Abstract
Several lines of evidences suggest that T cell/histiocyte-rich B-cell lymphoma (T/HRBCL) represents an aggressive variant of the clinically indolent entity nodular lymphocyte predominance Hodgkin’s lymphoma (LPHL). Still, this view has not yet been supported by firm genetic evidence. In this study, we analyzed 17 T/HRBCL cases using comparative genomic hybridization (CGH) combined with microdissection of single CD20+ neoplastic cells and DNA amplification by degenerate oligonucleotide primed-polymerase chain reaction, an approach we previously used in LPHL. Genomic imbalances were detected in all cases (in total, 80 changes). The most common imbalances included gain of Xq, 4q13q28, Xp21p11, and 18q21, and loss of 17p. Of note, a partial gain of 4q, a rare change in lymphoma, is also among the genomic imbalances most frequently encountered in LPHL. On the other hand, the CGH profiles of T/HRBCL and LPHL showed several distinct features, in particular with respect to the number of genomic imbalances (average of 4.7 in T/HRBCL versus 10.8 in LPHL) and their distribution (usually 1 to 5 in T/HRBCL versus 6 to 22 in LPHL). Altogether, our CGH findings of shared as well as distinctive cytogenetic features in both diseases suggest that T/HRBCL constitutes a separate lymphoma entity, possibly originating from the same precursor cell as LPHL.
T-cell/histiocyte-rich B-cell lymphoma (T/HRBCL) originally described by Delabie and colleagues, 1 is a controversial diffuse large B-cell lymphoma subtype of which the clinicopathological distinctiveness was recently demonstrated. 2,3 The disease typically affects middle-aged male patients who present with advanced-stage disease that is not adequately managed with current therapeutic strategies. Interestingly, both from a morphological and from an immunophenotypic perspective, T/HRBCL shows some resemblance to lymphocyte predominance Hodgkin’s lymphoma (LPHL), paragranuloma type. 1,2 In addition, both disorders are characterized by a striking male preponderance and may progress to a diffuse large B-cell lymphoma (DLBCL). 3 Based on these data it has been speculated that T/HRBCL merely represents a transformed LPHL, the more so as T/HRBCL patients tend to be slightly older on average than LPHL patients. Moreover occasionally, the occurrence of a T/HRBCL after an initial diagnosis of LPHL has been reported, 4 although it is uncertain whether these cases truly answer to the entity.
Notwithstanding the aforementioned similarities between T/HRBCL and LPHL, several noticeable differences usually do enable one to distinguish between T/HRBCL and LPHL. More convincingly, the completely distinct biological behavior of both disorders argues against a straightforward relationship.
A promising approach to resolving the controversy surrounding the presumed relationship between T/HRBCLand LPHL is to compare the overall cytogenetic profile characterizing both disorders. However, thus far, cytogenetic studies of both lymphomas have remained scarce as they are hampered by intrinsic technical obstacles in obtaining a sufficient number of evaluable karyotypes. This problem can be overcome by applying comparative genomic hybridization (CGH), a molecular cytogenetic approach that provides an overview of chromosomal imbalances in tumor cells without the need for metaphase chromosomes. 5 Even in tumors that are characterized by a paucity of neoplastic cells, this technique is effective when combined with microdissection of single neoplastic cells or cell clusters and subsequent DNA amplification by degenerate oligonucleotide-primed polymerase chain reaction (DOP-PCR) to ensure the availability of sufficient genetic material to perform CGH. Recently, the utility of the DOP-PCR-CGH approach has been unequivocally demonstrated by its successful application in Hodgkin’s lymphoma research. Whereas other groups studied the pattern of chromosomal imbalances characteristic of classical Hodgkin’s lymphoma, 6-8 our group has focused on LPHL. Not only did we demonstrate that it is feasible to detect DNA copy number changes in pools of four to five microdissected popcorn cells with high sensitivity and specificity, but we were also able to show that LPHL is characterized by the occurrence of consistent genomic alterations. 9 In the present study we applied the same molecular cytogenetic approach to investigate a pattern of chromosomal aberrations in T/HRBCL. Seventeen well-documented T/HRBCL cases were analyzed using DOP-PCR-CGH and the obtained CGH profiles were compared with those previously determined for LPHL.
Materials and Methods
Patients
Seventeen well-documented T/HRBCL cases with available frozen material were collected for CGH studies from the files of the Department of Pathology, University Hospitals, K.U. Leuven, Belgium.
