Abstract
Mammalian preimplantation embryos provide an excellent opportunity to study temporal and spatial gene expression in whole mount in situ hybridization (WISH). However, large-scale studies are made difficult by the size of the embryos (∼60 μm diameter) and their fragility. We have developed a chamber system that allows parallel processing of embryos without the aid of a microscope. We first selected 91 candidate genes that were transcription factors highly expressed in blastocysts, and more highly expressed in embryonic (ES) than in trophoblast (TS) stem cells. We then used the WISH to identify 48 genes expressed predominantly in the ICM and to follow several of these genes in all seven preimplantation stages. The ICM-predominant expressions of these genes suggest their involvement in the pluripotency of embryonic cells. This system provides a useful tool to a systematic genome-scale analysis of preimplantation embryos.
Keywords: preimplantation embryo, whole mount in situ hybridization, High-throughput screen, hybridization chamber, ICM, TE
Introduction
Preimplantation development encompasses the period from fertilization to implantation, and is marked by a number of critical events, including the degradation of maternally stored RNAs, zygotic genome activation (ZGA), compaction, and blastocyst formation (reviewed in (Edwards, 2003)). From the viewpoints of developmental potency (potential), fertilized eggs are the ultimate totipotent cells, giving rise to all cell types. The loss of totipotency occurs during preimplantation development, marked by the segregation of two distinct cell lineages in the blastocyst: the inner cell mass (ICM), which gives rise to the embryo proper and is thus pluripotent, and the trophectoderm (TE), which contributes to the trophoblast portion of the placenta and is thus lineage-restricted (Fig. 1B). Genes that are important for cellular pluripotency, such as Pou5f1/Oct4 (Pesce and Scholer, 2000) and Nanog (Chambers et al., 2003; Mitsui et al., 2003), are predominantly expressed in the ICM, and thus, the identification of genes expressed in the ICM will be an important first step towards understanding the cellular potency. Whether the emergence of such asymmetry between the ICM and TE originates from an earlier event, such as fertilization, is still controversial (Gardner, 2001; Hiiragi and Solter, 2004; Piotrowska et al., 2001).
Figure 1.
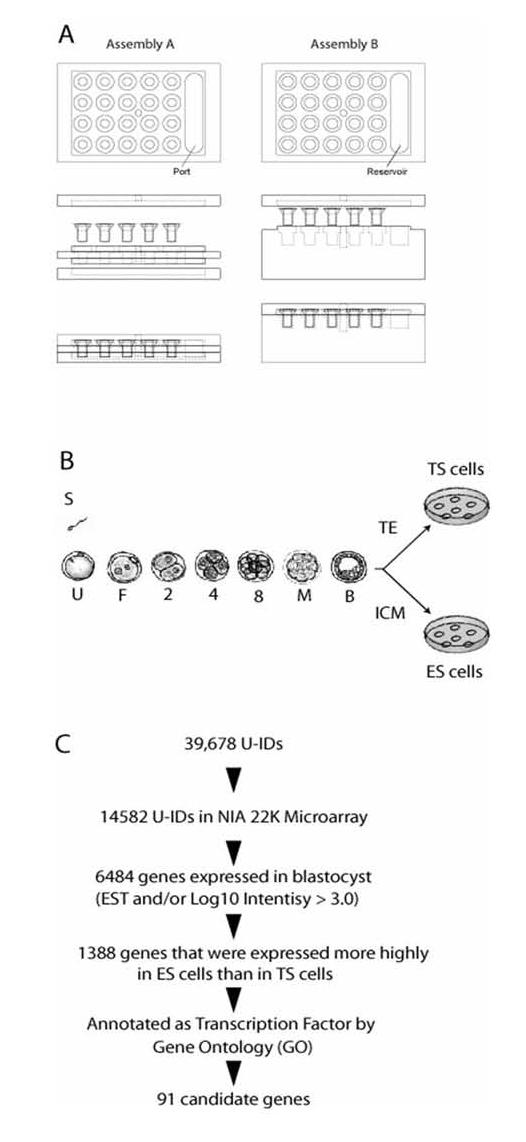
(A) Assembly A: Washing chamber. Assembly B: Hybridization chamber. (B) Schematic drawings of mouse preimplantation development. At blastocyst stage, there are two different cell types: Trophectoderm (TE) and Inner Cell Mass (ICM). ES cells are derived from the ICM, whereas TS cells are derived from the TE cells. (C) Bioinformatic selection of candidate genes.
Large-scale systematic analysis holds great promise for understanding preimplantation embryos as a whole (Ko, 2001). A large number of cDNA clones have been identified from mouse preimplantation embryos and mapped to the mouse genome (Ko et al., 2000; Sharov et al., 2003; Solter et al., 2002). Microarray analysis of the preimplantation embryos has provided global picture of expression changes during preimplantation mouse development (Hamatani et al., 2004; Tanaka and Ko, 2004; Wang et al., 2004; Zeng et al., 2004). The knowledge of genes expressed in preimplantation mouse embryos has increased dramatically. However, because RNA samples are taken from homogenized tissues, spatial information is lost, and thus, questions of their asymmetric expression cannot be directly addressed. WISH allows localization of gene transcripts in the individual cell, enabling the study of the heterogeneity of cells and/or their polarity at very early stages of the embryo, in which no morphological differences are seen among cells.
Large-scale in situ hybridizations have been performed on mouse intestine (Komiya et al., 1997), E9.5 embryos (Gitton et al., 2002; Neidhardt et al., 2000), and E9.5 and E10.5 embryos (Reymond et al., 2002), and mouse brain as well as on other species, such as Drosophila (Tomancak et al., 2002), Zebrafish (Kudoh et al., 2001), Xenopus (Gawantka et al., 1998), Medaka Fish (Quiring et al., 2004), Chick retina (Shintani et al., 2004), Ascidian (Mochizuki et al., 2003), Chicken embryos (Bell et al., 2004). A robotic workstation is available, but due to its larger filter pore size (35 μm) it cannot be used for small embryos, such as mammalian preimplantation embryos. Due to the technical difficulty of handling small embryos, WISH data for mouse preimplantation embryos is scarce even with small-scale methods based on individual genes. During the pipetting procedure, embryos are often lost. This has been addressed by using a microcentrifuge tube, which was cut at the bottom and attached to a 20 μm pore membrane (Newman-Smith and Werb, 1995). The method has successfully circumvented laborious micropipetting work, but the microtubes were made by hand each time and were not suited for parallel processing. While a pore size of 20 μm is necessary for achieving efficient drainage without special instruments, much smaller pores are preferable to maintain the best morphology of small samples. As a result, transwell with pore size 12 μm which are originally designed for cell culture were introduced into WISH (Hanna et al., 2002) to retain embryos. Although, solution changes were achieved by manually transferring the transwell from one well to another, it is difficult to have good buffer exchange through smaller pores without the assistance of a special device. Here we report the development of a chamber system that utilizes both the transwell inserts for parallel processing and capillary action for gentle buffer exchanges. Using this method, we have identified 48 genes that are expressed predominantly in the ICM.
