo replication and virulence of TGEV based on a recombinant PUR46-MAD virus (Ortego et al., 2003).
Due to lack of proof-reading activity of the virally encoded RNA-dependent RNA polymerase, coronaviruses introduce frequent mutations into their genomes during replication. In addition to randomly generated mutations, RNA recombination can also occur when multiple distinct coronavirus species infect the same host. The occurrence of frequent genomic changes leads to generation of new coronaviruses that can have altered pathogenicity, different tissue tropism or ability to cross host species barrier. An example is the SARS outbreak caused by a previously unidentified coronavirus (SARS-CoV). Recent studies identified bats as the likely natural reservoir of SARS-CoV. The SARS-CoV-like coronaviruses of bats may have potentially become adapted to humans through genomic mutation and recombination events either directly or via intermediate hosts (civet cats, etc.) (Hampton, 2005, Lau et al., 2005, Li et al., 2005, Normile, 2005). Although occurrence of SARS surprised the medical community, the relevance of RNA recombination and mutation to animal coronavirus evolution and tropism shift had been previously well recognized for TGEV and PRCV strains (Saif, 2004, Saif, 2005, Saif and Sestak, 2006, Sanchez et al., 1992). The independent appearance of PRCV in both Europe and the United States in the 1989s highlighted the possible emergence of new coronavirus strains with altered tissue tropism, virulence or host specificity due to genomic deletion and mutation events. The European PRCV strains characterized had a 672 nt deletion in the same position within the S gene, suggesting they evolved from the same predecessor (Callebaut et al., 1988, Sanchez et al., 1990, Sanchez et al., 1992). However, the initial PRCV strains detected in the United States had deletions of 681 nt in size (Kwon et al., 1998, Wesley et al., 1991). Subsequently other PRCV strains were isolated that varied in the size of the spike gene deletion (Halbur et al., 1993, Kim et al., 2000, Vaughn et al., 1994). Nevertheless, deletions in the S genes disrupt expression of antigenic sites on S proteins of both European and American PRCV strains. As a result, TGEV and PRCV can be serologically differentiated by monoclonal antibodies to the two antigenic sites (Callebaut et al., 1988, Simkins et al., 1993). TGEV and PRCV can also be genetically differentiated by nested RT-PCR using primers flanking the S gene deletion (Kim et al., 2000, Paton et al., 1997).
Phylogenetic analysis revealed a close genetic relationship between the PRCV-ISU-1 and the four TGEV strains, indicating that PRCV-ISU-1 shares a common ancestor with TGEV Miller and Purdue strains. Nevertheless, it remains unclear which strain of TGEV is the immediate ancestor of PRCV-ISU-1. It had been hypothesized that PRCV may have evolved from a live vaccine strain of TGEV such as the Purdue strain used for commercial vaccines in the US. However, our analysis suggests that PRCV-ISU-1 may have evolved from a TGEV Miller-like strain. Several lines of evidence support this conclusion. First, our results showed that PRCV-ISU-1 has a higher genomic sequence identity (98%) with Miller M6 than with Purdue strains and it has a closer relationship to Miller strains than to Purdue strains (including the complete Purdue strain genome available in GenBank) by phylogenetic analysis. Secondly, Miller strains have identical or similar deletions in the ORF3 gene as PRCV-ISU-1. The 29 nt deletion from nt 195 to 223 of the ORF3 gene of PRCV-ISU-1 is shared by Miller strains, but not by Purdue strains. The 117 nt deletion in ORF3b gene (nt 407 to 523) of PRCV-ISU-1 overlaps with the 531 nt deletion in ORF3b of Miller M60. This deletion is notable because it occurs at the same amino acid in each predicted polypeptide and at equivalent nucleotide coordinates in both M60 and PRCV-ISU-1. The fact that PRCV-ISU-1 has the closest relationship with Miller strains at both the genomic and protein levels and the ORF3 gene of Miller strains also have a high frequency of mutations and deletions suggests that a Miller-like strain is the ancestor of PRCV-ISU-1 and M60 may represent an analog of an intermediate species during PRCV evolution from a TGEV Miller-like strain.