The histological diagnosis was based on criteria that were established and validated in an earlier study: 2,3 1) a diffuse or vaguely nodular pattern of the neoplastic proliferation, 2) presence of scattered large atypical lymphoid cells that are of B-cell phenotype, do not express CD15, constitute only a minority of the overall cell population and occur singly or in very small clusters, 3) predominance of a reactive background infiltrate, composed of both T cells and nonepithelioid histiocytes, and 4) minimal presence of small B cells in the neoplastic areas. All cases but one (case 5) were included in this previously published series. 2,3 Lymph node or splenic biopsies taken either at the time of diagnosis (10 cases) or at the time of relapse (6 cases) were available for the study.
Microdissection, DOP-PCR, CGH
CGH analysis was performed according to the previously described protocol. 9 Briefly, frozen sections of 8 μm were cut, stained with CD20 (L26) antibody (DAKO, Glostrup, Denmark), and immunodetected by the avidin-biotin-peroxidase complex approach (Dakopatts, Glostrup, Denmark). Slides were pretreated with collagenase H (type II; σ-Aldrich, St. Louis, MO). Single CD20-positive cells were microdissected and harvested under a microscope (Leica, Wetzlar, Germany). Four to five microdissected cells were collected in a PCR tube and resuspended in proteinase K solution for digestion.
Tumor DNA and normal DNA were universally amplified by DOP-PCR. Before CGH the DOP-PCR products were purified, size-checked on a gel, and labeled with direct fluorochrome-conjugated nucleotides by nick-translation using fluorescein-12-dUTP (tumor) and lissamine-5-dUTP (normal) (NEN Life Science Products, Boston, MA). Equal amounts (1 μg) of labeled tumor and normal DNA were co-hybridized to slides with normal metaphase chromosomes. After 3 days of hybridization the specimens were washed and counterstained with 4,6-diamidino-2-phenylindole (blue) for chromosome identification. The image analysis was performed on an epifluorescence microscope (Leica DMRB, Wetzlar, Germany) with image analysis software (QUIPS; Vysis, Downers Grove, IL). The ratio of fluorescein/lissamine fluorescence intensities was calculated along each individual chromosome. Heterochromatic regions and the whole chromosome Y were excluded from analysis because of the abundance of high repetitive sequences. Chromosomes 19 and 22 results were excluded from the final calculation because they are known to be critical in CGH. In our control experiments, however, we did not observe abnormalities of chromosomes 19 and 22 and therefore imbalances affecting these chromosomes found in some of our patients are shown in brackets. By direct labeling ratios greater than 1.15 and less than 0.85 were considered to represent chromosomal gains and losses, respectively. Data are presented in accordance with the International System for Human Cytogenetic Nomenclature (ISCN) nomenclature. 10
Fluorescence in Situ Hybridization (FISH)
To confirm CGH results two-step CD20/FISH analysis was performed as described previously. 9 Initially, CD20-immunostained frozen sections from two cases were reviewed and several CD20+ large atypical cells were captured and positioned. Subsequently, specimens were subjected to FISH and re-evaluated. Applied probes included the α-satellite probe for chromosome 11 (Vysis) and LSI p53 for 17p13.1 (Vysis).
Results
A detailed description of the clinicopathological features of 16 of 17 analyzed T/HRBCL cases has been reported previously; 2,3 some of the most relevant clinical data are summarized in Table 1 ▶ . Of note, only two of four patients who obtained a complete remission after initial treatment had a continuous complete remission (cases 7 and 10). The remaining two patients (cases 1 and 3) were rescued with peripheral stem cell transplantation. Two patients who had a partial response after standard chemotherapy achieved a continuous complete remission after peripheral stem-cell transplantation or autologous bone marrow transplantation (cases 8 and 16).
Table 1.