1. Results
1.1. Design and fabrication of WISH chamber system
To perform a high-throughput WISH for preimplantation embryos (up to 100 μm diameter), we developed a chamber system that can run multiple probes in parallel without microscope-assistance (Fig. 1A). Embryos can be placed in plastic Transwell-inserts with 8 μm pore-size membrane on the bottom. Up to 20 inserts can be placed in one aluminum chamber, which allows analysis of up to 20 different probes in parallel. The small pore size helps maintain good embryo morphology while minimizing the chance of embryo loss during the WISH procedure. However, the small pore size makes it difficult to drain the solution through the bottom membrane. Initial design used negative air pressure by vacuum pump, resulting in poor morphology of embryos. We then devised a chamber system so that the distance between the bottom of the insert holder and the bottom of the solution container formed a small gap of 0.5 mm. This turned out to be the most effective way of draining the solution from the bottom of the transwell by capillary action. Solutions were exchanged through the port and the drainage was completed within seconds in every insert simultaneously.
One run of a WISH experiment takes only two days: one hour for the first day and six hours for the second day including the incubation time. Because multiple embryos of various stages can be processed in parallel, literally hundreds of embryos can be analyzed, and thus the collection of embryos becomes the actual rate-limiting step. Time-consuming microscopy and micropipetting are limited only to the step for transferring embryos into inserts. Overall, this new device and new protocol dramatically increased the throughput of WISH on preimplantation embryos over the conventional micropipetting method.
1.2. Test of the WISH chamber system
The WISH protocol was optimized by using antisense and sense cRNA probes for three representative genes. Pou5f1 (Oct3/4) is known to be expressed specifically in ICM of blastocysts, Krt2-8, in TE, and Actb, ubiquitously. The morphology of blastocyst was maintained relatively well and all results were consistent with previous reports (Fig. 2). It should be pointed out that because the ICM forms a dense clump inside the blastocysts, for genes that are ubiquitously expressed in both the ICM and TE, the staining of the ICM would look more intense compared to TE. On the other hand, genes that appear to be stained uniformly (e.g., Krt2-8) are in reality expressed predominantly in TE. This makes it difficult to distinguish genes that are predominantly expressed in ICM from genes that are ubiquitously expressed. After some trial and error, we decided to use a scoring system that utilized the staining patterns of Krt2-8, Pou5f1, and Actb as the standards. The pictures for individual genes were always compared to the pictures of these genes.
Figure 2.
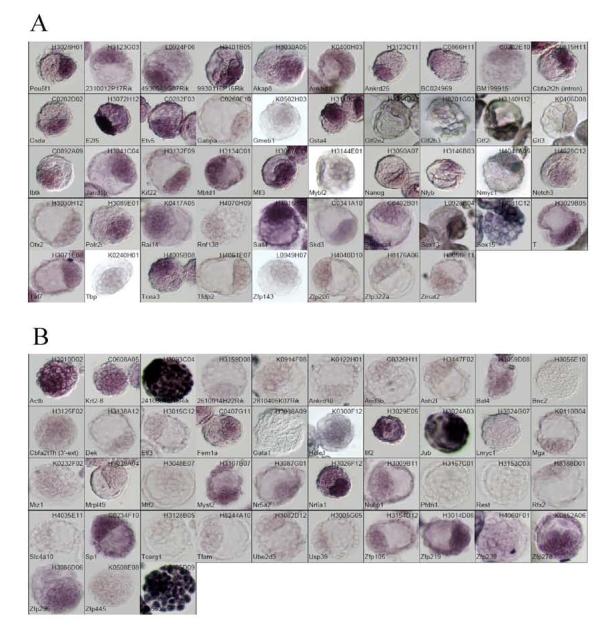
WISH results of 91 genes: Each image is accompanied with a gene symbol and a cDNA clone name used as a probe. Panel A, genes identified for their ICM-predominant expression (including Pou5f1). Panel B, genes that were not expressed in ICM-predominant manner (including Actb and Krt2-8).
1.3. Informatics selection of candidate genes
We used a series of informatic criteria to select candidate genes that are most likely expressed predominantly in the ICM (Fig. 1C). First, we identified 6484 genes that are highly expressed in mouse blastocysts by examining EST frequency data (Sharov et al., 2003) and microarray data (Hamatani et al., 2004). We then attempted to identify candidate genes that are expressed more highly in the ICM than in TE cells. Due to difficulty of collecting the ICM and TE from blastocysts separately, we exploited the cultured cells derived from these cell types in our previous study (Tanaka et al., 2002). Mouse ES cells are cultured from the ICM (Evans and Kaufman, 1981; Martin, 1981), whereas mouse TS cells are cultured from the TE (Tanaka et al., 1998) (Fig. 1B). Although these cells have been cultured in vitro, they represent the in vitro equivalents of the ICM and TE. Therefore, genes which are expressed more highly in ES than TS in microarray studies were good candidates for genes predominantly expressed in the ICM in blastocysts.
To supplement the data (Tanaka et al., 2002) and update to the latest microarray platform, we performed hybridizations of ES and TS RNAs onto the NIA 22K 60-mer oligonucleotide microarrays in triplicate (Carter et al., 2003a). This glass-slide microarray platform contains genes from preimplantation and stem cells (for experimental design, see ref (Tanaka et al., 2002); for all the array data set, see http://lgsun.grc.nia.nih.gov/data, and also Supplementary Material 3). By this criterion, we identified 1388 genes that were expressed more highly in ES cells than in TS cells. Because we were interested in transcription factors, we further narrowed down the list and identified 95 genes, which have “transcription factor activity” in GO terms (Ashburner et al., 2000). During the course of this work, however, the assignment and GO annotation of genes have been changed and at present 65 genes out of these 95 genes can be classified as transcription factors.
1.4. Genes expressed predominantly in ICMs
Out of 95 genes, we were able to find 91 cDNA clones from the NIA mouse cDNA collection (Sharov et al., 2003). We attempted to prepare probes for these 91 genes, but two probes did not pass the quality check. After adding Actb and Krt2-8 genes as controls, a total number of probes became 91. Results of the WISH using antisense probe for the 91 genes are shown in Fig. 2 and summarized in Table 1. The list included three genes (Nanog, Pou5f1, Otx2) which have been reported to be expressed specifically in ICM by WISH (Chambers et al., 2003; Kimura et al., 2001; Mitsui et al., 2003). When the chamber system was used on these genes, Nanog, and Pou5f1 had signals only in the ICM, whereas Otx2 showed weak, but rather ICM-predominant expression (Fig. 2).
Table 1.
