Abstract
Interferon-α therapy has been shown to be active in the treatment of mycosis fungoides although the individual response to this therapy is unpredictable and dependent on essentially unknown factors. In an effort to better understand the molecular mechanisms of interferon-α resistance we have developed an interferon-α resistant variant from a sensitive cutaneous T-cell lymphoma cell line. We have performed expression analysis to detect genes differentially expressed between both variants using a cDNA microarray including 6386 cancer-implicated genes. The experiments showed that resistance to interferon-α is consistently associated with changes in the expression of a set of 39 genes, involved in signal transduction, apoptosis, transcription regulation, and cell growth. Additional studies performed confirm that STAT1 and STAT3 expression and interferon-α induction and activation are not altered between both variants. The gene MAL, highly overexpressed by resistant cells, was also found to be expressed by tumoral cells in a series of cutaneous T-cell lymphoma patients treated with interferon-α and/or photochemotherapy. MAL expression was associated with longer time to complete remission. Time-course experiments of the sensitive and resistant cells showed a differential expression of a subset of genes involved in interferon-response (1 to 4 hours), cell growth and apoptosis (24 to 48 hours.), and signal transduction.
Interferons (IFNs) are members of the cytokine family of proteins with pleiotrophic cellular effects, such as antiproliferative, proapoptotic, and immunomodulatory effects in addition to possessing anti-viral and anti-tumoral properties. 1 Although the basic pathway through which IFNs act is only partially understood, there is an increasing flow of data concerning IFN targets. Thus IFN-α binds to its cognate receptor on the surface of cells, mainly lymphocytes, monocytes, and some neoplastic cells, 2 thereby initiating a signal cascade involving the JAK/STAT signaling pathway. 3 The JAK proteins are members of a family of tyrosine kinases, able to phosphorylate and thereby activate members of the STAT family of transcription factors. These phosphorylated STAT proteins translocate to the nucleus where they activate the transcription of a set of IFN-stimulated genes by binding to IFN-response elements in the DNA. 4
Apoptosis and growth inhibition are disparate responses of cells to IFN-α and the sensitivity toward these phenomena may vary independently between different cell types. 5 Some of the recognized IFN-α targets that explain its anti-tumoral role are hTERT, 6 bcl2, 7 c-MYC, 8 and other cell-cycle regulators such as p15, p21, and p27. 9
IFN-α is commonly used in the treatment of a variety of neoplasms including mycosis fungoides (MF), chronic myeloid leukemia, multiple myeloma, and malignant melanoma although the individual response to this therapy is essentially unpredictable. Despite the favorable effect in a large proportion of patients, a significant group of patients show initial or acquired resistance to this therapy. Although alterations in some of the IFN-α response genes, such as well-known members of the JAK/STAT signaling pathway, (STAT1, STAT3, SOCS1, or IFNR) have been claimed to play a role in IFN-α resistance, 10-14 this has not been able to explain the frequent clinical phenomenon of IFN-α resistance.
Here we have focused on mechanisms of resistance to IFN-α in MF, the most frequent type of cutaneous T-cell lymphoma (CTCL), a relatively frequent type of T-cell lymphoma in which IFN-α is commonly used as a first-line treatment. CTCLs comprise a heterogeneous group of neoplasms characterized by an accumulation of malignant T lymphocytes in skin and occasionally, as in Sezary syndrome, in skin and blood. 15 Because acquisition of clinical resistance to IFN-α is commonly observed in CTCL, it would be of great interest to determine the mechanism of clinical resistance to IFN-α at a molecular level, leading to development of tailored treatment strategies or alternative therapies.
As IFN-α exerts its action through different cell pathways involving a large number of genes and there may be cross-talk with other signal transduction pathways, we hypothesized that only a system of massive molecular analysis, such as cDNA microarray analysis, could allow the identification of a significant proportion of the genes eventually involved in IFN-α resistance, giving information about the diversity of the cell pathways involved.
cDNA microarrays are a powerful technique that allow the analysis of the expression of thousands of genes simultaneously. Gene expression is visualized by exciting fluorescein-labeled hybridized samples using a laser. A digital image of the array is captured using a confocal scanning laser microscope. 16 The raw data collected is a monochrome image, which is subsequently converted to numerical matrices, allowing analysis to be performed using bioinformatical techniques. Comparison of fluorescence intensities using microarrays is quite a robust method and correlates well to other methods used to quantify gene expression such as Northern blot hybridization and quantitative reverse transcriptase-polymerase chain reaction (PCR). 17,18
These cDNA microarrays have been used for monitoring the expression profile in CTCL cells selected for resistance to IFN-α. To confirm the results obtained, we have used quantitative PCR for the most relevant genes. Interestingly, this strategy has pinpointed to the existence of changes in the expression of a selected group of genes involved in signal transduction and apoptosis regulation. Time-course experiments to monitor differences in IFN-α response have confirmed that resistance appears to be caused by deregulation of cell-cycle control, apoptosis, and signal transduction.
Materials and Methods
Tumor Cell Lines
huT78 cells were obtained from the American Type Culture Collection (Rockville, MD). Cells were maintained in RPMI 1640 medium (Sigma Chemical Co., St. Louis, MO) supplemented with 20% fetal bovine serum (Sigma Chemical Co.), 2 mmol/L l-glutamine (Life Technologies, Inc., Grand Island, NY), 50 mg/ml penicillin (Life Technologies, Inc.), 50 mg/ml streptomycin (Life Technologies, Inc.), and 2.5 μg/ml fungizone (Life Technologies, Inc.) and grown at 37°C in a humidified atmosphere and 5% CO2.
The sensitivity to IFN-α was tested by culturing the cells for 72 hours to 96 hours in medium containing between 0 U/ml of IFN-α 2A (Hoffman La Roche, Nutley, NJ) and 1,000,000 U/ml of IFN-α. Cell viability was determined every 24 hours by trypan blue exclusion. The resistant variant of the huT78 cell line was obtained by culturing the cells in an increasing concentration of IFN-α 2A beginning with 100 U/ml and increasing to 200,000 U/ml loosely following a previously described protocol. 13 Cells were passaged once per week and viability determined by trypan blue exclusion. Resistant cells were retested after 1 month in the absence of treatment to ensure culture was nonrevertant.
For quantitative PCR studies huT78R and huT78S cells were stimulated with IFN-α (5000 U/ml) for 3 and 24 hours and the cells were collected along with pretreatment cells for RNA extraction. Time-course experiments were performed with the cell lines huT78R and huT78S. Cells were treated with 5000 U/ml of IFN-α and cells were collected before treatment and at time points 1 hour, 4 hours, 24 hours, and 48 hours after commencement of treatment.
Case Selection
A group of 20 samples from MF patients was selected from the medical records of the 12 de Octubre Hospital and the La Princesa Hospital, Madrid Spain [tissue samples were provided by the National Tumor Bank (CNIO), with the collaboration of the hospitals]. Diagnosis was based on generally accepted clinical-pathological criteria. Patients were included in a randomized clinical trial to compare the efficacy of combination psoralen and ultraviolet (UVA) irradiation (PUVA) versus PUVA + IFN-α in a group of stage Ia, Ib, or IIa MF patients. Ten patients were receiving PUVA alone and 10 patients were under PUVA + IFN-α treatment (9 μ, three times a week). Informed consent was obtained from the patients enrolled under the supervision of the local Ethical Committees.
RNA Isolation
Total RNA extraction was performed using the RNeasy kit (Qiagen Inc., Valencia, CA) and digested with RNase-free DNase I following the manufacturer’s instructions.
Quantitative PCR
The huT78R and huT78S cells were stimulated with IFN-α for 3 and 24 hours and the cells were collected along with pretreatment cells for RNA extraction for testing the levels of STAT1 and STAT3 mRNA expression. cDNA for use in quantitative PCR studies was synthesized using AMV reverse transcriptase (Access RT-PCR System; Promega, Madison, WI) and random hexamer primers (Promega). One μg of total RNA was transcribed. Primers and probes for quantitative PCR were designed using Primer express (Applied Biosystems, Foster City, CA) following whenever possible, the universal conditions of primer design recommended. In addition the probes were designed so that genomic DNA would not be detected during the PCR. Primers were synthesized by Applied Biosystems and/or Life Technologies, Inc. and all probes were synthesized by Applied Biosystems. Probes were labeled at the 5′ end with a reporter dye FAM (6-carboxy-fluorescein) and at the 3′ end with a quencher dye TAMRA (6-carboxy-tetramethylrhodamine). The sequences of primers and probes used for these studies are shown in Table 1 ▶ .
Table 1.