Clinical and CGH Features of 17 Patients with T/HRBCL
| Case no. | Sex/age* | Status† | Stage‡ | Follow-up | Os§ | CGH∥ Rev ish |
|---|---|---|---|---|---|---|
| 1 | M /53 | R | IA | CR | 112 | enh(11p11,14q11q24,Xq12q13) |
| 2 | F /46 | R | IVA | Died | 45 | enh(1q,2p16q37,3,4q13q31,5p14q32,6p25p22,7p21q22,10p15p13,13q,15q15q26),dim(4p16p15.3,8p23q21.3,12p,Xp) |
| 3 | F /16 | R | IIB | CR | 89 | enh(13q14q21,18q21),dim(17p,X) |
| 4 | M /59 | D | IVB | Died | 1 | enh(1q21q31,2p16p13,2q31q33,5p15q23,7p15p11,7q31,11q14q25,12q14q21,15q21q26,18q12q21,Xp21q28),dim(10p15q22,11p15p13,14q31q32,16,17p13q12) |
| 5 | F /75 | NA | NA | NA | NA | dim(6p25p23,8p23p21,Xp22q12,Xq21q28) |
| 6 | F /73 | D | IIIA | Died | 31 | enh(4q22q26,5q14q21) |
| 7 | M /30 | D | IIIA | CR | 34 | enh(12q12,Xq),dim(17) |
| 8 | M /31 | R | IVA | PR | 30 | enh(6p25p22,9q22q33,13q21q22,Xq12q22) |
| 9 | M /44 | D | IIIB | Died | 4 | enh(4p15q32) |
| 10 | M /31 | D | IVB | CR | 17 | enh(2q31q32,4q21q28,12p13,18p,20q12q13) |
| 11 | M /35 | D | IVB | Died | 12 | enh(Xq),dim(17p13q21) |
| 12 | M /42 | D | IIIB | Died | 14 | enh(9p21p13),dim(16q12q13) |
| 13 | M /48 | D | IVB | Died | 7 | enh(6q25q27,8q24,18q21q23,Xp22q21) |
| 14 | M /41 | R | IVB | Died | 25 | enh(1q24q25,3p22p24,4q25q27,9q21q22,14q13q24,18q12q21,X) |
| 15 | M /41 | D | IVB | Died | 10 | enh(4,6q24q25,Xp21q28) |
| 16 | M /55 | R | IVB | PR | 59 | enh(4q13q26,5p15.1p14,5q32q33,11p14,Xp22.1q24) |
| 17 | M /46 | D | IVB | Died | 8 | enh(Xq12q13) |
R, relapse; D, primary diagnosis; CR, complete remission; PR, partial remission; NA, not available.
*Age at time of primary diagnosis.
†Status at time of lymph node biopsy.
‡Clinical stage according to the Ann Arbor classification.
§Overall survival in months from time of diagnosis.
∥Described according to the ISCN nomenclature.
CGH studies were performed in all 17 cases. The results are summarized in Table 1 ▶ and in Figure 1 ▶ . Chromosomes 19 and 22 were finally excluded from the CGH analysis because of possible hybridization artifacts (discussed in Materials and Methods). Genomic imbalances, 80 in total, were found in all cases. Gains of genomic material were more frequently detected than losses (3.6 versus 1.0). The number of genomic imbalances ranged from 1 to 16 per case with an average of 4.7 chromosomal changes per tumor. The majority of cases (n = 14) displayed one to five imbalances, with two cases (cases 9 and 17) exhibiting single CGH aberrations (enh4p15q32 and enhXq12q13, respectively). In the remaining three cases (cases 2, 4 and 14) 7, 14 and 16 chromosomal changes were observed.
Figure 1.
Summary of CGH results obtained in 17 cases of T/HRBCL. Chromosomal gains are shown on the right of the ideogram and losses on the left. The numbers on top of each line refer to the case number (Table 1) ▶ . Chromosomal imbalances of critical regions of chromosomes 19 and 22 excluded from final calculation are shown here in brackets.
The specific pattern of gains and losses was as follows: 10 cases (cases 1, 6, 8, 9, 10, 13, 14, 15, 16, and 17) showed only gain of chromosomal material, three cases (cases 2, 4, and 7) had more gains than losses, in three cases (cases 3, 11, and 12) the number of gains and losses was equal, and in one case (case 5) only losses were observed. With the exception of chromosome 21, all chromosomes were affected (19, 22, and Y were excluded from analysis). Genetic material of 15 chromosomes (complete or partial), including 1q, 2, 3p, 4, 5, 6, 7p, 9q, 12q, 13q, 14q, 15q, 18q, and X, was recurrently gained but high-level amplifications were not detected. Distinct chromosomal regions of 8p, 16q, 17, and X were recurrently lost.
The following imbalances were found in at least four cases (23.5%): gain of Xq (10 cases), 4q (seven cases), Xp (five cases), and 18q (four cases) with the minimal overlapping region at Xq12q13, 4q25q26, Xp21p11, and 18q21, and a loss of 17p (four cases). The four most frequent changes as such comprised enhX, enh4q, enh18q, and dim17p. These were found in 15 tumors (all, except cases 5 and 12) but their number and composition varied (one imbalance in seven tumors, two imbalances in seven tumors, and three imbalances in one tumor). Interestingly, imbalances of chromosome X were unequally distributed among the genders: gain of Xq was found in 10 of 13 male patients (77%), whereas Xp was lost in three of four female patients (75%). These results cannot be considered a result of technical bias as hybridization was performed on 46,XY metaphases using male control DNA.