Summary of WISH results. Clone_ID indicates cDNA clones that were used for cRNA probe preparation. U_ID is the identification of U cluster, which represents individual genes in the NIA Mouse Gene Index (Sharov et al., Genome Res., in press; http://lgsun.grc.nia.nih.gov/geneindex4). Log intensity is the mean value of signal intensities at log-scale measured in triplicate, which represents the expression of level of genes in mouse blastocyst (from the data in ref. (Hamatani et al., 2004)). Fold (ES/TS) indicates the ratio of signal intensities for each gene between ES cell and TS cell, averaged among biological replicates. The intensity of staining in ICM and TE in WISH are shown arbitrarily in 5 grades (−, +/−, +, ++ and +++, with “−“ as no staining and “+++” as the highest intensity for the amount of probe, either 1 or 5 μl). Non specific staining is shown as NS.
| Clone_ID | U_ID | Gene Symbol | Annotation | Transcription factor? | Log Intensity (Blastocyst) | Fold (ES/TS) | Oligo ID | ICM (1 ul) | TE (1 ul) | ICM (5 ul) | TE (5 ul) | ICM predominant? |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H3028H01 | U017906 | Pou5f1 | POU domain, class 5, transcription factor 1 | Yes | 3.28 | 11.08 | Z04894 | ++ | − | ++ | +/− | Yes |
| H3123G03 | U042665 | 2310012P17Rik | RIKEN cDNA 2310012P17 gene | No | 3.13 | 2.40 | Z18099 | − | − | ++ | +/− | Yes |
| L0924F06 | U015681 | 4930548G07Rik | RIKEN cDNA 4930548G07 gene | Yes | 4.03 | 1.35 | Z12748 | ++ | − | ++ | + | Yes |
| H3101B05 | U036004 | 9930116P15Rik | Mus musculus ES cells cDNA, RIKEN full-length enriched library, clone:C330022M23 | No | 3.13 | 1.94 | Z10610 | − | − | ++ | +/− | Yes |
| H3030A05 | U037662 | Akap8 | A kinase (PRKA) anchor protein 8 | No | 2.95 | 1.58 | Z04916 | ++ | +/− | NS | NS | Yes |
| K0400H03 | U018590 | Ankhd1 | A062A08 GGTC Gene Trap Library GV03C04 Mus musculus cDNA clone A062A08 | Yes | 3.14 | 1.22 | Z15071 | +/− | − | ++ | +/− | Yes |
| H3123C11 | U030590 | Ankrd25 | ankyrin repeat domain 25 | Yes | 3.05 | 1.39 | Z11802 | + | − | ++ | +/− | Yes |
| C0866H11 | U038601 | BC024969 | MGC cDNA | Yes | 3.59 | 1.18 | Z04004 | − | − | ++ | +/− | Yes |
| C0202E10 | U089485,U013686,U119478 | BM199915 | Genbank number for EST | Yes | 3.98 | 1.21 | Z20771 | +/− | − | ++ | +/− | Yes |
| C0815H11 | U208874 | Cbfa2t2h (intron) | core-binding factor, runt domain, alpha subunit 2, translocated to, 2 homolog (human) | Yes | 2.91 | 1.46 | Z03694 | ++ | + | ++ | +/− | Yes |
| C0202D02 | U027935 | Csda | cold shock domain protein A | Yes | 3.72 | 1.26 | Z01041 | + | − | ++ | +/− | Yes |
| H3072H12 | U003038 | E2f5 | E2F transcription factor 5 | Yes | 3.12 | 1.74 | Z08851 | +/− | − | +++ | + | Yes |
| C0282F03 | U055077,U036911 | Etv5 | ets variant gene 5 | Yes | 3.60 | 2.93 | Z02758 | − | − | ++ | +/− | Yes |
| C0260F10 | U043544 | Gabpa | GA repeat binding protein, alpha | Yes | 3.34 | 1.70 | Z20218 | N/A | N/A | + | − | Yes |
| K0502H03 | U025592 | Gmeb1 | glucocorticoid modulatory element binding protein 1 | Yes | 3.74 | 1.17 | Z15720 | + | − | ++ | + | Yes |
| H3119G08 | U010777 | Gsta4 | glutathione S-transferase, alpha 4 | No | 3.69 | 21.43 | Z11620 | + | − | ++ | + | Yes |
| H3144B01 | U009246 | Gtf2e2 | general transcription factor II E, polypeptide 2 (beta subunit) | Yes | 3.27 | 1.46 | Z19185 | + | − | ++ | + | Yes |
| H8201G03 | U006190 | Gtf2h3 | general transcription factor IIH, polypeptide 3, 34kDa | Yes | 3.05 | 1.45 | Z19769 | + | − | + | +/− | Yes |
| H3140H12 | U026798 | Gtf2i | general transcription factor II I | Yes | 3.77 | 2.91 | Z19056 | ++ | − | +++ | + | Yes |
| K0406D08 | U030135 | Gtl3 | gene trap locus 3 | No | 3.64 | 1.47 | Z09177 | + | − | + | +/− | Yes |
| C0892A09 | U031153 | Ibtk | inhibitor of Bruton agammaglobulinemia tyrosine kinase | No | 3.85 | 1.49 | Z17931 | ++ | − | +/− | +/− | Yes |
| H3041C04 | U000832 | Jarid1b | jumonji, AT rich interactive domain 1B (Rbp2 like) | Yes | 3.27 | 2.22 | Z07056 | +/− | − | ++ | +/− | Yes |
| H3132F09 | U029275 | Kif22 | kinesin family member 22 | No | 2.67 | 1.75 | Z18589 | − | − | ++ | − | Yes |
| H3134C01 | U013196 | Mbtd1 | mbt domain containing 1 | No | 3.09 | 1.62 | Z18681 | − | − | ++ | +/− | Yes |
| H3057C11 | U026010 | Mll3 | myeloid/lymphoid or mixed-lineage leukemia 3 | Yes | 3.12 | 1.48 | Z07970 | − | − | ++ | +/− | Yes |
| H3144E01 | U002779,U095436,U165175 | Mybl2 | myeloblastosis oncogene-like 2 | Yes | 3.89 | 1.38 | Z19198 | ++ | − | +++ | + | Yes |
| H3050A07 | U007377,U125018 | Nanog | Nanog homeobox | Yes | 3.55 | 4.64 | Z07563 | ++ | − | ++ | − | Yes |
| H3146B03 | U032063 | Nfyb | nuclear transcription factor-Y beta | Yes | 3.40 | 1.41 | Z12544 | − | − | + | − | Yes |
| H4041A05 | U033813 | Nmyc1 | neuroblastoma myc-related oncogene 1 | Yes | 3.09 | 2.73 | Z21669 | + | − | NS | NS | Yes |