Primers and Probes Used for Quantitative PCR Studies
| Primer name | Accession number | Location | Primer sequence | Product size | ||||
|---|---|---|---|---|---|---|---|---|
| A: Primer and probe sets used for quantitative PCR using Taqman probes | ||||||||
| STAT1-F | XM_010893 | 722 | 5′-GCTGCGGTTCAGTGAGAGCT-3′ | |||||
| STAT1-R | 886 | 5′-GCCATGACTTTGTAATTGCGAAT-3′ | ||||||
| STAT1-P | 784 | 5′-AGAACGGAGGCGAACCTGACTTCCA-3′ | 165 bp | |||||
| STAT3-F | NM_003150 | 2311 | 5′-CCTGAAGCTGACCCAGGTAGC-3′ | |||||
| STAT3-R | 2458 | 5′-CACCTTCACCATTATTTCCAAACTG-3′ | ||||||
| STAT3-P | 2630 | 5′-TCTGTGTGACACCAACGACCTGCAGC-3′ | 148 bp | |||||
| MAL-F | NM_002371 | 92 | 5′-CCTGCCCAGTGGCTTCTC-3′ | |||||
| MAL-R | 191 | 5′-GGAGGAGGCCACCAGGAT-3′ | ||||||
| MAL-P | 126 | 5′-CCCGACTTGCTCTTCATCTTTGAGTTTAT-3′ | 100 bp | |||||
| BAG3-F | XM_055575 | 1969 | 5′-ACAGCAGCAGCGACTTCAAA-3′ | |||||
| BAG3-R | 2072 | 5′-ACACATCGGTTCCGAGTCTGA-3′ | ||||||
| BAG3-P | 2017 | 5′-AACCCAGCAGCACCGTAGCCTCTG-3′ | 104 bp | |||||
| SAMSN1-F | XM_012974 | 443 | 5′-AAAGCCAGTGACTCCATGGATAGT-3′ | |||||
| SAMSN1-R | 541 | 5′-TCGAAAGCTGTCCCGGTT-3′ | ||||||
| SAMSN1-P | 475 | 5′-CTTGTTATGCCACTTGATGAGCTCTGTCCA-3′ | 99 bp | |||||
| B: Primer pairs used for gene expression studies using SYBR green | ||||||||
| CAV1-F | NM_001753 | 431 | 5′-GGTCAACCGCGACCCTAA-3′ | |||||
| CAV1-R | 512 | 5′-TGTCCCTTCTGGTTCTGCAA-3′ | 82 bp | |||||
| TNFSF7-F | NM_001252 | 293 | 5′-TGCCGCTCGAGTCACTTG-3′ | |||||
| TNFSF7-R | 418 | 5′-CTGGTCCATGCAGGAAGGA-3′ | 126 bp | |||||
| P2Y5-F | AF000546 | 401 | 5′-CTACCCATTTAAGTCAAAGACTCTAAGAACC-3′ | |||||
| P2Y5-R | 548 | 5′-GCAGGCTTCTGAGGCATTGTT-3′ | 148 bp | |||||
In all cases a standard curve containing at least three concentrations (represented in duplicate or triplicate) of a control cDNA was constructed for both the endogenous control gene (GAPDH) and the gene of interest. In all cases standard curves had a coefficient of correlation >0.98. In each case all samples were represented in duplicate or triplicate for calculations.
For STAT1 and MAL 600 nmol/L of primers and probe were used. STAT3, BAG3, and SAMSN1 PCR reactions were performed using 200 nmol/L of primers and probe and 300 nmol/L of primers were used for all remaining genes. GAPDH PCR was performed using 20× pre-developed assay reagents (PDAR) primers and probe for GAPDH (Applied Biosystems). All PCRs were performed under the same conditions using the ABI prism 7700 (Applied Biosystems) and analysis and normalization of results were performed using the sequence detector software (Applied Biosystems). Principles of real time PCR have been described elsewhere. 19 The quantitative PCR control gene GAPDH was chosen based on initial experiments with three cell lines (huT78, EM2, Hs.895T) that were tested with GAPDH before and 24 hours after treatment with IFN-α. No significant changes were seen in any case. Of three endogenous control genes tested (cyclophilin, GAPDH, and 18S rRNA) GAPDH showed the lowest variability between samples and between treatment states (data not shown).
Construction and Analysis of cDNA Microarray
For all microarray studies the CNIO OncoChip was used. The CNIO OncoChip is a cDNA microarray that has been specially designed for looking at genes involved in cancer and includes a core of 2489 cancer-relevant genes in addition to genes involved in drug-response, tissue-specific genes, and control genes. There are a total of 6386 genes represented by 7237 clones. Human cDNA clones were purchased from Research Genetics (Huntsville, AL). The set consists of 7237 sequence validated I.M.A.G.E. clones, including 5253 clones representing known genes and the remaining 1984 clones representing expressed sequence tags (ESTs). Time-course experiments were performed using an extended version of the CNIO OncoChip in which cancer-related clones (n = 2489) were printed twice. The list of genes on the array can be found at: http://bioinfo.cnio.es/data/oncochip.
Microarray Fabrication
In brief, individual IMAGE clones were grown in 96-well plates at 37°C for 6 hours. The bacterial culture was used for PCR amplification and the products were purified. PCR amplification products were verified by agarose gel electrophoresis. c-DNAs were printed onto chemically activated glass slides (CMT-GAPS: Corning, Corning, NY) using the spotter Multigrid II (BioRobotics, Woburn, MA). After printing, the slides were stored in the dark at room temperature.
Target Preparation
T-7-based RNA amplifications and preparations of cDNA probes were performed as described previously. 20,21 Briefly, 5 μg of total RNA were converted to double-stranded cDNA using the superscript choice system (Life Technologies) using oligo-dT primer containing a T7 RNA polymerase promoter. The double-stranded cDNA was cleaned up, and T7 in vitro transcription was performed using Ampliscribe T7 in vitro transcription kit (Epicentre Technologies, Madison, WI) or Megascript T7 in vitro transcription kit (Ambion, Austin, TX) following the manufacturers’ instructions. Fluorescent first-strand cDNA was made with 5 μg of amplified RNA in the presence of 50 μmol/L of Cy5-dCTP or Cy3-dCTP (Amersham, Uppsala, Sweden), random hexamers (Promega), DTT, RNAsin, unlabeled dNTP (25 nmol of dCTP, dATP, and dGTP and 5 nmol of dDTP), Amp synthetic RNA (spiked cDNA synthesis control), and superscript reverse transcriptase. 22 Ten μg of Cot1 human DNA (Life Technologies, Inc.) was added to the labeled probe. The microarray slides were denaturalized by placing in boiling water for 3 minutes. Slides were incubated in a buffer containing 4× standard saline citrate, 1× bovine serum albumin, 2 μg/ml DNAs, and 0.1% sodium dodecyl sulfate for 45 minutes at 65°C followed by washing with water and filtered using a 45-μm filter. The probe was added to a solution containing 50% formamide, 20× standard saline citrate and 10% sodium dodecyl sulfate and incubated at 100°C for 3 minutes followed by 20 minutes at 42°C before applying to the microarray slide. Hybridization was performed at 42°C for 15 hours. Slides were scanned for Cy3 and Cy5 fluorescence using Scanarray 5000 XL (GSI Lumonics Kanata, Ontario, Canada) and quantified using the Quantarray software (GSI Lumonics) and/or GenePix Pro 4.0 software (Axon Instruments Inc., Union City, CA). The huT78S versus huT78R hybridizations were performed in duplicate using different target preparations. For the time-course experiments each sample (Cy5) was analyzed using the cDNA microarray CNIO OncoChip using the untreated cells as a control (Cy3).
Data Analysis
Fluorescence intensity measurements from each array element were compared with local background and background subtraction was performed. To normalize the data, the Cy3/Cy5 ratio was adjusted to a normalized factor equal to the median ratio value of all spots in the array. In addition, spots with background-subtracted signal intensities lower than 500 fluorescence units (sum of the two channels) were excluded from the analysis. Furthermore, bad spots or areas of the array with obvious defects were manually flagged. The Cy3:Cy5 ratios of the duplicated spots of the array were averaged. Genes were deemed to be up-regulated or down-regulated if the difference ratio was at least twofold.
For the time-course experiments, genes whose expression change was not significant (twofold) in at least one of the four time points were discarded from analysis. Analysis of the genes was divided into two subgroups with 1 and 4 hours being considered as early response genes, and 24 and 48 hours considered as late response genes. Within each category genes whose expression was not significantly different (twofold) between huT78S and huT78R at one or both time points were also discarded from analysis. For clustering analysis all ESTs and genes with unknown function were excluded. To classify the temporal profiles of gene expression cluster analysis was performed using the Cluster program SOTA (http://bioinfo.cnio.es/cgi-bin/tools/clustering/sotarray) based on Euclidean distances between genes. Biological functions were assigned by using the GENECARDS database 23 (http://bioinformatics.weizmann.ac.il/cards). Additionally each time point was considered individually. In this case genes whose expression was not significant (twofold) in either huT78S or huT78R for each time point were discarded from analysis. Genes for which the difference in expression was significantly different between huT78S and huT78R were retained. ESTs and hypothetical proteins were discarded. The raw data can be found in the supplementary information at http://bioinfo.cnio.es/data/interferon_resistance/.
Immunohistochemistry
Cell cytospins from sensitive and resistant huT78 cells were collected and fixed for 5 minutes using acetone. Immunohistochemistry assays were performed in the Immunohistochemistry Unit (CNIO) on the cell line cytospin preparations using monoclonal STAT1 and STAT3 antibodies (Santa Cruz, San Francisco, CA). Immunohistochemical techniques were also performed on paraffin-embedded tissue sections taken from the 20 patients included in the clinical trial before the start of the treatment using MAL antibody. 24
A heat-induced epitope retrieval step was performed in a solution of sodium citrate buffer with 0.01 mol/L of tri-sodium citrate solution, and heated for 2 minutes in a conventional pressure cooker. After heating, slides were rinsed in cool running water for 5 minutes. They were then quickly washed in Tris-buffered saline, pH 7.4, and incubated with mouse monoclonal IgG1 antibodies: STAT1 (C-136), STAT3 (F-2) (Santa Cruz), and MAL antibody, diluted 1:50, 1:500, and 1:250, respectively. After incubation with the primary antibody, immunodetection was performed with biotinylated anti-mouse Ig, followed by peroxidase-labeled streptavidin-biotin (LSAB-DAKO, Glostrup, Denmark) with diaminobenzidine chromogen as substrate. All immunostaining was performed using the TechMate 500 (DAKO) automatic immunostaining device. Incubations omitting the specific antibody, including incubation including unrelated antibodies, were used as a control of the technique.