Our sequence analysis revealed that genetic divergence most frequently occurs within the S gene and also between the S and M genes of TGEV in ORF3a/3b, suggesting that these regions are under the highest selective pressure during TGEV evolution. Mutations and deletions in these regions have been documented in TGEV variants that have altered tropisms, virulence or phenotype (Kim et al., 2000, Kwon et al., 1998, Page et al., 1991, Sanchez et al., 1992, Wesley et al., 1990a, Wesley et al., 1990b, Wesley et al., 1991). Our sequence analyses of the 4 TGEV strains and PRCV-ISU-1 have identified two common changes in the variable S and ORF3 genes, the T to G mutation at nt 1753 of the S genes of the attenuated M60 and P115 compared to the virulent counterparts and the deletions in ORF3b genes of M60 and PRCV-ISU-1 starting at a nearly identical position for each. These deletions may also be related to common mechanisms of attenuation between these related strains. We could not ascertain whether genetic changes at these two positions alone may have altered viral virulence; however, identification of these two common changes should help to localize sequences determining TGEV attenuation as confirmed by using reverse genetics.
Materials and methods
Viruses
The history of prototypic virulent TGEV Miller M6, attenuated TGEV Miller M60, virulent TGEV Purdue, attenuated TGEV Purdue P115, and PRCV-ISU-1 was summarized previously (Simkins et al., 1992). Briefly M6 and M60 are low and high tissue culture-passaged TGEV virulent and attenuated Miller strains, respectively (Saif and Sestak, 2006). The virulent Miller M6 was derived from field Miller isolate after 6 passages and 2 plaque purification steps in swine testicular (ST) cells in Ohio in 1965 (Bohl EH, 1965, Saif and Sestak, 2006, Welch and Saif, 1988). The Miller M60 was attenuated after 60 passages and 2 plaque purification steps in ST cells (Saif and Sestak, 2006, Woods, 1979). The virulent Purdue strain was originally isolated by Haelterman in Indiana and passed 8 times in pigs, with 2 subsequent cloning steps in ST cells (Haelterman, 1964). Continuous passage 115 times (115X), with numerous plaque purifications of the virulent Purdue strain led to the attenuated Purdue P115 strain (Bohl et al., 1972). The ISU-1 strain of PRCV originated from a herd isolate by Hill in Indiana in 1990. It was passaged 8× in ST cell culture and plaque purified 2× (Hill et al., 1990). The prototypic viruses were subjected to additional passages in ST cells or gnotobiotic pigs before sequencing in our lab. An additional 10, 4, 10, and 11 passages in ST cells were applied to Miller M6, Miller M60, PRCV-ISU-1, and Purdue P115, respectively. For virulent TGEV Purdue, 2 more passages were done in gnotobiotic pigs and the cell passaged M6 (10 additional passages) was also confirmed as virulent for the gnotobiotic pigs (Welch and Saif, 1988).
Virus purification and RNA extraction
Viral RNA was extracted from ST cell culture homogenates (TGEV M6, M60, P115 and PRCV-ISU-1) or infected gnotobiotic pig intestinal contents (virulent TGEV Purdue virus) after sucrose gradient purification as previously described (Kim et al., 2000, Paton et al., 1997). The total RNA was extracted from viral cell culture supernatants using TRIZOL LS reagent (Gibco, Life Tech, Grand Island, NY) according to the manufacturer's instructions. For virulent TGEV Purdue, the virus-containing gnotobiotic pig intestinal contents were purified by ultracentrifugation (112,700×g for 18 h) on 20% to 50% sucrose density gradients as described previously (Hasoksuz et al., 2002).