| H4028C12 | U037659 | Notch3 | Notch gene homolog 3 (Drosophila) | Yes | 4.00 | 1.22 | Z20375 | + | − | + | − | Yes |
| H3030H12 | U035481 | Otx2 | orthodenticle homolog 2 (Drosophila) | Yes | 3.25 | 4.59 | Z04978 | − | − | ++ | +/− | Yes |
| H3089E01 | U007921 | Polr2i | Mus musculus cDNA clone MGC:73656 IMAGE:3466417, complete cds. | Yes | 3.39 | 1.88 | Z10011 | − | − | ++ | +/− | Yes |
| K0417A05 | U036007 | Rai14 | retinoic acid induced 14 | Yes | 3.58 | 1.69 | Z19497 | − | − | ++ | +/− | Yes |
| H4070H09 | U018467 | Rnf138 | ring finger protein 138 | No | 3.43 | 1.32 | Z16641 | − | − | + | − | Yes |
| H4016B02 | U023697 | Sall4 | sal-like 4 (Drosophila) | No | 3.86 | 3.04 | Z13538 | +++ | + | +++ | + | Yes |
| C0341A10 | U008471 | Skd3 | suppressor of K+ transport defect 3 | Yes | 3.72 | 1.28 | Z02265 | +/− | − | + | − | Yes |
| C0402B01 | U010150 | Smarca4 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 | Yes | 3.55 | 1.67 | Z06449 | ++ | +/− | ++ | +/− | Yes |
| L0928B04 | U021771 | Sox13 | SRY-box containing gene 13 | Yes | 3.48 | 2.66 | Z05552 | ++ | +/− | +++ | ++ | Yes |
| H3081C12 | U012826 | Sox15 | SRY-box containing gene 15 | Yes | 3.49 | 2.27 | Z09444 | ++ | + | +++ | + | Yes |
| H3029B05 | U017472 | T | brachyury | Yes | 3.93 | 1.48 | Z06600 | ++ | +/− | ++ | + | Yes |
| H3071E08 | U038350 | Taf7 | TAF7 RNA polymerase II, TATA box binding protein (TBP)-associated factor | Yes | 3.47 | 2.56 | Z14353 | +/− | − | ++ | +/− | Yes |
| K0240H01 | U017529 | Tbp | TATA box binding protein | Yes | 3.20 | 1.21 | Z09055 | + | − | +/− | − | Yes |
| H4005B08 | U004965 | Tcea3 | transcription elongation factor A (SII), 3 | Yes | 3.08 | 6.22 | Z14874 | + | − | ++ | +/− | Yes |
| H4061E07 | U043020 | Tfdp2 | transcription factor Dp 2 | Yes | 2.58 | 1.66 | Z03781 | − | − | + | − | Yes |
| L0949H07 | U008625 | Zfp143 | zinc finger protein 143 | Yes | 3.09 | 1.29 | Z09829 | + | − | +/− | − | Yes |
| H4040D10 | U043572 | Zfp206 | zinc finger protein 206 | Yes | 4.41 | 1.21 | Z02924 | − | − | + | − | Yes |
| H8176A06 | U072774 | Zfp322a | zinc finger protein 322a | No | 3.52 | 1.73 | Z04195 | N/A | N/A | + | − | Yes |
| H3058F11 | U018598 | Zmat2 | zinc finger, matrin type 2 | No | 3.13 | 1.47 | Z14005 | − | − | + | − | Yes |
| H3010D02 | U026912 | Actb | actin, beta, cytoplasmic | No | +++ | ++ | N/A | N/A | No | |||
| C0608A05 | U036682 | Krt2-8 | cytokeratin endo A | No | 4.92 | 0.09 | Z06684 | No | ||||
| H3093C04 | U025552 | 2410081M15Rik | RIKEN cDNA 2410081M15 gene | No | 3.06 | 3.82 | Z10249 | NS | NS | NS | NS | No |
| H3159D08 | U028390 | 2610014H22Rik | Mus musculus cDNA clone IMAGE:5704432, partial cds. | Yes | 3.30 | 1.19 | Z19782 | − | − | +/− | − | No |
| K0914F08 | U023323 | 2810405K07Rik | RIKEN cDNA 2810405K07 gene | Yes | 3.32 | 1.56 | Z15697 | − | − | +/− | − | No |
| K0122H01 | U029585 | Ankrd10 | ankyrin repeat domain 10 | Yes | 3.50 | 1.91 | Z01774 | − | − | − | − | No |
| C0326H11 | U108385 | Arid3b | AT rich interactive domain 3B (Bright like) | No | 3.60 | 1.50 | Z02467 | − | − | +/− | − | No |
| H3147F02 | U029672 | Ash2l | ash2 (absent, small, or homeotic)-like (Drosophila) | Yes | 3.83 | 1.46 | Z02226 | − | − | +/− | − | No |
| H3059D08 | U065275,U017890 | Bat4 | Mus musculus adult male epididymis cDNA, RIKEN full-length enriched library, clone:9230110P17 | Yes | 3.56 | 1.16 | Z08070 | − | − | + | +/− | No |
| H3056E10 | U025224 | Bnc2 | basonuclin 2 | No | 3.14 | 1.31 | Z13933 | N/A | N/A | − | − | No |
| H3125F02 | U215470,U183035 | Cbfa2t1h (3′-ext) | CBFA2T1 identified gene homolog | Yes | 3.51 | 1.29 | Z18223 | − | − | +/− | +/− | No |
| H3138A12 | U034741,U134034 | Dek | DEK oncogene (DNA binding) | Yes | 3.65 | 1.90 | Z12228 | − | − | + | + | No |
| H3015C12 | U021793 | Elf3 | E74-like factor 3 | Yes | 3.55 | 1.37 | Z06047 | − | − | + | +/− | No |
| C0407G11 | U018129,U012414 | Fem1a | feminization 1 homolog a (C. elegans) | Yes | 3.70 | 1.23 | Z01940 | − | − | + | +/− | No |
| H3038A09 | U039236 | Gata1 | GATA binding protein 1 | Yes | 4.48 | 1.29 | Z13445 | − | − | N/A | N/A | No |
| K0300F12 | U039463 | Hcfc1 | host cell factor C1 | No | 3.69 | 1.33 | Z00572 | − | − | NS | NS | No |
| H3029E05 | U003494 | Ilf2 | interleukin enhancer binding factor 2 | No | 3.71 | 1.60 | Z06628 | − | − | ++ | + | No |
| H3024A03 | U035555 | Jub | ajuba | No | 3.19 | 2.43 | Z06379 | NS | NS | NS | NS | No |
| H3024G07 | U042574 | Lmyc1 | lung carcinoma myc related oncogene 1 | Yes | 3.41 | 1.25 | Z06413 | − | − | − | − | No |
| K0110B04 | U002325 | Mga | MAX gene associated | Yes | 3.85 | 1.77 | Z19295 | − | − | +/− | − | No |
| K0232F02 | U018914 | Miz1 | Mus musculus Msx-interacting-zinc finger (Miz1), mRNA | Yes | 2.66 | 1.92 | Z06850 | +/− | +/− | +/− | +/− | No |
| H3039A04 | U019023 | Mrpl49 | mitochondrial ribosomal protein L49 | No | 3.15 | 1.50 | Z06929 | − | − | +/− | − | No |