Clinical Data and Statistical Analysis
Rapid response is defined as complete remission within 16 weeks of treatment commencement whereas patients whose time to complete remission is longer than 16 weeks are considered to have a slow response. Sixteen weeks was the mean and the median of the time necessary to induce complete remission. The Fisher’s exact test was used to analyze the relationship between clinical outcome (rapid response or slow response) and MAL expression.
Results
Production of IFN-α Resistant huT78 Cell Line
huT78 is a cell line derived originally from a MF patient. 25 To test for sensitivity to IFN-α, cells were cultured for between 72 and 96 hours in medium containing between 0 U/ml of IFN-α-2a (Hoffman La Roche) and 1,000,000 U/ml IFN-α (Figure 1A) ▶ . The IC50 (the concentration of IFN-α required for 50% growth inhibition in 4 days) of huT78S cells was determined to be between 100 U/ml and 1000 U/ml.
Figure 1.
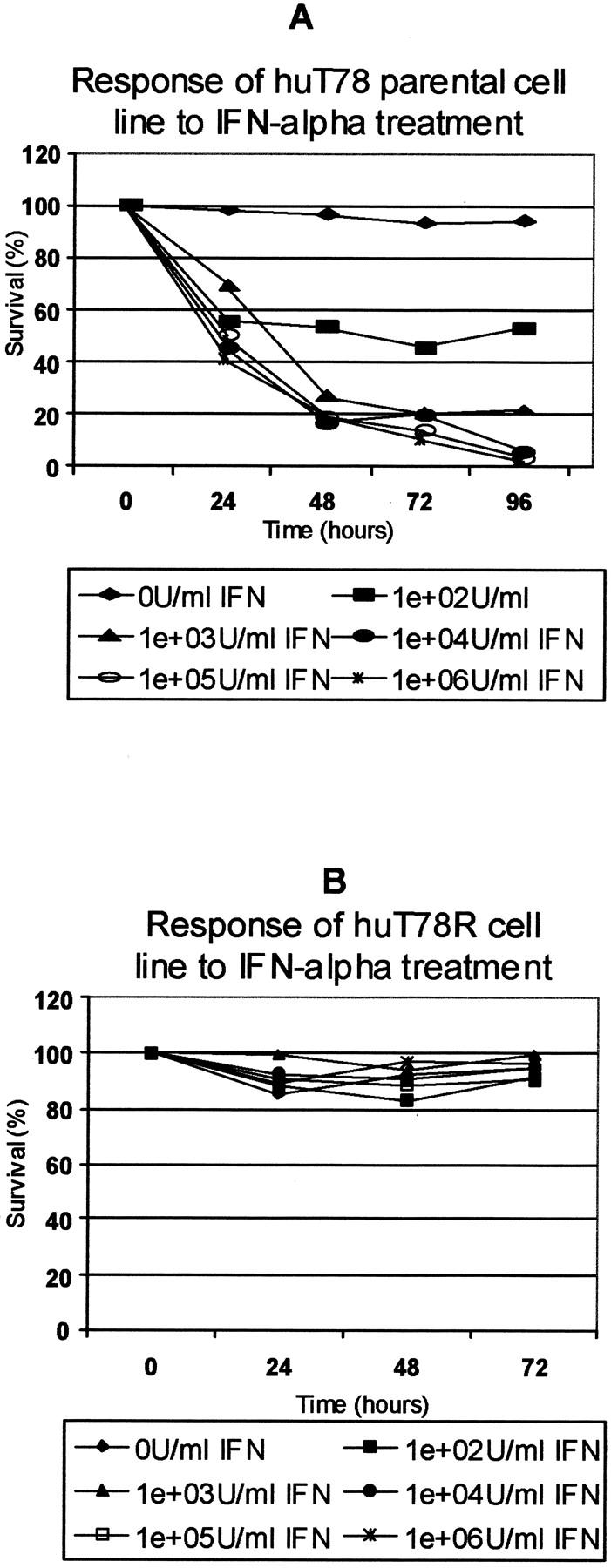
Comparison of the IFN-α sensitivity of huT78S and huT78R. A: The parental huT78 (huT78S) was tested for its sensitivity to IFN-α by culturing cells throughout 96 hours in concentrations of between 0 U/ml IFN-α and 1,000,000 U/ml IFN-α. The viability of cells was determined every 24 hours by trypan blue exclusion. These cells are sensitive to IFN-α at concentrations as low as 100 U/ml. The IC50 for this cell line at 96 hours was calculated to be between 100 U/ml and 1000 U/ml. B: The huT78R cell line produced was tested for its IFN-α sensitivity after 1 month of growth in the absence of IFN-α. As can be seen above this variant strain is extremely resistant to IFN-α even at concentrations of IFN-α of 1,000,000 U/ml.
With the purpose of producing a resistant variant of the huT78 cell line, the cells were cultured in an increasing concentration of IFN-α, beginning with 100 U/ml and increasing to 200,000 U/ml IFN-α, following a previously described protocol. 13 Cells were passaged once per week and viability determined by trypan blue exclusion. Resistant cells were retested after 1 month in the absence of treatment to ensure culture was nonrevertant (Figure 1B) ▶ .
This resistant cell line is characterized by high levels of proliferation even in the presence of elevated IFN-α levels. In fact, cell proliferation is increased in the presence of IFN-α. This resistant cell culture was used as a model system for the study of IFN-α resistance.
IFN-α Induces STAT1 and Activates STAT1 and STAT3 in Both the Resistant and Sensitive huT78 Cell Lines
Expression of STAT1 and STAT3 were studied in the sensitive parental huT78S line and its resistant huT78R partner. In both cell lines STAT1 mRNA expression shows a rapid and dramatic increase after 3 and 24 hours of treatment with 5000 U/ml of IFN-α, with the basal level of expression being similar between both cell lines (Figure 2, A and B) ▶ . STAT3 mRNA was also measured in the same cell lines using cells without treatment and cells harvested at 3 and 24 hours after treatment with 5000 U/ml of IFN-α. The basal level of STAT3 is much more similar between sensitive and resistant cells (Figure 2A) ▶ . However in response to treatment with IFN-α, sensitive cells show some up-regulation of STAT3 shortly after treatment (3 hours). This observed increase is bordering on significance showing slightly less than a twofold increase in expression. This IFN-α-dependent induction of STAT3 is not observed in the resistant variant (Figure 2C) ▶ .
Figure 2.
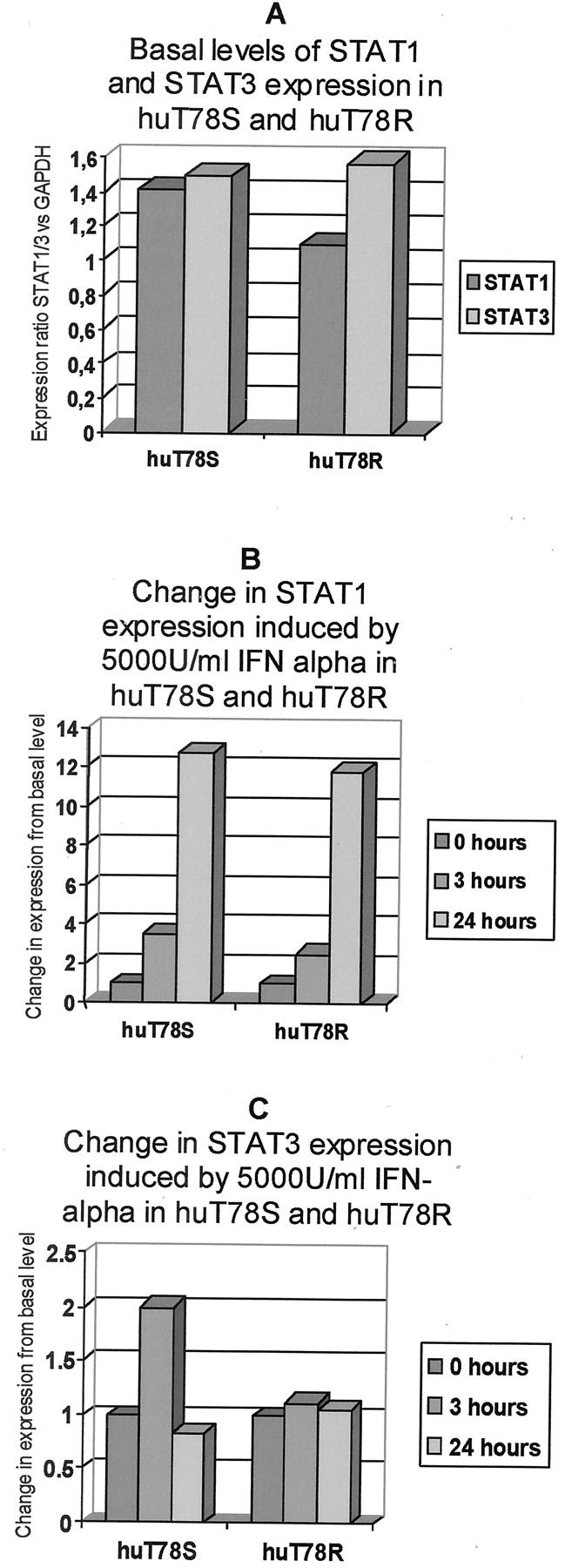
Basal expression of STAT1 and STAT3 in huT78S and huT78R and the changes induced in the expression of these genes by IFN-α (5000 U/ml) after 3 and 24 hours. A: Quantitative PCR results showing that the basal levels of STAT1 and STAT3 are not significantly different between huT78S and huT78R variants. B: Cells from huT78S and huT78R were harvested before commencement of treatment and 3 and 24 hours after commencement of treatment of the cultures with 5000 U/ml IFN-α. Quantitative PCR for STAT1 showed a rapid and significant increase in STAT1 mRNA levels in the case of both huT78S and huT78R. C: STAT3 was also studied under the same conditions. In the case of huT78S there is a slight up-regulation of STAT3 in response to IFN-α. There is no significant change in expression observed in the huT78R variant.