Viral genome sequencing
Specific oligonucleotide primers were designed using attenuated TGEV strain PUR46-MAD (NC_002306) as a reference genome. Primers were designed at every 500 bp along the genome. An M13 sequence tag was added to the 5′ end of each primer to be used for sequencing (forward primer: TGTAAAACGACGGCCAGT; reverse primer: CAGGAAACAGCTATGACC). Oligonucleotide primers were purchased from Invitrogen (Carlsbad, California, USA). Primer sequences are included in Supplementary Table S1. Reverse transcription and polymerase chain reactions (RT-PCR) were performed with 50–200 ng of coronavirus RNA supplemented with ribonuclease inhibitor (RNASEOUT, Invitrogen, Carlsbad, California, USA) using OneStep RT-PCR according to the manufacturers instructions (OneStep RT-PCR Kit, Qiagen, Valencia, CA, USA). Duplicate reactions were analyzed for quality control purposes by agarose gel electrophoresis. Amplicons were prepared for sequencing by incubation at 37 °C for 60 min with 0.5 U of Shrimp Alkaline Phosphatase (USB, Cleveland, Ohio) and 1 U of Exonuclease I (USB, Cleveland, Ohio) to inactivate remaining dNTPs and to digest the single-stranded primers. The enzymes were inactivated by incubation at 72 °C for 15 min. Sequencing reactions were performed on a standard high-throughput sequencing system using Big Dye Terminator chemistry (Applied Biosystems) with 2 μl of template cDNA. Each amplicon was sequenced from each end using M13 forward and reverse primers listed above. Sequencing reactions were analyzed on a 3730 ABI sequencer.
Sequence alignment and phylogenetic analyses
Alignment of nucleotide and amino acid sequences was performed using ClustalW program in Lasergene Software (DNASTAR Inc. Madison, WI). The phylogenetic tree (Fig. 3) was based on the entire genome alignment using multiple sequence comparison by log-expectation (MUSCLE) software (http://www.drive5.com/muscle/). The tree search was conducted with equally weighted parsimony method with gaps treated as a fifth state as implemented in TNT (Goloboff et al., 2005). Tree search was conducted using new technology parameters until a stable consensus was discovered. This search procedure produced a single tree of 7360 steps. Bootstrap values were calculated for 1000 resampling replicates.
Acknowledgments
This work was supported by grants R01 AI060739 and R21 AI062763 from the NIAID, NIH. Salaries and research support were provided by state and federal funds provided to the Ohio Agricultural Research and Development Center, The Ohio State University. For the contributions of Daniel Janies, the support of the Department of Biomedical Informatics and the Ohio State University Medical Center and the support by, or in part by, the U.S. Army Research Laboratory and the U.S. Army Research Office under contract/grant number W911NF-05-1-0271 is acknowledged.
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.virol.2006.08.051.
Appendix A. Supplementary Data
Primers used to amplify TGEV isolates
References
- Bae I., Jackwood D.J., Benfield D.A., Saif L.J., Wesley R.D., Hill H. Differentiation of transmissible gastroenteritis virus from porcine respiratory coronavirus and other antigenically related coronaviruses by using cDNA probes specific for the 5′ region of the S glycoprotein gene. J. Clin. Microbiol. 1991;29(1):215–218. doi: 10.1128/jcm.29.1.215-218.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesteros M.L., Sanchez C.M., Enjuanes L. Two amino acid changes at the N-terminus of transmissible gastroenteritis coronavirus spike protein result in the loss of enteric tropism. Virology. 1997;227(2):378–388. doi: 10.1006/viro.1996.8344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohl E.H., Kumagai T. The use of cell culture for the study of TGE virus of swine. Proc. U. S. Livestock San Assoc. 1965;69:343–350. [Google Scholar]