The validity of the methods was confirmed previously by performing a series of control experiments. 9 In addition, the reliability of the CGH results was evaluated in two cases that were characterized by loss of chromosome 17 material (cases 7 and 11). A two-step CD20/FISH analysis on frozen sections using probes for cent-11 (control in green) and p53/17p13.1 (test in red) was performed. Seven CD20+ neoplastic cells in case 7 and four selected cells from case 11 were evaluated. All cells showed a lower number of p53 signals in comparison with cent-11 (eg, 1R2G, 2R3G, 2R4G).
In 16 cases with clinical information available, the CGH findings were compared with disease presentation and outcome. The number of imbalances did not correlate with lymphoma stage, age, or response to therapy, although on average the highest number of imbalances was found in cases presenting with stage IV disease (6.6 versus 2.7 in I to III). Advanced stage patients were also more likely to succumb to their disease (7 of 10 patients with stage IV have died), but a direct relationship between the number of genomic imbalances and outcome was not evident. On the one hand, the three cases displaying the highest number of imbalances (cases 7, 14, and 16) were indeed diagnosed with stage IV disease and they did die of their lymphoma. On the other hand two other advanced-stage patients who succumbed to their disease showed only one imbalance. With regard to virtually all commonly observed specific chromosomal imbalances, the numbers of surviving and deceased patients were similar.
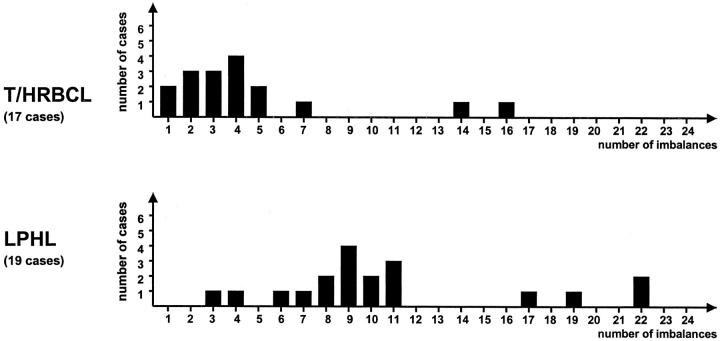
A comparison of the CGH profile observed in T/HRBCL with the pattern of genomic imbalances that we have previously identified in a series of 19 LPHL cases 9 showed several distinct features, listed in Table 2 ▶ . The most striking dissimilarity concerns the number of detected genomic imbalances (80 in total, with an average of 4.7 in T/HRBCL versus 205 with an average of 10.8 in LPHL) and their distribution (82% of T/HRBCL cases exhibit 1 to 5 imbalances per tumor whereas 89% of LPHL cases have 6 to 22 imbalances per tumor) (Figure 2) ▶ . In addition, the range of genomic regions targeted by the DNA copy number imbalances proved to be different in both lymphomas: chromosomal gains and losses in LPHL tended to affect whole chromosomes or entire chromosome arms, whereas genomic imbalances in T/HRBCL predominantly involved chromosomal subregions. Imbalances of chromosome 4q, X, and 17p were frequently observed in both T/HRBCL and LPHL. The CGH pattern of two of the cases included in the present study corresponds more closely to the profile typically observed in LPHL, in particular with respect to the complexity of the observed anomalies. On review, a notable high number of CD20+ B cells was observed in distinct areas of both biopsies, which were otherwise characteristic of T/HRBCL. Among these cells, both smaller, reactive-appearing lymphocytes as well as large atypical elements were found, thus suggesting that these cases may in fact represent cases of LPHL evolving into a DLBCL.
Table 2.
Some Relevant CGH Findings in T/HRBCL and LPHL
| Characteristics | T/HRBCL* | LPHL* |
|---|---|---|
| Number of genomic imbalances (average) | 80 (4.7) | 205 (10.8) |
| Range of genomic imbalances per tumor | 1–16 | 3–22 |
| Number of cases with 1 to 5 imbalances | 14 | 2 |
| Number of cases with more than 6 imbalances | 3 | 17 |
| Percentage of imbalances involving whole chromosome/arm | 34.9 | 78.7 |
| Percentage of imbalances involving chromosomal subregions | 65.1 | 21.3 |
| Frequently overrepresented regions | 4q, 18q, Xq, Xp, | 1, 2q, 3, 4q, 5q, 6, 8q, 11q, 12q, X |
| Frequently lost regions | 17p, [19] | 17, [19] |
*Chromosome 19 imbalances excluded from final calculations are shown here in brackets.