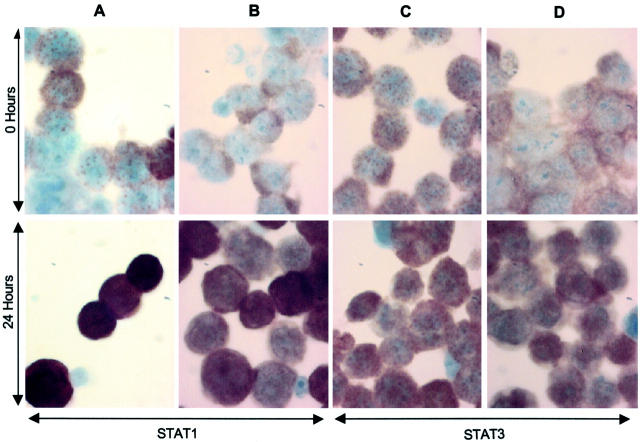
To determine whether IFN-α induces activation of STAT1 and STAT3 in resistant and sensitive huT78 cells after treatment with 5000 U/ml IFN-α at 0, 3, and 24 hours, immunohistochemistry studies were performed to detect the nuclear translocation associated with STAT activation. STAT1 and STAT3 proteins were observed in the cytoplasm before treatment, moving to the nucleus after 3 hours, and more dramatically after 24 hours of treatment. The nuclear localization of both proteins was observed in both sensitive and resistant cell lines and there were no obvious differences between the huT78S and huT78R patterns (Figure 3) ▶ .
Figure 3.
STAT1 and STAT3 expression and activation by IFN-α in huT78S and huT78R. Cells from huT78S and huT78R were collected by cytospin and fixed in acetone before commencement of treatment and 3 and 24 hours after commencement of treatment of the cultures with 5000 U/ml IFN-α (0 and 24 hours only shown here). Cytospins were stained with both STAT1 and STAT3 antibodies. In huT78S (A) and huT78R (B) initially STAT1 protein is found in the cytoplasm. At 24 hours STAT1 has been activated with protein clearly visible and concentrated in the nucleus. In huT78S (C) and huT78R (D) STAT3 is also initially located in the cytoplasm. The protein is activated and translocated to the nucleus after 24 hours.
Genes Differentially Expressed in huT78R and huT78S
To identify genes that may be of interest in contributing to IFN-α resistance, c-DNA microarray studies were performed to compare the resistant and sensitive strains. Total RNA was extracted from 3 million cells in each case, amplified by T7 in vitro transcription, labeled with Cy3 (huT78S) or Cy5 (huT78R), and hybridized simultaneously to the CNIO OncoChip v.1 microarray to compare expression levels of 6386 genes between sensitive and resistant strains. The hybridization was performed in duplicate using RNAs extracted and amplified independently to ensure result reproducibility. Taking into account the results of both experiments, a total of 39 genes were found to be consistently and significantly changed in expression level between the two samples. Of the 39 genes whose expression was found to be changed, the resistant cells overexpress 6 genes while the other 33 genes are repressed. These genes can be seen in Table 2 ▶ . The raw data can be found in the supplementary information at http://bioinfo.cnio.es/data/interferon_resistance/.
Table 2.
Genes Differentially Expressed between huT78S and huT78R Found Using Microarray Analysis
| Unigene cluster ID | Accession number | Gene symbol | Gene name | Ratio | Function | |
|---|---|---|---|---|---|---|
| A. Genes downregulated in huT78R | ||||||
| Hs.3268 | G4738473 | HSPA6 | Heat shock 70kD protein 6 (HSP70B′) | 5.23 | Stress response | |
| Hs.24633 | G1557522 | SAMSN1 | SAM domain, SH3 domain and nuclear localisation signals, 1 | 4.69 | Signal transduction | |
| Hs.74034 | G1548237 | CAV1 | Caveolin 1, caveolae protein, 22kD | 4.66 | Signal transduction, structural protein, lipid rafts | |
| Hs.189999 | G1444110 | P2Y5 | Purinergic receptor (family A group 5) | 3.62 | IFN pathway | |
| Hs.103839 | G2229156 | EPB41L3 | Erythrocyte membrane protein band 4.1-like 3 | 3.48 | Structural protein, cell shape and size control | |
| Hs.342849 | G2103538 | ARL5 | ADP-ribosylation factor-like 5 | 3.30 | GTP binding, intracellular protein traffic | |
| Hs.296398 | G2433839 | LC27 | Putative integral membrane transporter | 2.91 | Putative integral membrane transporter | |
| Hs.46 | G3777678 | PTAFR | Platelet-activating factor receptor | 2.88 | Growth factor | |
| Hs.151544 | G4075718 | SH2D1A | SH2 domain protein 1A, Duncan’s disease (lymphoproliferative syndrome) | 2.88 | Signal transduction | |
| Hs.9754 | G722754 | ATF5 | Activating transcription factor 5 | 2.70 | Transcription factor | |
| Hs.82890 | G2178057 | DAD1 | Defender against cell death 1 | 2.68 | Apoptosis inhibitor | |
| Hs.78225 | G1017878 | ANXA1 | Annexin A1 | 2.67 | Drug response | |
| Hs.149923 | G1406118 | XBP1 | X-box binding protein 1 | 2.66 | Transcription factor, stress response | |
| Hs.44396 | G3162290 | CORO2A | Coronin, actin-binding protein, 2A | 2.64 | Structural protein | |
| Hs.94360 | G1242830 | MT1L | Metallothionein 1L | 2.59 | Cell proliferation | |
| Hs.74647 | G2111484 | TRA@ | T cell receptor alpha locus | 2.59 | Immune response | |
| Hs.76272 | G2185876 | RBBP2 | Retinoblastoma-binding protein 2 | 2.42 | Cell cycle | |
| Hs.76917 | G2241018 | FBXO8 | F-box only protein 8 | 2.34 | Signal transduction | |
| Hs.799 | G768936 | DTR | Diphtheria toxin receptor (heparin-binding epidermal growth factor-like growth factor) | 2.32 | IFN pathway, cellular proliferation | |
| Hs.73793 | G774590 | VEGF | Vascular endothelial growth factor | 2.31 | IFN pathway, growth factor | |
| Hs.301956 | G4687355 | ZNF-U69274 | Zinc finger protein | 2.28 | Transcription factor | |
| Hs.343214 | G2177640 | Homo sapiens, clone MGC:19762 IMAGE:3636045, mRNA, complete cds | 2.22 | Unknown-EST | ||
| Hs.252587 | G4114487 | PTTG1 | Pituitary tumor-transforming 1 | 2.20 | Transcription factor | |
| Hs.274404 | G2167397 | PLAT | Plasminogen activator, tissue | 2.19 | Cell migration | |
| Hs.173936 | G993268 | IL10RB | Interleukin 10 receptor, beta | 2.17 | Immune response | |
| Hs.3446 | G774572 | MAP2K1 | Mitogen-activated protein kinase kinase 1 | 2.16 | IFN pathway, signal transduction | |
| Hs.169611 | G1388704 | SMAC | Second mitochondria-derived activator of caspase | 2.16 | Apoptosis regulator | |
| Hs.79070 | G2189484 | MYC | v-myc avian myelocytomatosis viral oncogene homolog | 2.14 | IFN pathway, protoncogene, apoptosis inducer | |
| Hs.80887 | G928713 | LYN | v-yes-1 Yamaguchi sarcoma viral related oncogene homolog | 2.1 | Oncogene | |