- Bohl E.H., Gupta R.K., Olquin M.V., Saif L.J. Antibody responses in serum, colostrum, and milk of swine after infection or vaccination with transmissible gastroenteritis virus. Infect. Immun. 1972;6(3):289–301. doi: 10.1128/iai.6.3.289-301.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callebaut P., Correa I., Pensaert M., Jimenez G., Enjuanes L. Antigenic differentiation between transmissible gastroenteritis virus of swine and a related porcine respiratory coronavirus. J. Gen. Virol. 1988;69:1725–1730. doi: 10.1099/0022-1317-69-7-1725. [DOI] [PubMed] [Google Scholar]
- Correa I., Jimenez G., Sune C., Bullido M.J., Enjuanes L. Antigenic structure of the E2 glycoprotein from transmissible gastroenteritis coronavirus. Virus Res. 1988;10(1):77–93. doi: 10.1016/0168-1702(88)90059-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas B., Gelfi J., Laude H. Antigenic structure of transmissible gastroenteritis virus: II. Domains in the peplomer glycoprotein. J. Gen. Virol. 1986;67:1405–1418. doi: 10.1099/0022-1317-67-7-1405. [DOI] [PubMed] [Google Scholar]
- Delmas B., Rasschaert D., Godet M., Gelfi J., Laude H. Four major antigenic sites of the coronavirus transmissible gastroenteritis virus are located on the amino-terminal half of spike glycoprotein S. J. Gen. Virol. 1990;71:1313–1323. doi: 10.1099/0022-1317-71-6-1313. [DOI] [PubMed] [Google Scholar]
- Delmas B., Gelfi J., L'Haridon R., Vogel L.K., Sjostrom H., Noren O., Laude H. Aminopeptidase N is a major receptor for the entero-pathogenic coronavirus TGEV. Nature. 1992;357(6377):417–420. doi: 10.1038/357417a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas B., Gelfi J., Sjostrom H., Noren O., Laude H. Further characterization of aminopeptidase-N as a receptor for coronaviruses. Adv. Exp. Med. Biol. 1993;342:293–298. doi: 10.1007/978-1-4615-2996-5_45. [DOI] [PubMed] [Google Scholar]
- Doyle L.P. Transmissible gastroenteritis of pigs. North Am. Vet. 1951;32(7):477–478. [PubMed] [Google Scholar]
- Doyle L.P., Hutchings L.M. A transmissible gastroenteritis in pigs. J. Am. Vet. Med Assoc. 1946;108:257–259. [PubMed] [Google Scholar]
- Furuuchi S., Shimizu M., Shimizu Y. Field trials on transmissible gastroenteritis live virus vaccine in newborn piglets. Natl. Inst. Anim. Health Q (Tokyo) 1978;18(3–4):135–142. [PubMed] [Google Scholar]
- Godet M., Grosclaude J., Delmas B., Laude H. Major receptor-binding and neutralization determinants are located within the same domain of the transmissible gastroenteritis virus (coronavirus) spike protein. J. Virol. 1994;68(12):8008–8016. doi: 10.1128/jvi.68.12.8008-8016.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goloboff, P., Farris, S., Nixon, K., 2005. TNT: Tree Analysis Using New Technologies. Software package distributed by the authors and available at:http://www.zmuc.dk/public/phylogeny/TNT.
- Haelterman E.O. Epidemiological studies of transmissible gastroenteritis of swine. Proc. U. S. Livestock San Assoc. 1964;68:305–314. [Google Scholar]
- Halbur P.G., Paul P.S., Vaughn E.M., Andrews J.J. Experimental reproduction of pneumonia in gnotobiotic pigs with porcine respiratory coronavirus isolate AR310. J. Vet. Diagn. Invest. 1993;5(2):184–188. doi: 10.1177/104063879300500207. [DOI] [PubMed] [Google Scholar]
- Hampton T. Bats may be SARS reservoir. JAMA. 2005;294(18):2291. doi: 10.1001/jama.294.18.2291. [DOI] [PubMed] [Google Scholar]
- Harada K., Furuuchi S., Kumagai T., Sasahara J. Pathogenicity, immunogenicity and distribution of transmissible gastroenteritis virus in pigs. Natl. Inst. Anim. Health Q (Tokyo) 1969;9(4):185–192. [PubMed] [Google Scholar]
- Hasoksuz M., Hoet A.E., Loerch S.C., Wittum T.E., Nielsen P.R., Saif L.J. Detection of respiratory and enteric shedding of bovine coronaviruses in cattle in an Ohio feedlot. J. Vet. Diagn. Invest. 2002;14(4):308–313. doi: 10.1177/104063870201400406. [DOI] [PubMed] [Google Scholar]
- Hill H., Wood B.J. Proceedings, 21st Annu. Meet Am. Assoc. Swine Proct. 1990. Porcine respiratory coronavirus isolated from two US swine herds; pp. 333–335. [Google Scholar]