| H3048E07 | U005938 | Mtf2 | metal response element binding transcription factor 2 | Yes | 4.03 | 4.22 | Z07461 | − | − | − | − | No |
| H3107B07 | U033300 | Myst2 | MYST histone acetyltransferase 2 | Yes | 3.45 | 1.23 | Z11023 | − | − | ++ | + | No |
| H3087G01 | U021824 | Nr5a2 | nuclear receptor subfamily 5, group A, member 2 | Yes | 2.94 | 3.28 | Z09889 | − | − | + | +/− | No |
| H3026F12 | U022570 | Nr6a1 | nuclear receptor subfamily 6, group A, member 1 | Yes | 3.50 | 2.49 | Z06529 | + | +/− | +++ | ++ | No |
| H3009B11 | U015867 | Nufip1 | nuclear fragile X mental retardation protein interacting protein | No | 3.29 | 1.58 | Z05791 | − | − | + | +/− | No |
| H3157C01 | U038334 | Pfdn1 | prefoldin 1 | Yes | 3.67 | 1.33 | Z13037 | − | − | − | − | No |
| H3153C03 | U042648 | Rest | RE1-silencing transcription factor | Yes | 3.05 | 2.21 | Z19589 | − | − | − | − | No |
| H8188D01 | U037958 | Rfx2 | regulatory factor X, 2 (influences HLA class II expression) | Yes | 3.77 | 1.60 | Z14719 | − | − | +/− | − | No |
| H4035E11 | U001816,U067822 | Slc4a10 | solute carrier family 4, sodium bicarbonate cotransporter-like, member 10 | Yes | 3.30 | 1.31 | Z21184 | − | − | − | − | No |
| C0234F10 | U043484 | Sp1 | trans-acting transcription factor 1 | Yes | 3.49 | 1.87 | Z01040 | − | − | ++ | + | No |
| H3128B05 | U043672 | Tcerg1 | transcription elongation regulator 1 (CA150) | Yes | 4.04 | 1.37 | Z18367 | − | − | − | − | No |
| H8244A10 | U031911 | Tfam | transcription factor A, mitochondrial | Yes | 2.66 | 1.22 | Z02399 | − | − | +/− | − | No |
| H3082D12 | U003889 | Ube2d3 | UBIQUITIN-CONJUGATING ENZYME E2-17 KDA 3 (EC 6.3.2.19) | No | 2.28 | 1.39 | Z09504 | − | − | + | +/− | No |
| H3005G05 | U027470 | Usp39 | ubiquitin specific protease 39 | No | 3.20 | 1.33 | Z05697 | − | − | +/− | − | No |
| H3154D12 | U089830,U011188 | Zfp105 | zinc finger protein 105 | Yes | 3.40 | 2.58 | Z19647 | +/− | − | ++ | + | No |
| H3014D06 | U035530 | Zfp219 | zinc finger protein 219 | Yes | 3.58 | 1.77 | Z05989 | − | − | +++ | ++ | No |
| H4060F01 | U043940 | Zfp239 | zinc finger protein 239 | Yes | 3.02 | 2.90 | Z03653 | − | − | + | +/− | No |
| K0852A06 | U012241 | Zfp278 | zinc finger protein 278 | Yes | 3.45 | 2.02 | Z04975 | +/− | − | ++ | + | No |
| H3086D06 | U007769 | Zfp296 | zinc finger protein 296 | No | 3.82 | 3.71 | Z09785 | +/− | − | ++ | + | No |
| K0508E08 | U031463 | Zfp445 | zinc finger protein 445 | Yes | 3.27 | 1.71 | Z21112 | +/− | − | +/− | +/− | No |
| C0405D09 | U008795 | Zfp553 | zinc finger protein 553 | No | 3.18 | 1.56 | Z02818 | NS | NS | NS | NS | No |
Overall, out of 91 genes, 48 genes were scored for higher signals in the ICM than the TE. However, there was no gene that showed clear-cut ICM-dominant expression like Pou5f1 (Oct3/4). Rather, they showed differential expression, with some expression still detectable in TE. Among these genes, 14 genes showed particularly high-contrast signals between the ICM and TE: Csda, E2f5, Gmeb1, Gtf2e2, Gtf2i, Kif22, Mybl2, Nanog, Nmyc1, Smarca4, Sox13, Sox15, Tbp, and Zfp143.
1.5. All stage analysis of selected genes
To test whether this chamber system can be applied to all preimplantation stages, we selected seven genes with apparent ICM-predominant expression. We performed WISH on embryos from unfertilized eggs to blastocysts. All-stage WISH confirmed that the seven genes that we selected indeed showed the ICM-predominant expression. The expression patterns of Pou5f1 (Hanna et al., 2002; Pelton et al., 2002) and Nanog (Chambers et al., 2003; Mitsui et al., 2003) confirmed previously published results. Two new genes identified here were Mybl2 and Gtf2e2. Mybl2 (also called B-Myb) is myeloblastosis oncogene-like 2 and is known to play a major role during S phase (Joaquin and Watson, 2003). In the microarray analysis of preimplantation mouse development, Mybl2 gene is grouped in Cluster 2 together with Nanog, Lefty, Pcaf, and Dnmt3a (Hamatani et al., 2004). These WISH results confirmed this microarray-based finding. It is reported that Mybl2−/− mice die at around E4.5 - E6.5 and in vitro culture of Mybl2−/− blastocyst indicates that Mybl2 is required for ICM formation (Tanaka et al., 1999). The ICM-predominant expression of Mybl2 is thus consistent with this null phenotype.
Gtf2e2 encodes one of the two (beta) subunits of general transcription factor IIE (TFIIE), which recruits TFIIH to the initiation complex and modulates its kinase and helicase activities (Enkhmandakh et al., 2004). Interestingly, one of the subunits of the TFIIH (Gtf2h3), TATA box binding protein (Tbp/TFIID), one of the subunits of RNA polymerase II (Polr2i), transcription elongation factor A (SII) 3 (Tcea3), and one of the TATA box binding protein (TBP)-associated factor (Taf7) were also identified here for ICM-predominant expression.