| Hs.2375 | G676421 | EMR1 | EGF-like module containing, mucin-like, hormone receptor-like sequence 1 | 2.08 | Hormone receptor | |
| Hs.179881 | G1775265 | CBFB | Core-binding factor, beta subunit | 2.06 | IFN pathway, transcription factor | |
| Hs.77768 | G2189901 | DNAJB2 | DnaJ (Hsp40) homolog, subfamily B, member 2 | 2.06 | IFN pathway, heat shock, stress response | |
| Hs.15259 | G3889125 | BAG3 | BCL2-associated athanogene 3 | 2.02 | Apoptosis inhibitor | |
| B. Genes upregulated in huT78R | ||||||
| Hs.281434 | g2229865 | Homo sapiens cDNA FLJ14028 fis, clone HEMBA1003838 | 2.12 | 0.47 | Unknown—EST | |
| Hs.99899 | g4084828 | TNFSF7 | Tumor necrosis factor (ligand) superfamily, member 7 | 2.22 | 0.45 | CD27 ligand, CD70, Hodgkin’s cells |
| Hs.75596 | g1549615 | IL2RB | Interleukin 2 receptor, beta | 2.32 | 0.43 | Signal transduction/immune response |
| Hs.184640 | g2837105 | C11orf9 | Chromosome 11 open reading frame 9 | 2.56 | 0.39 | Cell growth and/or maintenance |
| Hs.285754 | g2069697 | MET | met proto-oncogene (hepatocyte growth factor receptor) | 2.7 | 0.37 | Growth factor receptor, protoncogene |
| Hs.80395 | g1849138 | MAL | mal, T-cell differentiation protein | 5.5 | 0.18 | Signal transduction, lipid rafts |
Validation of Microarray Results
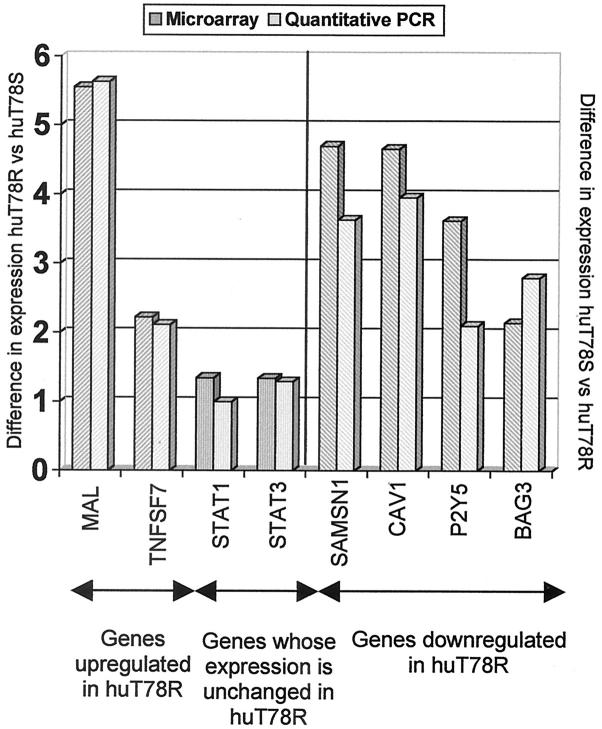
To confirm the differential expression observed in the cDNA microarray analysis, quantitative PCR was performed. In the case of five genes (MAL, BAG3, SAMSN1, STAT1, and STAT3), TaqMan probes were used and SYBR green was used in the case of three further genes (CAV1, TNFSF7, and P2Y5). The primers and probes used are shown in Table 1 ▶ . As shown in Figure 4 ▶ the changes in gene expression detected were similar using both techniques and although in some cases there is a slight difference between results, these differences are not significant in any case.
Figure 4.
Comparison of the results achieved using microarray analysis and quantitative PCR between the two variants. To validate the microarray result, quantitative PCR was performed for eight genes using TaqMan probes (five genes; MAL, SAMSN1, BAG3, STAT1, and STAT3) or SYBR green (three genes; TNFSF7, CAV1, and P2Y5). The genes studied included: ▨, genes up-regulated huT78R (MAL, SAMSN1), ▥, genes whose expression was unchanged between the two variants (STAT1, STAT3), and ▧, genes down-regulated in huT78R (BAG3, TNFSF7, CAV1, P2Y5). For all eight genes studied the microarray results were confirmed and in most cases the results obtained from both techniques were extremely similar and no differences observed were significant.
Expression of MAL Protein in Patient Samples: Correlation with Clinical Data
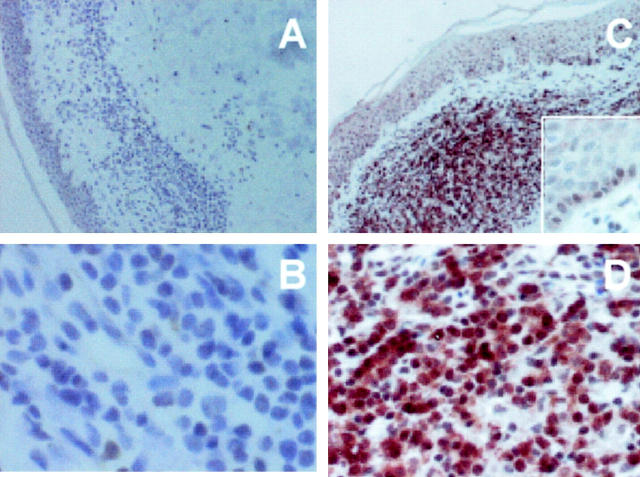
MAL protein expression was analyzed in the samples of 20 patient treated with PUVA and/or IFN-α and variability in expression was observed (Figure 5 ▶ and Table 3 ▶ ). The relationship between MAL expression and the response to therapy was examined using the Fisher test (Table 3) ▶ . Of the 10 patients that achieved complete remission within 16 weeks of treatment (rapid response), 70% do not express the MAL protein whereas 80% of the 10 patients who showed a slow response do express MAL protein. The association between MAL expression and absence of complete remission within 16 weeks was significant (Fisher’s exact test, P = 0.03) (Table 3A) ▶ .
Figure 5.
Immunohistochemical study of MAL protein in paraffin sections of MF tumoral tissue from patients treated with IFN-α. A and B: A case of MF in a patient in which MAL is not detected. B and D: A case of a MF in a patient positive for MAL and who shows a slow response in the case of IFN-α treatment.
Table 3.
MAL Protein Expression and Clinical Response
| MAL +ve | MAL −ve | Total | ||||
|---|---|---|---|---|---|---|
| A: Relationship between MAL protein expression and clinical response to IFN-α and/or PUVA (Fisher’s exact test, P = 0.03) | ||||||
| RR | 3 | 7 | 10 | |||
| SR | 8 | 2 | 10 | |||
| Total | 11 | 9 | 20 | |||
| B: Relationship between MAL protein expression and clinical response to IFN-α and PUVA (Fisher’s exact test, P = 0.04) | ||||||
| RR | 1 | 5 | 6 | |||
| SR | 4 | 0 | 4 | |||
| Total | 5 | 5 | 10 | |||
RR, Rapid response: patients that achieved complete remission within 16 weeks of treatment commencement; SR, slow response: patients that needed more than 16 weeks of treatment to achieve complete remission; MAL +ve, MAL = positive; MAL −ve, MAL = negative.
Taking into consideration exclusively the 10 patients treated with IFN-α + PUVA, 6 of them presented rapid response and the remaining, slow response. Among the rapid response patients, only 1 of 6 expressed MAL protein. All four patients with slow response were MAL-positive. Fisher’s test showed a statistically significant difference (P = 0.04) (Table 3B) ▶ .
Time-Course Experiments
A time-course experiment has been performed to identify genes expressed after IFN-α treatment in IFN-α-resistant and -sensitive cells using the CNIO OncoChip v.1.1. Details of the genes co-ordinately up- or down-regulated by IFN at the various times can be found in the supplementary information at http://bioinfo.cnio.es/data/interferon_resistance/. Results have been analyzed at 1 and 4 hours (early times) and 24 and 48 hours (late times). The SOTA cluster program organized the differentially regulated genes into four main clusters in each case (Figure 6) ▶ . Forty-nine early response genes differentially expressed have been identified, including mainly genes involved in signal transduction, drug metabolism, and cell-cycle control (Table 4) ▶ . Fifty-one late response differentially expressed genes have been observed, mainly composed by genes involved in cell growth and maintenance, signal transduction, transcription regulation, cell cycle, and apoptosis (Table 4) ▶ .
Figure 6.