Figure 2.
Distribution of genomic imbalances in series of T/HRBCL and LPHL analyzed by DOP-PCR-CGH (present study and Franke et al 9 ).
Discussion
Very few data are currently available with respect to the genetic and molecular features of T/HRBCL. To characterize the cytogenetic profile of T/HRBCL and to define critical chromosomal subregions associated with the pathogenesis of this lymphoma we performed a CGH study of 17 well-documented cases. All these cases were included in the previous series described by Achten and colleagues 2,3 and answer to the strict disease definition put forward in these studies. These criteria are slightly more stringent than those proposed in the World Health Organization classification, 11 in which T/HRBCL is listed as a subtype of DLBCL. 12
By means of a combined DOP-PCR-CGH approach using a pool consisting of no more than four to five microdissected malignant cells, a characteristic pattern of chromosomal imbalances in T/HRBCL emerged. The most frequently detected imbalances included gain of Xq (59%, minimal overlapping region Xq12q13), 4q (41%, minimal overlapping region 4q25q26), Xp (29%, minimal overlapping region Xp21p11), and 18q (24%, minimal overlapping region 18q21), as well as loss of 17p (24%). These chromosomal regions may harbor genes that are involved in the etiology of this disease but thus far, BCL2 (18q21), MLT (18q21), and p53 (17p13) are the only lymphoma-related genes that have been identified in these regions. 13 Whether any of these genes is associated with the pathogenesis of T/HRBCL remains unknown.
Of note, overrepresentation of the X chromosome has frequently been observed in a wide spectrum of NHL (Mitelman Database of Chromosome Aberrations in Cancer. Mitelman F, Johansson B, and Mertens, F. http://cgap. nci. nih. gov/Chromosomes/Mitelman. 2002). 14 Moreover, its involvement in several translocations with breakpoints located at Xp22, Xq28, and Xq13, 15 suggests that chromosome X comprises a number of putative, as yet unidentified, lymphoma-associated oncogenes.
As opposed to overrepresentation of chromosomes X and 18, and loss of the short arm of chromosome 17 observed in various subtypes of lymphoma, 16-19 gain of 4q is a rare event in NHL in general (Mitelman Database of Chromosome Aberrations in Cancer. Mitelman F, Johansson B, and Mertens, F. http://cgap. nci. nih. gov/Chromosomes/Mitelman. 2002). Interestingly, we found an overrepresentation of the 4q sequences not only in 41% of T/HRBCL cases (including one case with enh4p15q32 as a sole chromosomal aberration) but also in ∼50% of the LPHL cases recently analyzed by DOP-PCR-CGH 9 which suggests a role of 4q-associated gene(s) in pathogenesis of both disorders. Still, only detailed additional cytogenetic and molecular studies of T/HRBCL may pinpoint the crucial lymphoma-related gene(s) among the at least 44 known genes including CASP6, LEF1, PITX2, and CCNA2 (http://cgap.nci.nih.gov), located at this particular genomic site.
The recurrent loss of chromosome 19/19p sequences detected in T/HRBCL and LPHL represents another noteworthy observation. It was decided to exclude these aberrations from the final calculations of CGH imbalances in both instances because chromosome 19 has been regarded as a critical genomic region for CGH. Yet we have failed to identify imbalances of chromosome 19 either in our control experiments or in lymphoma samples analyzed by DOP-PCR-CGH, suggesting that the loss of 19/19p detected in ∼50% of T/HRBCL and LPHL may represent an authentic finding and therefore a genomic feature characteristic of both disorders.