2. Discussion
The high-throughput WISH system described here has provided spatial and temporal expression patterns of many genes during preimplantation development. This rather simple system is expandable to increase the number of probes tested in one session. The device can be used for embryos or organs in a similar size range (∼100 μm diameter) without any modification. Materials with larger size ranges, such as postimplantation mammalian embryos, Xenopus embryos, Zebrafish embryos, can also be done, probably with slight modification. Of course, the WISH will tell us only the expression patterns of RNAs, but not proteins, and there are differences between the localizations of RNAs and proteins. For example, Smarca4 showed ICM-predominant expression of RNAs in this study, but an earlier immunohistochemical study showed the protein localizing in both ICM and TE (LeGouy et al., 1998). Similarly, immunohistochemistry showed that the protein GTF2I2 is localized in both ICM and TE (Enkhmandakh et al., 2004), but our in situ study showed ICM-predominant RNA localization. However, RNA localization provides a valuable entry point for further study. The in situ images have been incorporated into the Open Microscopy Environment (OME), which will allow web access of these images (Swedlow et al., 2003). An automatic image classification system that is being developed (I.G., unpublished) can eventually facilitate the automatic analysis of these images. The genes that have been identified for the ICM-predominant expression will be good candidates for further analysis of their role in preimplantation development and cellular pluripotency.
3. Experimental Procedures
3.1. Gene Selection and Annotation
We combined the following public database's Gene Ontology terms and eliminated redundancy in each ‘U’ cluster member sequences to generate Gene Ontology terms for each ‘U’ cluster. 1. Based on Fantom2 sequence membership in NIA Mouse Gene Index (version 1). 2. Based on InterPro domain names. 3. Based on LocusLink. We searched the above databases in April 2003.
3.2. Design and fabrication of aluminum chamber
Parallel micro WISH system consists of four aluminum parts and disposable Transwell-inserts (Corning) (Fig. 1A, 1B). Our device follows standard 24-well format and can hold up to 20 inserts. Solutions were exchanged through “port” shown in the figure. Exact dimensions of the chamber are available for its fabrication (Supplementary Material 1 or http://lgsun.grc.nia.nih.gov/data). Transwell-inserts were placed in the insert-holder, which was then placed in the solution container. There is a 0.5 mm gap between the bottom of the insert and the bottom of the solution container. We add 20–25 ml solution through the port of the insert-holder. The solution fills the space between the insert-holder and the solution container and then enters each transwell-insert through the microporous membrane on the bottom of inserts. Embryos in the insert were suspended in about 100 - 200 μl solution. The whole chamber is agitated on a shaker during washing steps. We drain solutions by suction through the same port of the insert-holder. Because of the capillary pressure generated in the gap, the same negative pressure was applied to all Transwell-inserts and the drainage was completed within seconds simultaneously in all inserts.
3.3. Embryo collection
We collected one-cell embryos and blastocysts from super-ovulated mice at 0.5 dpc and 3.5 dpc, respectively. Oocytes were collected from unmated super-ovulated females. We collected in vitro cultured two-, four-, eight-cell stage embryos, and morula.
3.4. Whole Mount In Situ Hybridization (WISH)
For each gene in the list, we identified a cDNA clone that contains approximately 1 kb 3′-end sequence from the NIA mouse cDNA collection (Carter et al., 2003b) (http://lgsun.grc.nia.nih.gov/cDNA). All the EST clones are available through ATCC (individual clones) and the designated academic centers (Tanaka et al., 2000; VanBuren et al., 2002). In this mouse cDNA collection, cDNAs were cloned at the SalI/NotI site of vector pSPORT1 or pCMV-SPORT6 (Invitrogen). We prepared the DIG-labeled RNA probes by digesting plasmids followed by in vitro transcription using SP6 or T7 RNA polymerase.
Preimplantation embryos were kept in a Transwell-insert (Corning, Catalogue #3422) during the entire WISH procedure. It has a polycarbonate membrane with pore size of 8.0 μm at the bottom, and thus can retain embryos and allow sufficient solution exchange with the aid of the Chamber System. The insert (6.5 mm diameter) fits to a well of a 24-well plate and a well of our custom-made aluminum chamber device (see below for more details). This chamber device enables us to change solutions simultaneously for all inserts placed in the chamber. WISH was performed essentially as described (Wilkinson and Nieto, 1993). Detailed step-by-step WISH protocol is available (Supplementary Material 2; http://lgsun.grc.nia.nih.gov/data). Here we highlight major steps. (1) Fix embryos in 4% paraformaldehyde in PBT (phosphate buffered saline with 0.1% Tween-20) at 4 °C for 30 min. – overnight. (2) Treat embryos with Proteinase K. (3) Fix embryos again in 4% paraformaldehyde, 0.2% EM-grade glutaraldehyde in PBT at room temperature for 20 minutes. (4) Treat embryos with prehybridization buffer (4X SSC[pH7.0], 50% deionized Formamide, 100 μg/ml Heparin, 250 μg/ml Yeast tRNA, 100 μg/ml Salmon Sperm DNA, 2X Denhardt's solution, 0.1% Tween-20) typically at 60 °C (or up to ∼70 °C) for 3 – 8 hrs. (5) Treat embryos with DIG-RNA probe in hybridization buffer for overnight at the same temperature used for prehybridization. (6) Wash embryos with a buffer (50% Formamide, 2X SSC, 0.1% Tween-20) at least three times at the hybridization temperature. (7) Follow the manufacturer's recommended procedure to detect DIG-labeled probes. (8) Transfer embryos suspended in PBS (phosphate buffered saline) containing 1 mM EDTA and 20% glycerol to a 24-well plate for brightfield photographing with 20X objective lens.
Supplementary Material
Figure 3.
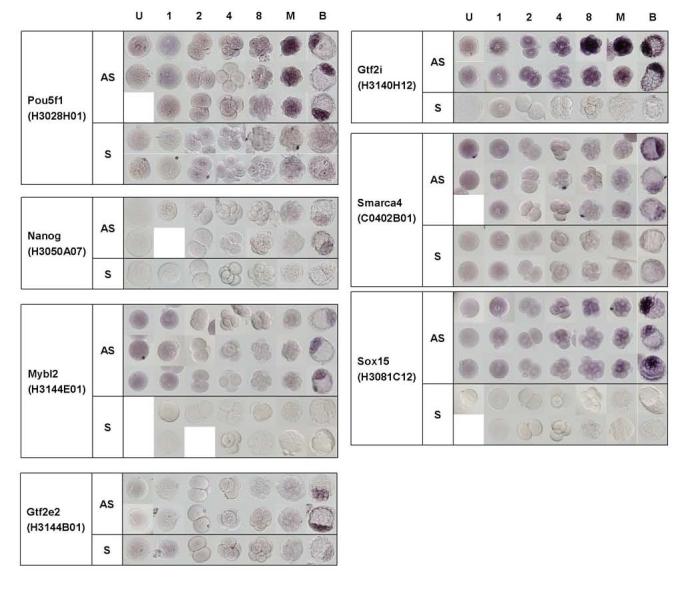
WISH results of 7 genes during preimplantation development. AS; antisense probe, S; sense probe
Acknowledgments
We would like to thank Drs. Vincent VanBuren and Tetsuya S. Tanaka for discussion. TY was supported by the postdoctoral fellowships from the Uehara Memorial Foundation and the Japan Society for the Promotion of Science (JSPS). We would like to thank Mr. Richard Zichos for his excellent work in fabricating the aluminum block portion of the device. We would like to thank Drs. Janet Rossant and Tilo Kunath for providing RNAs from ES and TS cells.