- Hodgson T., Cavanagh B.P. Neither the RNA nor the proteins of open reading frames 3a and 3b of the coronavirus infectious bronchitis virus are essential for replication. J. Virol. 2006;80(1):296–305. doi: 10.1128/JVI.80.1.296-305.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izeta A., Smerdou C., Alonso S., Penzes Z., Mendez A., Plana-Duran J., Enjuanes L. Replication and packaging of transmissible gastroenteritis coronavirus-derived synthetic minigenomes. J. Virol. 1999;73(2):1535–1545. doi: 10.1128/jvi.73.2.1535-1545.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemeny L.J., Woods R.D. Quantitative transmissible gastroenteritis virus shedding patterns in lactating sows. Am. J. Vet. Res. 1977;38(3):307–310. [PubMed] [Google Scholar]
- Kim L., Hayes J., Lewis P., Parwani A.V., Chang K.O., Saif L.J. Molecular characterization and pathogenesis of transmissible gastroenteritis coronavirus (TGEV) and porcine respiratory coronavirus (PRCV) field isolates co-circulating in a swine herd. Arch. Virol. 2000;145(6):1133–1147. doi: 10.1007/s007050070114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon H.M., Saif L.J., Jackwood D.J. Field isolates of transmissible gastroenteritis virus differ at the molecular level from the Miller and Purdue virulent and attenuated strains and from porcine respiratory coronaviruses. J. Vet. Med. Sci. 1998;60(5):589–597. doi: 10.1292/jvms.60.589. [DOI] [PubMed] [Google Scholar]
- Lai M.M., Cavanagh D. The molecular biology of coronaviruses. Adv. Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K., Woo P.C., Li K.S., Huang Y., Tsoi H.W., Wong B.H., Wong S.S., Leung S.Y., Chan K.H., Yuen K.Y. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. U.S.A. 2005;102(39):14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laude H., Gelfi J., Lavenant L., Charley B. Single amino acid changes in the viral glycoprotein M affect induction of alpha interferon by the coronavirus transmissible gastroenteritis virus. J. Virol. 1992;66(2):743–749. doi: 10.1128/jvi.66.2.743-749.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H., Zhang J., McEachern J., Field H., Daszak P., Eaton B.T., Zhang S., Wang L.F. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310(5748):676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. The genome sequence of the SARS-associated coronavirus. Science. 2003;300(5624):1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- Martin Alonso J.M., Balbin M., Garwes D.J., Enjuanes L., Gascon S., Parra F. Antigenic structure of transmissible gastroenteritis virus nucleoprotein. Virology. 1992;188(1):168–174. doi: 10.1016/0042-6822(92)90746-C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGoldrick A., Lowings J.P., Paton D.J. Characterisation of a recent virulent transmissible gastroenteritis virus from Britain with a deleted ORF3a. Arch. Virol. 1999;144(4):763–770. doi: 10.1007/s007050050541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normile D. Virology. Researchers tie deadly SARS virus to bats. Science. 2005;309(5744):2154–2155. doi: 10.1126/science.309.5744.2154. [DOI] [PubMed] [Google Scholar]
- Ortego J., Sola I., Almazan F., Ceriani J.E., Riquelme C., Balasch M., Plana J., Enjuanes L. Transmissible gastroenteritis coronavirus gene 7 is not essential but influences in vivo virus replication and virulence. Virology. 2003;308(1):13–22. doi: 10.1016/S0042-6822(02)00096-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page K.W., Mawditt K.L., Britton P. Sequence comparison of the 5′ end of mRNA 3 from transmissible gastroenteritis virus and porcine respiratory coronavirus. J. Gen. Virol. 1991;72:579–587. doi: 10.1099/0022-1317-72-3-579. [DOI] [PubMed] [Google Scholar]
- Paton D., Ibata G., Sands J., McGoldrick A. Detection of transmissible gastroenteritis virus by RT-PCR and differentiation from porcine respiratory coronavirus. J. Virol. Methods. 1997;66(2):303–309. doi: 10.1016/S0166-0934(97)00055-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul P.S., Vaughn E.M., Halbur P.G. Pathogenicity and sequence analysis studies suggest potential role of gene 3 in virulence of swine enteric and respiratory coronaviruses. Adv. Exp. Med. Biol. 1997;412:317–321. doi: 10.1007/978-1-4899-1828-4_52. [DOI] [PubMed] [Google Scholar]