On the other hand, genomic regions frequently overrepresented in LPHL, in particular chromosome 1, 2q, 3, 5q, 6 (specific for LPHL), 8q, 11q, and 12q were only sporadically affected in T/HRBCL. Moreover, we noticed several further aberrations distinct for either T/HRBCL or LPHL, the most striking of which refers to the general pattern of genomic imbalances observed in both disorders. As such, the inherent complexity of the genomic imbalances detected in LPHL reflected by a high average number of chromosomal gains and losses (10.8) contrasts with the markedly less complex pattern of genomic imbalances in T/HRBCL (average 4.7 imbalances per case) that remains in agreement with the only one single reported karyotype of the T/HRBCL case. 20 Not only did both lymphomas differ in the average number of chromosomal gains and losses, the distribution of these imbalances was also quite dissimilar: whereas the vast majority of T/HRBCL cases (82%) featured only 1 to 5 imbalances per tumor, most of LPHL cases (89%) had 6 to 22 imbalances per tumor (Figure 2) ▶ . Altogether, these results challenge the contention that T/HRBCL simply represents an evolutionary stage of LPHL. Indeed, accepting the universal concept that karyotypic evolution of malignant cells is associated with progressive acquisition of additional genomic aberrations and increasing complexity of chromosomal changes, 21 one would expect to find a more complex genetic profile in T/HRBCL if it had actually evolved from LPHL. Furthermore, assuming a straightforward progression from LPHL to T/HRBCL, the chromosomal regions involved in both disorders should have been approximately comparable.
Rather, and in view of recent phenotypic and molecular genetic evidence suggesting that the tumor cells in both conditions are derived from a germinal center ancestor 22-27 we speculate that the initial transforming events in LPHL and T/HRBCL may indeed be similar. As such these oncogenic incidents probably operate on a germinal center cell and, considering the presence of common chromosomal imbalances in the present study, likely involve one or more thus far unspecified gene(s) located on chromosomes 4q and/or chromosome 19. Depending on the nature of subsequent oncogenic hits, the tumor cells acquire either a popcorn cell phenotype, resulting in an indolent biological behavior, or they are transformed into atypical B cells embedded in a considerably less well-structured T-cell-rich, histiocyte-rich background, heralding a fulminant downhill clinical course. Direct support for this hypothesis might be derived from molecular studies of cases of LPHL and T/HRBCL that have evolved successively in the same patient. However, a review of the literature 28 and a survey of our files have shown that the consecutive occurrence of these lymphomas is uncommon, when relying on strict diagnostic criteria. Not only does the discovery of clear-cut differences in the cytogenetic profile of T/HRBCL as compared to that of LPHL, offer a valid explanation for the contrasting clinical behavior of both disorders, it also clarifies and supports this obvious morphological distinctiveness. Essentially, and in contrast with T/HRBCL, typical LPHL tend to retain a rudimentary germinal center environment, as exemplified by a clearly nodular structure, reflecting a supporting follicular dendritic cell network, by the presence of an important nonneoplastic B-cell component, and by a prominent CD57+ T-cell population, often arranged in rosettes surrounding L&H cells.
Although the vast majority of T/HRBCL cases are reliably distinguished from LPHL, it cannot be denied that in sporadic cases, a differential diagnosis by morphology alone is problematic. This diagnostic limitation might be carried forward to the genetic level as two of the cases included in the present study (cases 2 and 4), in contrast to the remaining 15 cases, show more resemblance to LPHL with respect to the complexity of the observed cytogenetic aberrations. Therefore, awaiting further clarification of the crucial events determining the ultimate fate of a transformed germinal center cell, the establishment of an accurate diagnosis in such borderline cases may rely above all on a sound communication between pathologists and clinicians.
In summary, by applying a DOP-PCR-CGH approach on a pool of few microdissected single tumor cells from a series of 17 cases, we elucidated for the first time the cytogenetic profile of genomic imbalances characteristic of T/HRBCL that showed to be in clear-cut contrast with the inherent complexity of the genomic imbalances observed in LPHL. Although both disorders may originate from a similar precursor B cell, our molecular cytogenetic findings suggest that T/HRBCL constitutes a distinct lymphoma entity rather than an evolutionary stage of LPHL.
Footnotes
Address reprint requests to Dr. Anne Hagemeijer, Center for Human Genetics, University of Leuven, Campus Gasthuisberg O&N6, Herestraat 49, B-3000 Leuven, Belgium. E-mail: anne.hagemeijer@med.kuleuven.ac.be.
Supported by grant G025298 from the Fund for Scientific Research, Flanders, Belgium.
This text presents research results of the Belgian program of Interuniversity Poles of attraction initiated by the Belgian State, Prime Minister’s Office, Science Policy Programming. The scientific responsibility is assumed by the authors.
Peter Vandenberghe is a clinical investigator and Ruth Achten is a Fellow of the Fund for Scientific Research.