Footnotes
Supplementary Materials
(1) A blueprint of WISH Aluminum Chamber.
(2) Step-by-step WISH protocol.
(3) ES versus TS microarray data.
References
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell GW, Yatskievych TA, Antin PB. GEISHA, a whole-mount in situ hybridization gene expression screen in chicken embryos. Dev Dyn. 2004;229:677–87. doi: 10.1002/dvdy.10503. [DOI] [PubMed] [Google Scholar]
- Carter MG, Hamatani T, Sharov AA, Carmack CE, Qian Y, Aiba K, Ko NT, Dudekula DB, Brzoska PM, Hwang SS, Ko MSH. In situ-synthesized novel microarray optimized for mouse stem cell and early developmental expression profiling. Genome Res. 2003a;13:1011–21. doi: 10.1101/gr.878903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter MG, Piao Y, Dudekula DB, Qian Y, VanBuren V, Sharov AA, Tanaka TS, Martin PR, Bassey UC, Stagg CA, Aiba K, Hamatani T, Matoba R, Kargul GJ, Ko MS. The NIA cDNA project in mouse stem cells and early embryos. C R Biol. 2003b;326:931–40. doi: 10.1016/j.crvi.2003.09.008. [DOI] [PubMed] [Google Scholar]
- Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–55. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- Edwards RG. Aspects of the molecular regulation of early mammalian development. Reprod Biomed Online. 2003;6:97–113. doi: 10.1016/s1472-6483(10)62061-5. [DOI] [PubMed] [Google Scholar]
- Enkhmandakh B, Bitchevaia N, Ruddle F, Bayarsaihan D. The early embryonic expression of TFII-I during mouse preimplantation development. Gene Expr Patterns. 2004;4:25–8. doi: 10.1016/s1567-133x(03)00155-8. [DOI] [PubMed] [Google Scholar]
- Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–6. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Gardner RL. Specification of embryonic axes begins before cleavage in normal mouse development. Development. 2001;128:839–47. doi: 10.1242/dev.128.6.839. [DOI] [PubMed] [Google Scholar]
- Gawantka V, Pollet N, Delius H, Vingron M, Pfister R, Nitsch R, Blumenstock C, Niehrs C. Gene expression screening in Xenopus identifies molecular pathways, predicts gene function and provides a global view of embryonic patterning. Mech Dev. 1998;77:95–141. doi: 10.1016/s0925-4773(98)00115-4. [DOI] [PubMed] [Google Scholar]
- Gitton Y, Dahmane N, Baik S, Ruiz i Altaba A, Neidhardt L, Scholze M, Herrmann BG, Kahlem P, Benkahla A, Schrinner S, Yildirimman R, Herwig R, Lehrach H, Yaspo ML. A gene expression map of human chromosome 21 orthologues in the mouse. Nature. 2002;420:586–90. doi: 10.1038/nature01270. [DOI] [PubMed] [Google Scholar]
- Hamatani T, Carter MG, Sharov AA, Ko MSH. Dynamics of global gene expression changes during mouse preimplantation development. Dev Cell. 2004;6:117–31. doi: 10.1016/s1534-5807(03)00373-3. [DOI] [PubMed] [Google Scholar]
- Hanna LA, Foreman RK, Tarasenko IA, Kessler DS, Labosky PA. Requirement for Foxd3 in maintaining pluripotent cells of the early mouse embryo. Genes Dev. 2002;16:2650–61. doi: 10.1101/gad.1020502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiiragi T, Solter D. First cleavage plane of the mouse egg is not predetermined but defined by the topology of the two apposing pronuclei. Nature. 2004;430:360–4. doi: 10.1038/nature02595. [DOI] [PubMed] [Google Scholar]
- Joaquin M, Watson RJ. Cell cycle regulation by the B-Myb transcription factor. Cell Mol Life Sci. 2003;60:2389–401. doi: 10.1007/s00018-003-3037-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura C, Shen MM, Takeda N, Aizawa S, Matsuo I. Complementary functions of Otx2 and Cripto in initial patterning of mouse epiblast. Dev Biol. 2001;235:12–32. doi: 10.1006/dbio.2001.0289. [DOI] [PubMed] [Google Scholar]
- Ko MSH. Embryogenomics: developmental biology meets genomics. Trends Biotechnol. 2001;19:511–518. doi: 10.1016/s0167-7799(01)01806-6. [DOI] [PubMed] [Google Scholar]
- Ko MSH, Kitchen JR, Wang X, Threat TA, Hasegawa A, Sun T, Grahovac MJ, Kargul GJ, Lim MK, Cui Y, Sano Y, Tanaka T, Liang Y, Mason S, Paonessa PD, Sauls AD, DePalma GE, Sharara R, Rowe LB, Eppig J, Morrell C, Doi H. Large-scale cDNA analysis reveals phased gene expression patterns during preimplantation mouse development. Development. 2000;127:1737–49. doi: 10.1242/dev.127.8.1737. [DOI] [PubMed] [Google Scholar]
- Komiya T, Tanigawa Y, Hirohashi S. A large-scale in situ hybridization system using an equalized cDNA library. Anal Biochem. 1997;254:23–30. doi: 10.1006/abio.1997.2399. [DOI] [PubMed] [Google Scholar]
- Kudoh T, Tsang M, Hukriede NA, Chen X, Dedekian M, Clarke CJ, Kiang A, Schultz S, Epstein JA, Toyama R, Dawid IB. A gene expression screen in zebrafish embryogenesis. Genome Res. 2001;11:1979–87. doi: 10.1101/gr.209601. [DOI] [PubMed] [Google Scholar]
- LeGouy E, Thompson EM, Muchardt C, Renard JP. Differential preimplantation regulation of two mouse homologues of the yeast SWI2 protein. Dev Dyn. 1998;212:38–48. doi: 10.1002/(SICI)1097-0177(199805)212:1<38::AID-AJA4>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci U S A. 1981;78:7634–8. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–42. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- Mochizuki Y, Satou Y, Satoh N. Large-scale characterization of genes specific to the larval nervous system in the ascidian Ciona intestinalis. Genesis. 2003;36:62–71. doi: 10.1002/gene.10199. [DOI] [PubMed] [Google Scholar]
- Neidhardt L, Gasca S, Wertz K, Obermayr F, Worpenberg S, Lehrach H, Herrmann BG. Large-scale screen for genes controlling mammalian embryogenesis, using high-throughput gene expression analysis in mouse embryos. Mech Dev. 2000;98:77–94. doi: 10.1016/s0925-4773(00)00453-6. [DOI] [PubMed] [Google Scholar]