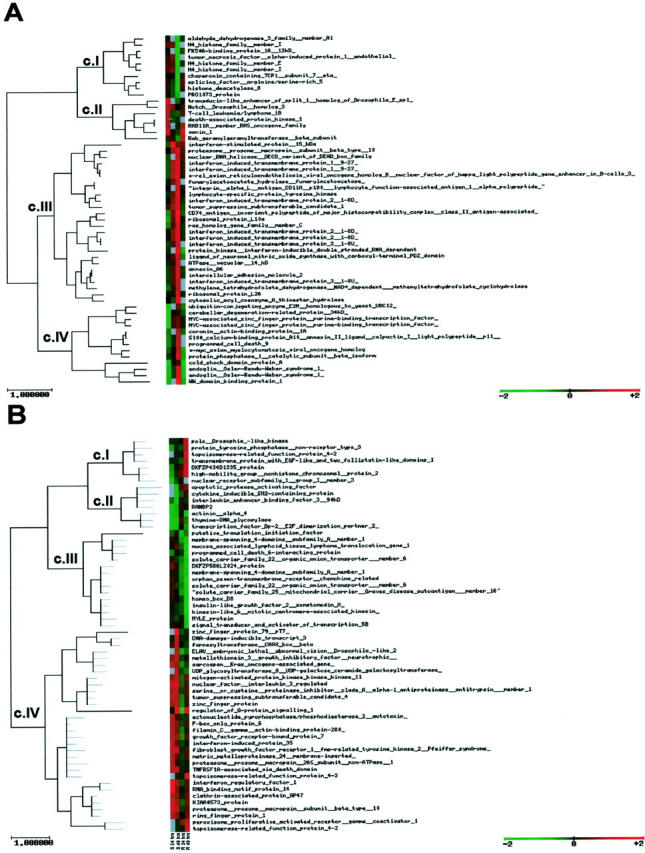
A: Cluster of IFN-α early response genes (1 and 4 hours) in huT78S and huT78R. CI: Genes down-regulated in huT78R, expression in huT78S does not change. CII-A: Genes up-regulated in huT78S, expression in huT78R does not change or is up-regulated but delayed. CIII: Genes up-regulated in huT78R, expression change in huT78S variable (unchanged, down-regulated or up-regulated but delayed). CIV: Genes down-regulated in huT78S and either up-regulated in huT78R and/or down-regulated but delayed. B: Cluster of IFN-α late response genes (24 and 48 hours) in huT78S and huT78R. CI: Genes up-regulated in the huT78R, expression in huT78S does not change or down-regulation is observed. CII: Genes down-regulated in huT78S, expression in huT78R does not change. CIII: Genes down-regulated in huT78R, expression in huT78S does not change. CIV: Genes up-regulated in huT78S and expression does not change in huT78R.
Table 4.
Function/Biological Process of the Differentially Regulated Genes Induced by IFN-α [Clusters (c) Defined by Figure 6 ▶ ]
| Function/biological process | Early | Late | Total | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| c. I | c. II | c. III | c. IV | Total | c. I | c. II | c. III | c. IV | Total | ||
| Signal transduction | 1 | 1 | 3 | 5 | 10 | 1 | 4 | 5 | 15 | ||
| Cell cycle | 4 | 3 | 1 | 8 | 1 | 1 | 2 | 2 | 6 | 14 | |
| Cell growth/maintenance | 3 | 1 | 4 | 2 | 3 | 7 | 12 | 16 | |||
| Developmental process | 1 | 1 | 1 | 2 | 3 | 4 | |||||
| IFN induced | 2 | 5 | 7 | 3 | 3 | 10 | |||||
| Transport | 1 | 1 | 1 | 1 | 1 | 3 | 4 | ||||
| Cell adhesion | 1 | 1 | 1 | ||||||||
| Apoptosis | 1 | 1 | 2 | 1 | 2 | 5 | 6 | ||||
| Metabolism | 3 | 5 | 1 | 9 | 1 | 2 | 3 | 12 | |||
| Transcription regulation | 2 | 2 | 4 | 1 | 3 | 4 | 8 | 12 | |||
| Immune response | 1 | 1 | 1 | 3 | 1 | 2 | 3 | 6 | |||
| Total | 9 | 8 | 21 | 11 | 49 | 8 | 6 | 10 | 27 | 51 | 100 |
Additionally we have performed analysis of each time point on an individual basis comparing genes differentially expressed after IFN treatment between huT78S and huT78R (see supplementary information at http://bioinfo.cnio.es/data/interferon_resistance/.). At 1 hour we have observed genes implicated in signal transduction, apoptosis, stress response, and oncogenes. At 4 hours we have found a very small number of genes, which are differentially regulated, most of which are implicated in IFN response. At 24 hours the differentially regulated genes are involved in a large variety of functions mainly signal transduction and cell-cycle control, apoptosis, stress response, immune response, and oncogenes. At 48 hours we have found that the majority of differentially regulated genes are implicated in signal transduction, IFN signaling, apoptosis, and cell-cycle control.
Discussion
The huT78 cell line used in these studies is a MF-derived cell line, in which initial studies using trypan blue exclusion to test cell viability indicated a high sensitivity to IFN-α treatment, even at an IFN-α concentrations as low as 100 U/ml. The culture of cells throughout a period of 9 months by gradually increasing the concentration of IFN-α in the growth medium has produced a cell line resistant to IFN-α concentrations of up to 1,000,000 U/ml. The cells were retested after 1 month in the absence of IFN-α and resistance was persistent. Thus this huT78R cell line was deemed to be a good model for the study of IFN-α resistance.
Previous studies have shown that IFN-α resistance could be dependent on alterations in the level of expression of STAT1 13 or defects in STAT3 activation 14 presumably secondary to the presence of undetected STAT1 null mutations or STAT3 active mutations. The data obtained in our experimental model do not confirm these results, because all of the expression analysis performed here (c-DNA microarray, quantitative PCR, and immunohistochemistry) show that the resistant cells preserve STAT1 and STAT3 expression. Similarly both cell lines retain STAT1 induction by IFN-α. Additionally both cell lines show similar activation of STAT1 and STAT3 by IFN-α as shown by the nuclear expression of STAT1 and STAT3, which has been demonstrated to be a necessary step for STAT activity, secondary to phosphorylation. Thus, instead of unique alterations in genes closely related with the IFN receptor pathway, our results point toward the implication of a relatively large number of genes involved in different cell pathways as responsible for IFN-α resistance.
To gain a better understanding of the intrinsic factors that may give rise to IFN-α resistance, we used the CNIO OncoChip to look at differences in gene expression between the two variants of huT78. c-DNA microarray experiments identified that in the huT78 cell line the resistance to IFN-α is associated with significant changes in the expression of a total of 39 genes. Thus, six genes or ESTs (MAL, MET, C11ORF9, IL2RB, TNFSF7, and 1 EST) are expressed at a higher level by the IFN-α resistant cells, whereas the expression of a set of 33 genes and ESTs is down-regulated in the resistant cell line. The most significant of these genes were HSPA6, SAMSN1, CAV1, P2Y5, EPB41L3, ARL5, and SH2D1A. Additionally the c-DNA microarray study confirmed that STAT1 and STAT3 basal levels are not significantly different between the two variants.
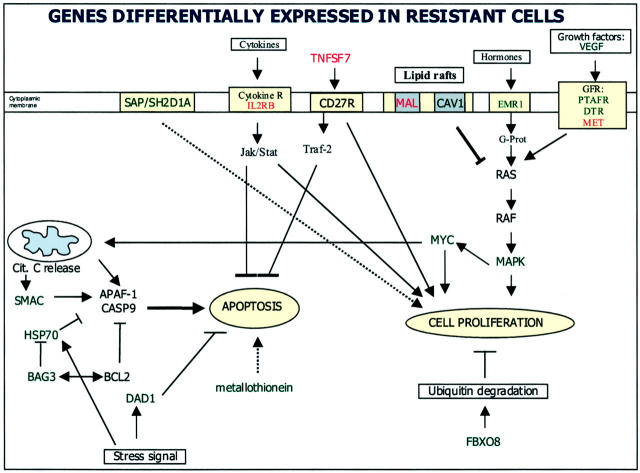
The genes identified as being implicated in intrinsic resistance of cells to IFN-α code for proteins involved in a variety of functions including membrane transport and signal transduction (MAL, CAV1, ARL5, SAMSN1, IL2RB, MAP2K1, SH2D1A), apoptosis (BAG3, SMAC, DAD1), stress-response (HSPA6, DNAJB2), transcription regulation (ATF5, PTTG1, CBFB), and cell growth (MET, VEGF, TNFSF7), or play a role as multifunctional genes (MYC). Notably all of the genes overexpressed by the resistant code for membrane proteins, all of them having been described as being involved in signal transduction and cell growth.
The functional significance of these changes, and their relationship with the response to IFN-α can be partially explained taking into account the existing information concerning the function of all these genes. Thus, although some of the genes identified by this strategy appear to belong to the IFN pathway (P2Y5, DTR, VEGF) the presence of the majority of them can only be explained taking into account the relevant role that they play in the control of key gene functions, such as signal transduction, cell growth, or apoptosis.
MAL, the gene up-regulated in resistant cells to the highest degree, is a component of membrane rafts. 26 These rafts are membrane microdomains that play a central role in signal transduction acting as a scaffold in which molecules of signal transduction pathways can interact. Indeed ∼10% of STAT3, and other STAT molecules such as STAT1 and STAT5, are found in such membrane rafts. 27 Membrane rafts, in addition to STATs and other signaling molecules, contain membrane-bound receptors such as gp130 and IFN receptors (IFNAR1 and IFNGR2) in the case of caveolin-containing rafts. 28 A potential role for MAL protein in IFN-α and PUVA resistance has indeed been identified in the group of patients analyzed here, thus underscoring the critical role that this protein seems to play in signal transduction, and extending the results of the experimental model chosen to a group of patients.
CAV1, a caveolin underexpressed by IFN-resistant cells, is also a membrane raft protein. However MAL and CAV1 are found in distinct membrane microdomains/rafts, 29 thus suggesting that they may have specialized functions. CAV1 is a tumor suppressor gene, localized on chromosome 7q31, for which a role as a negative regulator of the Ras-p42/44 MAP kinase cascade has been proposed (Figure 7) ▶ . The down-regulation of caveolin in the resistant cell line suggests that these caveolin-containing membrane rafts may be essential for IFN signaling and particularly for IFN-α signaling because it is known that these rafts do contain IFN receptors. 28 The up-regulation of MAL in the resistant cells suggests that, in the absence or depletion of CAV1, MAL-containing membrane rafts become more important for maintaining normal membrane trafficking and signaling.