A. H. and C. D.W.-P. share senior authorship.
References
- 1.Delabie J, Vandenberghe E, Kennes C, Verhoef G, Foschini MP, Stul M, Cassiman JJ, De Wolf-Peeters C: Histiocyte-rich B-cell lymphoma. A distinct clinicopathologic entity possibly related to lymphocyte predominant Hodgkin’s disease, paragranuloma subtype. Am J Surg Pathol 1992, 16:37-48 [PubMed] [Google Scholar]
- 2.Achten R, Verhoef G, Vanuytsel L, De Wolf-Peeters C: Histiocyte-rich, T-cell-rich B-cell lymphoma: a distinct diffuse large B-cell lymphoma subtype showing characteristic morphologic and immunophenotypic features. Histopathology 2002, 40:31-45 [DOI] [PubMed] [Google Scholar]
- 3.Achten R, Verhoef G, Vanuytsel L, De Wolf-Peeters C: T-cell/histiocyte-rich large B-cell lymphoma: a distinct clinicopathologic entity. J Clin Oncol 2002, 20:1269-1277 [DOI] [PubMed] [Google Scholar]
- 4.De Jong D, Van Gorp J, Sie-Go D, Van Heerde P: T-cell rich B-cell non-Hodgkin’s lymphoma: a progressed form of follicle centre cell lymphoma and lymphocyte predominance Hodgkin’s disease. Histopathology 1996, 28:15-24 [DOI] [PubMed] [Google Scholar]
- 5.Kallioniemi A, Kallioniemi OP, Sudar D, Rutovitz D, Gray JW, Waldman F, Pinkel D: Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science 1992, 258:818-821 [DOI] [PubMed] [Google Scholar]
- 6.Joos S, Kupper M, Ohl S, von Bonin F, Mechtersheimer G, Bentz M, Marynen P, Moller P, Pfreundschuh M, Trumper L, Lichter P: Genomic imbalances including amplification of the tyrosine kinase gene JAK2 in CD30+ Hodgkin cells. Cancer Res 2000, 60:549-552 [PubMed] [Google Scholar]
- 7.Joos S, Menz CK, Wrobel G, Siebert R, Gesk S, Ohl S, Mechtersheimer G, Trumper L, Moller P, Lichter P, Barth TF: Classical Hodgkin lymphoma is characterized by recurrent copy number gains of the short arm of chromosome 2. Blood 2002, 99:1381-1387 [DOI] [PubMed] [Google Scholar]
- 8.Ohshima K, Ishiguro M, Ohgami A, Sugihara M, Haraoka S, Suzumiya J, Kikuchi M: Genetic analysis of sorted Hodgkin and Reed-Sternberg cells using comparative genomic hybridization. Int J Cancer 1999, 82:250-255 [DOI] [PubMed] [Google Scholar]
- 9.Franke S, Wlodarska I, Maes B, Vandenberghe P, Delabie J, Hagemeijer A, De Wolf-Peeters C: Lymphocyte predominance Hodgkin disease is characterized by recurrent genomic imbalances. Blood 2001, 97:1845-1853 [DOI] [PubMed] [Google Scholar]
- 10.: ISCN: Mitelman F eds. Guidelines for Cancer Cytogenetics: Supplement to an International System for Human Cytogenetic Nomenclature. 1995. Karger, Basel
- 11.De Wolf-Peeters C, Achten R: “T cell rich large B cell lymphoma.” Will we ever see the wood for the trees? Histopathology 2002, 4:269-271 [DOI] [PubMed] [Google Scholar]
- 12.Gatter KC, Warnke RA: Diffuse large B-cell lymphoma. Jaffe ES Harris NL Vardiman JW eds. Pathology and Genetics of Tumours of Haematopoietic and Lymphoid Tissues. 2001:pp 171-174 IARC Press, Lyon
- 13.Willis TG, Dyer MJ: The role of immunoglobulin translocations in the pathogenesis of B-cell malignancies. Blood 2000, 96:808-822 [PubMed] [Google Scholar]
- 14.McDonald HL, Gascoyne RD, Horsman D, Brown CJ: Involvement of the X chromosome in non-Hodgkin lymphoma. Genes Chromosom Cancer 2000, 28:246-257 [PubMed] [Google Scholar]
- 15.Callet-Bauchu E, Rimokh R, Gazzo S, Pages J, Bastion Y, Berger F, Coeur P, Felman P: Unbalanced X;autosome translocation (X;18)(q13;p11) in a case of aggressive natural killer non-Hodgkin lymphoma. Cancer Genet Cytogenet 1997, 98:16-19 [DOI] [PubMed] [Google Scholar]