- Pensaert M., Callebaut P., Vergote J. Isolation of a porcine respiratory, non-enteric coronavirus related to transmissible gastroenteritis. Vet. Q. 1986;8(3):257–261. doi: 10.1080/01652176.1986.9694050. [DOI] [PubMed] [Google Scholar]
- Penzes Z., Gonzalez J.M., Calvo E., Izeta A., Smerdou C., Mendez A., Sanchez C.M., Sola I., Almazan F., Enjuanes L. Complete genome sequence of transmissible gastroenteritis coronavirus PUR46-MAD clone and evolution of the Purdue virus cluster. Virus Genes. 2001;23(1):105–118. doi: 10.1023/A:1011147832586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard G.C. Transmissible gastroenteritis in endemically infected breeding herds of pigs in East Anglia, 1981–85. Vet. Rec. 1987;120(10):226–230. doi: 10.1136/vr.120.10.226. [DOI] [PubMed] [Google Scholar]
- Rasschaert D., Laude H. The predicted primary structure of the peplomer protein E2 of the porcine coronavirus transmissible gastroenteritis virus. J. Gen. Virol. 1987;68:1883–1890. doi: 10.1099/0022-1317-68-7-1883. [DOI] [PubMed] [Google Scholar]
- Rasschaert D., Gelfi J., Laude H. Enteric coronavirus TGEV: partial sequence of the genomic RNA, its organization and expression. Biochimie. 1987;69(6–7):591–600. doi: 10.1016/0300-9084(87)90178-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300(5624):1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- Saif L.J. Animal coronaviruses: what can they teach us about the severe acute respiratory syndrome? Rev. Sci. Tech. 2004;23(2):643–660. doi: 10.20506/rst.23.2.1513. [DOI] [PubMed] [Google Scholar]
- Saif L.J. Comparative biology of animal coronaviruses: lessons for SARS. In: Peiris M., Anderson L.J., Osterhaus A.D.M.E., Klaus Stohr K.Y., editors. Severe Acute Respiratory Syndrome. Blackwell Pub; Oxford, UK: 2005. pp. 84–99. [Google Scholar]
- Saif L.J., Sestak K. Transmissible gastroenteritis virus and porcine respiratory coronavirus. In: Zimmerman J.J., editor. Diseases of Swine. 9th ed. Iowa State University Press; Ames, Iowa: 2006. pp. 489–516. [Google Scholar]
- Sanchez C.M., Jimenez G., Laviada M.D., Correa I., Sune C., Bullido M., Gebauer F., Smerdou C., Callebaut P., Escribano J.M. Antigenic homology among coronaviruses related to transmissible gastroenteritis virus. Virology. 1990;174(2):410–417. doi: 10.1016/0042-6822(90)90094-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez C.M., Gebauer F., Sune C., Mendez A., Dopazo J., Enjuanes L. Genetic evolution and tropism of transmissible gastroenteritis coronaviruses. Virology. 1992;190(1):92–105. doi: 10.1016/0042-6822(92)91195-Z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez C.M., Izeta A., Sanchez-Morgado J.M., Alonso S., Sola I., Balasch M., Plana-Duran J., Enjuanes L. Targeted recombination demonstrates that the spike gene of transmissible gastroenteritis coronavirus is a determinant of its enteric tropism and virulence. J. Virol. 1999;73(9):7607–7618. doi: 10.1128/jvi.73.9.7607-7618.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki S.G., Sawicki D.L. Coronavirus transcription: subgenomic mouse hepatitis virus replicative intermediates function in RNA synthesis. J. Virol. 1990;64(3):1050–1056. doi: 10.1128/jvi.64.3.1050-1056.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S., Wen Z.L., Liu D.X. Emergence of a coronavirus infectious bronchitis virus mutant with a truncated 3b gene: functional characterization of the 3b protein in pathogenesis and replication. Virology. 2003;311(1):16–27. doi: 10.1016/S0042-6822(03)00117-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simkins R.A., Saif L.J., Weilnau P.A. Epitope mapping and the detection of transmissible gastroenteritis viral proteins in cell culture using biotinylated monoclonal antibodies in a fixed-cell ELISA. Arch. Virol. 1989;107(3–4):179–190. doi: 10.1007/BF01317915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simkins R.A., Weilnau P.A., Bias J., Saif L.J. Antigenic variation among transmissible gastroenteritis virus (TGEV) and porcine respiratory coronavirus strains detected with monoclonal antibodies to the S protein of TGEV. Am. J. Vet. Res. 1992;53(7):1253–1258. [PubMed] [Google Scholar]