Figure 7.
Apoptotic and signal transduction pathways disrupted in the IFN-α resistant cell line detected by microarray analysis. Expression of several genes acting in concert to regulate the balance between proliferation and programmed cell death is altered in the resistance variant (hut78R) as compared to the sensitive variant (hut78S). Up-regulated (red text label) and down-regulated genes (green text label) in huT78R versus huT78S are noted. Changes in the expression of several genes could possibly be the cause of the resistance phenotype (as the up-regulation of TNFSF7 and MAL and the down-regulation of SHD2H1A and CAV1) although some others may be a consequence.
Interleukin2 receptor B is another of the genes that is up-regulated in resistant cells. The receptor for interleukin 2 is composed of three subunits α, β, and γ. The β subunit (shown here to be overexpressed by huT78R) of the interleukin receptor is critical for receptor-mediated signaling leading to the proliferative and survival effect of interleukin activation. 30 Hence the resistant cells, which overexpress this subunit, may be more responsive to interleukin-induced proliferation and survival giving them a survival advantage over sensitive cells.
The SH2D1A gene (also known as SAP, DSHP, and XLP) encodes a small protein consisting of a single SH2 domain. In vitro and in vivo analysis of gene expression suggest that this gene is an intrinsic component of lymphocyte activation pathways being present in activated lymphocytes and high-grade lymphomas. 31 On the other hand, deletions and nonsense mutations have been described in SAP/SH2D1A in XLP patients 32 and those patients present severe EBV-associated illness. 33 Although the function of this protein is not completely characterized, the absence of mRNA in the resistance cell line suggests a possible role in the resistance phenotype. However this gene is found to be differentially regulated in response to IFN treatment as shown by the time-course experiments (1 hour), which raises some doubt about the role that this gene may play in the resistance to IFN-α.
MET, a transforming gene up-regulated in the resistant cells codes for a membrane protein containing a SH2 domain. It has features characteristic of the tyrosine kinase family of growth factor receptors. It has been shown to be capable of activating some signal transduction pathways including the RAS/MAPK pathway. Additionally the MET gene is activated by IFN-γ and other cytokines, and can also activate STAT proteins. 34
TNFSF7 (tumor necrosis factor ligand superfamily member 7) (cd27 ligand, CD70) is a surface antigen found on activated, but not resting, B and T cells. This gene has been found to be up-regulated in resistant cells. It induces cell proliferation 35 and it has been described as an initiator of anti-apoptotic signaling via TRAF proteins, 36 although a description as an apoptotic inducer molecule has also been described (Figure 7) ▶ . 37
Heat shock protein 70B (HSPA6), a gene regulated via JAK/STAT signaling, is the most down-regulated gene in resistant cells. HSP70 has binding sites in its promoter that can bind both STAT1 and STAT3, and is known to be induced by IFN-γ in a STAT1-dependent manner. 38 The down-regulation of this gene in resistant cells could be derived from the loss of some upstream regulators in these cells. A relation between HSP70 expression and apoptosis sensitivity has been previously provided by the findings of Gerner and colleagues, 39 in which HSP70 expression was associated with apoptosis dependent on fas-signaling (Figure 7) ▶ .
The time-course experiments reveal that indeed the different behavior exhibited by the resistant and sensitive cell line seems to be the result of deregulation of cell-cycle control, cell growth and maintenance, and signal transduction pathways (Table 4) ▶ . Although the group of apoptosis genes differentially expressed is low 8 (8 genes) they could be critical in the process of resistance to IFN-α.
One interesting gene, which is differentially regulated between the sensitive and resistant variants, is the oncogene MYC. This gene is up-regulated in huT78R in response to IFN-α whereas expression in huT78S appears to be down-regulated. In fact this gene is known to be regulated by IFN. 8 Conflicting data with that observed in the analysis of basal levels of c-myc expression in huT78S and huT78R possibly reflects the pleiotrophic effects of c-myc on cell growth, proliferation, and control of apoptosis. 40 Another gene MAZ [MYC-associated zinc finger protein (purine-binding transcription factor)], is also found in the same cluster as MYC. It plays an important role in transcription initiation and binds two sites in the MYC promoter region. 41 Up-regulation of both genes in huT78R indicates that transcription of MYC is active in huT78R while being suppressed in huT78S giving the resistant cells a growth advantage over their sensitive counterparts, because MYC is responsible for the transcription of genes involved in cell proliferation and growth. 40
Another gene that could be implicated in the resistance is RANBP2 (RAN-binding protein 2), which plays an important role in cell-cycle progression. The down-regulation detected in huT78S may imply that cells are not able to progress through the cell cycle while in huT78R, because no down-regulation is observed, cell-cycle progression continues. Another potential gene involved in apoptosis sensitivity in huT78S cells is TRADD (TNFRSF1A-associated via death domain) which encodes a proapoptotic death receptor. 42 The lack of induction of this gene in huT78R may help explain the resistance observed.
By examining the differences in the basal expression of genes between huT78S and huT78R the factors responsible for the intrinsic resistance of huT78R to the effects of IFN can be identified, whereas the time-course experiments that we have performed, illustrate the downstream signal transduction pathway deregulation caused by these basal changes. Thus the changes observed here in the signal transduction pathways after IFN-α challenge could be secondary to the malfunction of the membrane raft microdomains, linked with the abnormal expression of MAL and CAV1, as detected in the basal cell lines.
In summary, these results show that resistance to IFN-α in CTCL cells is dependent on changes in the expression of a selected number of genes, including genes implicated in signal transduction, cell cycle, cell growth, and apoptosis. The most up-regulated gene, MAL, was found to be a prediction factor for treatment outcome of IFN-α in a clinical series. Nevertheless it seems plausible that to fully explain the clinical phenomena of IFN-α resistance, a larger number of genes need to be taken into account.
Acknowledgments
We thank I. Fernandez for her invaluable help with the cell culture, M. Lopez for excellent technical assistance with molecular assays, the Immunohistochemistry Unit (CN10) for assistance afforded for immunohistochemical assays, and Andreas Rosenwald for his comments and discussion on the time-course experiments.
Footnotes
Address reprint requests to Dr. Miguel A. Piris, Programa de Patología Molecular, Centro Nacional de Investigaciones Oncológicas, Melchor Fernández Almagro, 3, Madrid 28029 Spain. E-mail: mapiris@cnio.es.
Supported by the Ministerio de Ciencia y Tecnología (grants 1FD97-0431, BIO2000-0275-C02/01 and/02) and the Centro Nacional de Investigaciones Oncológicas (to L. T. and J. H.).