- 16.Avet-Loiseau H, Vigier M, Moreau A, Mellerin MP, Gaillard F, Harousseau JL, Bataille R, Milpied N: Comparative genomic hybridization detects genomic abnormalities in 80% of follicular lymphomas. Br J Haematol 1997, 97:119-122 [DOI] [PubMed] [Google Scholar]
- 17.Bentz M, Barth TF, Bruderlein S, Bock D, Schwerer MJ, Baudis M, Joos S, Viardot A, Feller AC, Muller-Hermelink HK, Lichter P, Dohner H, Moller P: Gain of chromosome arm 9p is characteristic of primary mediastinal B-cell lymphoma (MBL): comprehensive molecular cytogenetic analysis and presentation of a novel MBL cell line. Genes Chromosom Cancer 2001, 30:393-401 [DOI] [PubMed] [Google Scholar]
- 18.Dierlamm J, Rosenberg C, Stul M, Pittaluga S, Wlodarska I, Michaux L, Dehaen M, Verhoef G, Thomas J, de-Kelver W, Bakker ST, Cassiman JJ, Raap AK, De Wolf-Peeters C, Van den Berghe H, Hagemeijer A: Characteristic pattern of chromosomal gains and losses in marginal zone B cell lymphoma detected by comparative genomic hybridization. Leukemia 1997, 11:747-7589180302 [Google Scholar]
- 19.Monni O, Joensuu H, Franssila K, Klefstrom J, Alitalo K, Knuutila S: BCL2 overexpression associated with chromosomal amplification in diffuse large B-cell lymphoma. Blood 1997, 90:1168-1174 [PubMed] [Google Scholar]
- 20.La Starza R, Aventin A, Falzetti D, Stul M, Martelli MF, Falini B, Mecucci C: 14q+ chromosome marker in a T-cell-rich B-cell lymphoma. J Pathol 1996, 178:227-231 [DOI] [PubMed] [Google Scholar]
- 21.Johansson B, Mertens F, Mitelman F: Secondary chromosomal abnormalities in acute leukemias. Leukemia 1994, 8:953-962 [PubMed] [Google Scholar]
- 22.Braeuninger A, Kuppers R, Strickler JG, Wacker HH, Rajewsky K, Hansmann ML: Hodgkin and Reed-Sternberg cells in lymphocyte predominant Hodgkin disease represent clonal populations of germinal center-derived tumor B cells. Proc Natl Acad Sci USA 1997, 94:9337-9342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brauninger A, Kuppers R, Spieker T, Siebert R, Strickler JG, Schlegelberger B, Rajewsky K, Hansmann ML: Molecular analysis of single B cells from T-cell-rich B-cell lymphoma shows the derivation of the tumor cells from mutating germinal center B cells and exemplifies means by which immunoglobulin genes are modified in germinal center B cells. Blood 1999, 93:2679-2687 [PubMed] [Google Scholar]
- 24.Brauninger A, Hansmann ML, Strickler JG, Dummer R, Burg G, Rajewsky K, Kuppers R: Identification of common germinal-center B-cell precursors in two patients with both Hodgkin’s disease and non-Hodgkin’s lymphoma. N Engl J Med 1999, 340:1239-1247 [DOI] [PubMed] [Google Scholar]
- 25.Falini B, Bigerna B, Pasqualucci L, Fizzotti M, Martelli MF, Pileri S, Pinto A, Carbone A, Venturi S, Pacini R, Cattoretti G, Pescarmona E, Lo CF, Pelicci PG, Anagnastopoulos I, Dalla-Favera R, Flenghi L: Distinctive expression pattern of the BCL-6 protein in nodular lymphocyte predominance Hodgkin’s disease. Blood 1996, 87:465-471 [PubMed] [Google Scholar]
- 26.Marafioti T, Hummel M, Anagnostopoulos I, Foss HD, Falini B, Delsol G, Isaacson PG, Pileri S, Stein H: Origin of nodular lymphocyte-predominant Hodgkin’s disease from a clonal expansion of highly mutated germinal-center B cells. N Engl J Med 1997, 337:453-458 [DOI] [PubMed] [Google Scholar]
- 27.Ohno T, Stribley JA, Wu G, Hinrichs SH, Weisenburger DD, Chan WC: Clonality in nodular lymphocyte-predominant Hodgkin’s disease. N Engl J Med 1997, 337:459-465 [DOI] [PubMed] [Google Scholar]
- 28.Bodis S, Kraus MD, Pinkus G, Silver B, Kadin ME, Canellos GP, Shulman LN, Tarbell NJ, Mauch PM: Clinical presentation and outcome in lymphocyte-predominant Hodgkin’s disease. J Clin Oncol 1997, 15:3060-3066 [DOI] [PubMed] [Google Scholar]