- Simkins R.A., Weilnau P.A., Van Cott J., Brim T.A., Saif L.J. Competition ELISA, using monoclonal antibodies to the transmissible gastroenteritis virus (TGEV) S protein, for serologic differentiation of pigs infected with TGEV or porcine respiratory coronavirus. Am. J. Vet. Res. 1993;54(2):254–259. [PubMed] [Google Scholar]
- Sola I., Alonso S., Zuniga S., Balasch M., Plana-Duran J., Enjuanes L. Engineering the transmissible gastroenteritis virus genome as an expression vector inducing lactogenic immunity. J. Virol. 2003;77(7):4357–4369. doi: 10.1128/JVI.77.7.4357-4369.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sune C., Jimenez G., Correa I., Bullido M.J., Gebauer F., Smerdou C., Enjuanes L. Mechanisms of transmissible gastroenteritis coronavirus neutralization. Virology. 1990;177(2):559–569. doi: 10.1016/0042-6822(90)90521-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underdahl N.R., Mebus C.A., Torres-Medina A. Recovery of transmissible gastroenteritis virus from chronically infected experimental pigs. Am. J. Vet. Res. 1975;36(10):1473–1476. [PubMed] [Google Scholar]
- VanCott J.L., Brim T.A., Simkins R.A., Saif L.J. Isotype-specific antibody-secreting cells to transmissible gastroenteritis virus and porcine respiratory coronavirus in gut- and bronchus-associated lymphoid tissues of suckling pigs. J. Immunol. 1993;150(9):3990–4000. [PubMed] [Google Scholar]
- Vaughn E.M., Halbur P.G., Paul P.S. Three new isolates of porcine respiratory coronavirus with various pathogenicities and spike (S) gene deletions. J. Clin. Microbiol. 1994;32(7):1809–1812. doi: 10.1128/jcm.32.7.1809-1812.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch S.K., Saif L.J. Monoclonal antibodies to a virulent strain of transmissible gastroenteritis virus: comparison of reactivity with virulent and attenuated virus. Arch. Virol. 1988;101(3–4):221–235. doi: 10.1007/BF01311003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley R.D., Cheung A.K., Michael D.D., Woods R.D. Nucleotide sequence of coronavirus TGEV genomic RNA: evidence for 3 mRNA species between the peplomer and matrix protein genes. Virus Res. 1989;13(2):87–100. doi: 10.1016/0168-1702(89)90008-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley R.D., Woods R.D., Cheung A.K. Genetic basis for the pathogenesis of transmissible gastroenteritis virus. J. Virol. 1990;64(10):4761–4766. doi: 10.1128/jvi.64.10.4761-4766.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley R.D., Woods R.D., Hill H.T., Biwer J.D. Evidence for a porcine respiratory coronavirus, antigenically similar to transmissible gastroenteritis virus, in the United States. J. Vet. Diagn. Invest. 1990;2(4):312–317. doi: 10.1177/104063879000200411. [DOI] [PubMed] [Google Scholar]
- Wesley R.D., Woods R.D., Cheung A.K. Genetic analysis of porcine respiratory coronavirus, an attenuated variant of transmissible gastroenteritis virus. J. Virol. 1991;65(6):3369–3373. doi: 10.1128/jvi.65.6.3369-3373.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood E.N., Pritchard G.C., Gibson E.A. Transmissible gastroenteritis of pigs. Vet. Rec. 1981;108(2):41. doi: 10.1136/vr.108.2.41. [DOI] [PubMed] [Google Scholar]
- Woods R.D. Leukocyte-aggregation assay for transmissible gastroenteritis of swine. Am. J. Vet. Res. 1976;37(12):1405–1408. [PubMed] [Google Scholar]
- Woods R.D. Humoral and cellular responses in swine exposed to transmissible gastroenteritis virus. Am. J. Vet. Res. 1979;40(1):108–110. [PubMed] [Google Scholar]
- Woods R.D., Wesley R.D. Immune response in sows given transmissible gastroenteritis virus or canine coronavirus. Am. J. Vet. Res. 1986;47(6):1239–1242. [PubMed] [Google Scholar]
- Yoo D., Deregt D. A single amino acid change within antigenic domain II of the spike protein of bovine coronavirus confers resistance to virus neutralization. Clin. Diagn. Lab. Immunol. 2001;8(2):297–302. doi: 10.1128/CDLI.8.2.297-302.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used to amplify TGEV isolates