- Newman-Smith ED, Werb Z. Stem cell defects in parthenogenetic peri-implantation embryos. Development. 1995;121:2069–77. doi: 10.1242/dev.121.7.2069. [DOI] [PubMed] [Google Scholar]
- Pelton TA, Sharma S, Schulz TC, Rathjen J, Rathjen PD. Transient pluripotent cell populations during primitive ectoderm formation: correlation of in vivo and in vitro pluripotent cell development. J Cell Sci. 2002;115:329–39. doi: 10.1242/jcs.115.2.329. [DOI] [PubMed] [Google Scholar]
- Pesce M, Scholer HR. Oct-4: control of totipotency and germline determination. Mol Reprod Dev. 2000;55:452–7. doi: 10.1002/(SICI)1098-2795(200004)55:4<452::AID-MRD14>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Piotrowska K, Wianny F, Pedersen RA, Zernicka-Goetz M. Blastomeres arising from the first cleavage division have distinguishable fates in normal mouse development. Development. 2001;128:3739–48. doi: 10.1242/dev.128.19.3739. [DOI] [PubMed] [Google Scholar]
- Quiring R, Wittbrodt B, Henrich T, Ramialison M, Burgtorf C, Lehrach H, Wittbrodt J. Large-scale expression screening by automated whole-mount in situ hybridization. Mech Dev. 2004;121:971–6. doi: 10.1016/j.mod.2004.03.031. [DOI] [PubMed] [Google Scholar]
- Reymond A, Marigo V, Yaylaoglu MB, Leoni A, Ucla C, Scamuffa N, Caccioppoli C, Dermitzakis ET, Lyle R, Banfi S, Eichele G, Antonarakis SE, Ballabio A. Human chromosome 21 gene expression atlas in the mouse. Nature. 2002;420:582–6. doi: 10.1038/nature01178. [DOI] [PubMed] [Google Scholar]
- Sharov AA, Piao Y, Matoba R, Dudekula DB, Qian Y, VanBuren V, Falco G, Martin PR, Stagg CA, Bassey UC, Wang Y, Carter MG, Hamatani T, Aiba K, Akutsu H, Sharova L, Tanaka TS, Kimber WL, Yoshikawa T, Jaradat SA, Pantano S, Nagaraja R, Boheler KR, Taub D, Hodes RJ, Longo DL, Schlessinger D, Keller J, Klotz E, Kelsoe G, Umezawa A, Vescovi AL, Rossant J, Kunath T, Hogan BL, Curci A, D'Urso M, Kelso J, Hide W, Ko MSH. Transcriptome analysis of mouse stem cells and early embryos. PLoS Biol. 2003;1:E74. doi: 10.1371/journal.pbio.0000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T, Kato A, Yuasa-Kawada J, Sakuta H, Takahashi M, Suzuki R, Ohkawara T, Takahashi H, Noda M. Large-scale identification and characterization of genes with asymmetric expression patterns in the developing chick retina. J Neurobiol. 2004;59:34–47. doi: 10.1002/neu.10338. [DOI] [PubMed] [Google Scholar]
- Solter D, de Vries WN, Evsikov AV, Peaston AE, Chen FH, Knowles BB. Fertilization and activation of the embryonic genome. In: Rossant J, Tam PPL, editors. Mouse Development: Patterning, Morphogenesis, and Organogenesis. Academic Press; San Diego: 2002. pp. 5–19. [Google Scholar]
- Swedlow JR, Goldberg I, Brauner E, Sorger PK. Informatics and quantitative analysis in biological imaging. Science. 2003;300:100–2. doi: 10.1126/science.1082602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, Rossant J. Promotion of trophoblast stem cell proliferation by FGF4. Science. 1998;282:2072–5. doi: 10.1126/science.282.5396.2072. [DOI] [PubMed] [Google Scholar]
- Tanaka TS, Jaradat SA, Lim MK, Kargul GJ, Wang X, Grahovac MJ, Pantano S, Sano Y, Piao Y, Nagaraja R, Doi H, Wood WH, 3rd, Becker KG, Ko MSH. Genome-wide expression profiling of mid-gestation placenta and embryo using a 15,000 mouse developmental cDNA microarray. Proc Natl Acad Sci U S A. 2000;97:9127–32. doi: 10.1073/pnas.97.16.9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka TS, Ko MSH. A global view of gene expression in the preimplantation mouse embryo: morula versus blastocyst. Eur J Obstet Gynecol Reprod Biol. 2004;115(Suppl 1):S85–91. doi: 10.1016/j.ejogrb.2004.01.026. [DOI] [PubMed] [Google Scholar]
- Tanaka TS, Kunath T, Kimber WL, Jaradat SA, Stagg CA, Usuda M, Yokota T, Niwa H, Rossant J, Ko MSH. Gene expression profiling of embryo-derived stem cells reveals candidate genes associated with pluripotency and lineage specificity. Genome Res. 2002;12:1921–8. doi: 10.1101/gr.670002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Patestos NP, Maekawa T, Ishii S. B-myb is required for inner cell mass formation at an early stage of development. J Biol Chem. 1999;274:28067–70. doi: 10.1074/jbc.274.40.28067. [DOI] [PubMed] [Google Scholar]
- Tomancak P, Beaton A, Weiszmann R, Kwan E, Shu S, Lewis SE, Richards S, Ashburner M, Hartenstein V, Celniker SE, Rubin GM. Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-12-research0088. RESEARCH0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanBuren V, Piao Y, Dudekula DB, Qian Y, Carter MG, Martin PR, Stagg CA, Bassey UC, Aiba K, Hamatani T, Kargul GJ, Luo AG, Kelso J, Hide W, Ko MSH. Assembly, verification, and initial annotation of the NIA mouse 7.4K cDNA clone set. Genome Res. 2002;12:1999–2003. doi: 10.1101/gr.633802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QT, Piotrowska K, Ciemerych MA, Milenkovic L, Scott MP, Davis RW, Zernicka-Goetz M. A genome-wide study of gene activity reveals developmental signaling pathways in the preimplantation mouse embryo. Dev Cell. 2004;6:133–44. doi: 10.1016/s1534-5807(03)00404-0. [DOI] [PubMed] [Google Scholar]
- Wilkinson DG, Nieto MA. Detection of messenger RNA by in situ hybridization to tissue sections and whole mounts. Methods Enzymol. 1993;225:361–73. doi: 10.1016/0076-6879(93)25025-w. [DOI] [PubMed] [Google Scholar]
- Zeng F, Baldwin DA, Schultz RM. Transcript profiling during preimplantation mouse development. Dev Biol. 2004;272:483–96. doi: 10.1016/j.ydbio.2004.05.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