References
- 1.Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD: How cells respond to interferons. Annu Rev Biochem 1998, 67:227-264 [DOI] [PubMed] [Google Scholar]
- 2.Platanias LC: Interferons: laboratory to clinic investigations. Curr Opin Oncol 1995, 7:560-565 [PubMed] [Google Scholar]
- 3.Ihle JN, Witthuhn BA, Quelle FW, Yamamoto K, Thierfelder WE, Kreider B, Silvennoinen O: Signaling by the cytokine receptor superfamily: JAKs and STATs. Trends Biochem Sci 1994, 19:222-227 [DOI] [PubMed] [Google Scholar]
- 4.Darnell JR JE, Kerr IM, Stark GR: Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 1994, 264:1415-1421 [DOI] [PubMed] [Google Scholar]
- 5.Sangfelt O, Erickson S, Castro J, Heiden T, Einhorn S, Grander D: Induction of apoptosis and inhibition of cell growth are independent responses to interferon-alpha in hematopoietic cell lines. Cell Growth Differ 1997, 8:343-352 [PubMed] [Google Scholar]
- 6.Xu D, Erickson S, Szeps M, Gruber A, Sangfelt O, Einhorn S, Pisa P, Grander D: Interferon alpha down-regulates telomerase reverse transcriptase and telomerase activity in human malignant and nonmalignant hematopoietic cells. Blood 2000, 96:4313-4318 [PubMed] [Google Scholar]
- 7.Jewell AP, Worman CP, Lydyard PM, Yong KL, Giles FJ, Goldstone AH: Interferon-alpha up-regulates bcl-2 expression and protects B-CLL cells from apoptosis in vitro and in vivo. Br J Haematol 1994, 88:268-274 [DOI] [PubMed] [Google Scholar]
- 8.Matikainen S, Sareneva T, Ronni T, Lehtonen A, Koskinen PJ, Julkunen I: Interferon-alpha activates multiple STAT proteins and upregulates proliferation-associated IL-2Ralpha, c-myc, and pim-1 genes in human T cells. Blood 1999, 93:1980-1991 [PubMed] [Google Scholar]
- 9.Sangfelt O, Erickson S, Castro J, Heiden T, Gustafsson A, Einhorn S, Grander D: Molecular mechanisms underlying interferon-alpha-induced G0/G1 arrest: CKI-mediated regulation of G1 Cdk-complexes and activation of pocket proteins. Oncogene 1999, 18:2798-2810 [DOI] [PubMed] [Google Scholar]
- 10.Abramovich C, Shulman LM, Ratovitski E, Harroch S, Tovey M, Eid P, Revel M: Differential tyrosine phosphorylation of the IFNAR chain of the type I interferon receptor and of an associated surface protein in response to IFN-alpha and IFN-beta. EMBO J 1994, 13:5871-5877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McKendry R, John J, Flavell D, Muller M, Kerr IM, Stark GR: High-frequency mutagenesis of human cells and characterization of a mutant unresponsive to both alpha and gamma interferons. Proc Natl Acad Sci USA 1991, 88:11455-11459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sakamoto H, Yasukawa H, Masuhara M, Tanimura S, Sasaki A, Yuge K, Ohtsubo M, Ohtsuka A, Fujita T, Ohta T, Furukawa Y, Iwase S, Yamada H, Yoshimura A: A Janus kinase inhibitor, JAB, is an interferon-gamma-inducible gene and confers resistance to interferons. Blood 1998, 92:1668-1676 [PubMed] [Google Scholar]
- 13.Sun WH, Pabon C, Alsayed Y, Huang PP, Jandeska S, Uddin S, Platanias LC, Rosen ST: Interferon-alpha resistance in a cutaneous T-cell lymphoma cell line is associated with lack of STAT1 expression. Blood 1998, 91:570-576 [PubMed] [Google Scholar]
- 14.Yang CH, Murti A, Pfeffer LM: STAT3 complements defects in an interferon-resistant cell line: evidence for an essential role for STAT3 in interferon signaling and biological activities. Proc Natl Acad Sci USA 1998, 95:5568-5572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siegel RS, Pandolfino T, Guitart J, Rosen S, Kuzel TM: Primary cutaneous T-cell lymphoma: review and current concepts. J Clin Oncol 2000, 18:2908-2925 [DOI] [PubMed] [Google Scholar]
- 16.Zhang L, Zhou W, Velculescu VE, Kern SE, Hruban RH, Hamilton SR, Vogelstein B, Kinzler KW: Gene expression profiles in normal and cancer cells. Science 1997, 276:1268-1272 [DOI] [PubMed] [Google Scholar]
- 17.DeRisi J, Penland L, Brown PO, Bittner ML, Meltzer PS, Ray M, Chen Y, Su YA, Trent JM: Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nat Genet 1996, 14:457-460 [DOI] [PubMed] [Google Scholar]
- 18.Iyer VR, Eisen MB, Ross DT, Schuler G, Moore T, Lee JC, Trent JM, Staudt LM, Hudson J, Jr, Boguski MS, Lashkari D, Shalon D, Botstein D, Brown PO: The transcriptional program in the response of human fibroblasts to serum. Science 1999, 283:83-87 [DOI] [PubMed] [Google Scholar]
- 19.Heid CA, Stevens J, Livak KJ, Williams PM: Real time quantitative PCR. Genome Res 1996, 6:986-994 [DOI] [PubMed] [Google Scholar]
- 20.Van Gelder RN, von Zastrow ME, Yool A, Dement WC, Barchas JD, Eberwine JH: Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proc Natl Acad Sci USA 1990, 87:1663-1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eberwine J: Amplification of mRNA populations using aRNA generated from immobilized oligo(dT)-T7 primed cDNA. Biotechniques 1996, 20:584-591 [DOI] [PubMed] [Google Scholar]
- 22.Hegde P, Qi R, Abernathy K, Gay C, Dharap S, Gaspard R, Hughes JE, Snesrud E, Lee N, Quackenbush J: A concise guide to cDNA microarray analysis. Biotechniques 2000, 29:548-556 [DOI] [PubMed] [Google Scholar]
- 23.Rebhan M, Chalifa-Caspi V, Prilusky J, Lancet D: GeneCards: a novel functional genomics compendium with automated data mining and query reformulation support. Bioinformatics 1998, 14:656-664 [DOI] [PubMed] [Google Scholar]
- 24.Copie-Bergman C, Gaulard P, Maouche-Chretien L, Briere J, Haioun C, Alonso MA, Romeo PH, Leroy K: The MAL gene is expressed in primary mediastinal large B-cell lymphoma. Blood 1999, 94:3567-3575 [PubMed] [Google Scholar]
- 25.Gazdar AF, Carney DN, Bunn PA, Russell EK, Jaffe ES, Schechter GP, Guccion JG: Mitogen requirements for the in vitro propagation of cutaneous T-cell lymphomas. Blood 1980, 55:409-417 [PubMed] [Google Scholar]
- 26.Alonso MA, Millan J: The role of lipid rafts in signalling and membrane trafficking in T lymphocytes. J Cell Sci 2001, 114:3957-3965 [DOI] [PubMed] [Google Scholar]
- 27.Ndubuisi MI, Guo GG, Fried VA, Etlinger JD, Sehgal PB: Cellular physiology of STAT3: where’s the cytoplasmic monomer? J Biol Chem 1999, 274:25499-25509 [DOI] [PubMed] [Google Scholar]
- 28.Takaoka A, Mitani Y, Suemori H, Sato M, Yokochi T, Noguchi S, Tanaka N, Taniguchi T: Cross talk between interferon-gamma and -alpha/beta signaling components in caveolar membrane domains. Science 2000, 288:2357-2360 [DOI] [PubMed] [Google Scholar]
- 29.Millan J, Puertollano R, Fan L, Alonso MA: Caveolin and MAL, two protein components of internal detergent-insoluble membranes, are in distinct lipid microenvironments in MDCK cells. Biochem Biophys Res Commun 1997, 233:707-712 [DOI] [PubMed] [Google Scholar]
- 30.Zamorano J, Wang HY, Wang R, Shi Y, Longmore GD, Keegan AD: Regulation of cell growth by IL-2: role of STAT5 in protection from apoptosis but not in cell cycle progression. J Immunol 1998, 160:3502-3512 [PubMed] [Google Scholar]
- 31.Nichols KE, Harkin DP, Levitz S, Krainer M, Kolquist KA, Genovese C, Bernard A, Ferguson M, Zuo L, Snyder E, Buckler AJ, Wise C, Ashley J, Lovett M, Valentine MB, Look AT, Gerald W, Housman DE, Haber DA: Inactivating mutations in an SH2 domain-encoding gene in X-linked lymphoproliferative syndrome. Proc Natl Acad Sci USA 1998, 95:13765-13770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sumegi J, Huang D, Lanyi A, Davis JD, Seemayer TA, Maeda A, Klein G, Seri M, Wakiguchi H, Purtilo DT, Gross TG: Correlation of mutations of the SH2D1A gene and Epstein-Barr virus infection with clinical phenotype and outcome in X-linked lymphoproliferative disease. Blood 2000, 96:3118-3125 [PubMed] [Google Scholar]
- 33.Sumazaki R, Kanegane H, Osaki M, Fukushima T, Tsuchida M, Matsukura H, Shinozaki K, Kimura H, Matsui A, Miyawaki T: SH2D1A mutations in Japanese males with severe Epstein-Barr virus-associated illnesses. Blood 2001, 98:1268-1270 [DOI] [PubMed] [Google Scholar]
- 34.Nagahori T, Dohi M, Matsumoto K, Saitoh K, Honda ZI, Nakamura T, Yamamoto K: Interferon-gamma upregulates the c-Met/hepatocyte growth factor receptor expression in alveolar epithelial cells. Am J Respir Cell Mol Biol 1999, 21:490-497 [DOI] [PubMed] [Google Scholar]
- 35.Goodwin RG, Alderson MR, Smith CA, Armitage RJ, VandenBos T, Jerzy R, Tough TW, Schoenborn MA, Davis-Smith T, Hennen K: Molecular and biological characterization of a ligand for CD27 defines a new family of cytokines with homology to tumor necrosis factor. Cell 1993, 73:447-456 [DOI] [PubMed] [Google Scholar]
- 36.Gravestein LA, Amsen D, Boes M, Calvo CR, Kruisbeek AM, Borst J: The TNF receptor family member CD27 signals to Jun N-terminal kinase via Traf-2. Eur J Immunol 1998, 28:2208-2216 [DOI] [PubMed] [Google Scholar]
- 37.Prasad KV, Ao Z, Yoon Y, Wu MX, Rizk M, Jacquot S, Schlossman SF: CD27, a member of the tumor necrosis factor receptor family, induces apoptosis and binds to Siva, a proapoptotic protein. Proc Natl Acad Sci USA 1997, 94:6346-6351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stephanou A, Isenberg DA, Nakajima K, Latchman DS: Signal transducer and activator of transcription-1 and heat shock factor-1 interact and activate the transcription of the Hsp-70 and Hsp-90beta gene promoters. J Biol Chem 1999, 274:1723-1728 [DOI] [PubMed] [Google Scholar]
- 39.Gerner C, Frohwein U, Gotzmann J, Bayer E, Gelbmann D, Bursch W, Schulte-Hermann R: The Fas-induced apoptosis analysed by high throughput proteome analysis. J Biol Chem 2000, 275:39018-39026 [DOI] [PubMed] [Google Scholar]
- 40.Amati B, Alevizopoulos K, Vlach J: Myc and the cell cycle. Front Biosci 1998, 3:D250-D268 [DOI] [PubMed] [Google Scholar]
- 41.Bossone SA, Asselin C, Patel AJ, Marcu KB: MAZ, a zinc finger protein, binds to c-MYC and C2 gene sequences regulating transcriptional initiation and termination. Proc Natl Acad Sci USA 1992, 89:7452-7456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hsu H, Xiong J, Goeddel DV: The TNF receptor 1-associated protein TRADD signals cell death and NF-kappa B activation. Cell 1995, 81:495-504 [DOI] [PubMed] [Google Scholar]