Abstract
Synovial sarcoma is an aggressive spindle cell sarcoma with two major histological subtypes, biphasic and monophasic, defined respectively by the presence or absence of areas of glandular epithelial differentiation. It is characterized by a specific chromosomal translocation, t(X;18)(p11.2;q11.2), which juxtaposes the SYT gene on chromosome 18 to either the SSX1 or the SSX2 gene on chromosome X. The chimeric SYT-SSX products are thought to function as transcriptional proteins that deregulate gene expression, thereby providing a putative oncogenic stimulus. We investigated the pattern of gene expression in synovial sarcoma using cDNA microarrays containing 6548 sequence-verified human cDNAs. A tissue microarray containing 37 synovial sarcoma samples verified to bear the SYT-SSX fusion was constructed for complementary analyses. Gene expression analyses were performed on individual tumor samples; 14 synovial sarcomas, 4 malignant fibrous histiocytomas, and 1 fibrosarcoma. Statistical analysis showed a distinct expression profile for the group of synovial sarcomas as compared to the other soft tissue sarcomas, which included variably high expression of ERBB2, IGFBP2, and IGF2 in the synovial sarcomas. Immunohistochemical analysis of protein expression in tissue microarrays of 37 synovial sarcomas demonstrated strong expression of ERBB2 and IGFBP2 in the glandular epithelial component of biphasic tumors and in solid epithelioid areas of some monophasic tumors. Fluorescence in situ hybridization analysis indicated that the ERBB2 overexpression was not because of gene amplification. Differentially expressed genes were also found in a comparison of the expression profiles of the biphasic and monophasic histological subgroups of synovial sarcoma, notably several keratin genes, and ELF3, an epithelial-specific transcription factor gene. Finally, we also noted differential overexpression of several neural- or neuroectodermal-associated genes in synovial sarcomas relative to the comparison sarcoma group, including OLFM1, TLE2, CNTNAP1, and DRPLA. Our high-throughput studies of gene expression patterns, complemented by tissue microarray studies, confirm the distinctive expression profile of synovial sarcoma, provide leads for the study of glandular morphogenesis in this tumor, and identify a new potential therapeutic target, ERBB2, in a subset of cases.
Synovial sarcoma accounts for 5 to 10% of all soft tissue sarcomas and are primarily located in the extremities, most frequently affecting young adults. The primary treatment is surgery supplemented with radiotherapy. At present, the 5-year survival for patients who present with localized disease is only ∼60% because of the subsequent appearance of metastatic disease, which is almost uniformly fatal. 1-3 Although some response to certain chemotherapy regimens has been observed, 4 there is a need for new therapeutic options for disseminated disease.
Synovial sarcomas predominantly occur in two major histological subtypes. Biphasic tumors contain epithelial cells arranged in glandular structures in a background of spindle cells, whereas monophasic tumors are entirely composed of spindle cells with or without scattered solid epithelioid areas, and lack well-developed glandular spaces. The t(X;18)(p11.2;q11.2) translocation is characteristic of synovial sarcomas and fuses the SYT gene on chromosome 18 to either the SSX1 or the SSX2 gene (or rarely, SSX4) on chromosome X. 5-8 The resulting SYT-SSX1 or SYT-SSX2 fusion transcript is clinically useful in the molecular diagnosis of these tumors. 9
Current data suggest that the SYT-SSX fusion protein, which lacks a DNA-binding domain, acts as a transcriptional co-factor, with a repression domain, SSX-RD, contributed by the C-terminus of SSX and a strong activation domain, the QPGY domain, within the SYT portion of the fusion protein. Transactivation by the latter appears mediated at least in part through interactions with the BRM protein, whereas repression by the SSX-RD domain seems to be because of interactions with Polycomb group proteins. 10,11 A more recent study also found an association of SYT with SNF/SWI complexes and of the C-terminal region of SSX1 with core histones. 12 The downstream transcriptional targets of SYT, SSX, or SYT-SSX are presently unknown.
The differentiation lineage of synovial sarcoma is unclear and the regulation of epithelial differentiation in these tumors is a biological issue of some interest. Recent studies have confirmed that the spindle and epithelial components of synovial sarcoma are clonally related based on the detection of the t(X;18) chromosomal rearrangement (or the SYT-SSX fusion transcript) in both components. 13-15 The ability of some synovial sarcoma cell lines to exhibit immunohistochemical and ultrastructural epithelial features supports the notion that synovial sarcoma arises from a mesenchymal stem cell with a capacity for epithelial differentiation. 16,17 Features of neuroectodermal differentiation have also been rarely reported. 18
In the present study we used cDNA microarrays to investigate gene expression profiles in synovial sarcomas. A comparison group of other soft tissue sarcoma samples consisting mainly of malignant fibrous histiocytomas (MFHs) was included, because these tumors have similar locations as synovial sarcomas and may also share similar contaminating stromal elements. The tumor profiling on cDNA microarrays was followed by immunohistochemical analysis of the expression of selected proteins on a tissue microarray (TMA) of 37 synovial sarcomas. Our data provide new leads for the investigation of epithelial morphogenesis in this tumor and identify new targets for experimental therapeutic approaches.
Materials and Methods
Tumors and Reference Cell Line
Tumor tissues from 10 synovial sarcomas were collected at Memorial Sloan-Kettering Cancer Center under approved IRB protocols. All were confirmed as being positive for the SYT-SSX1 or SYT-SSX2 fusion transcript by reverse transcriptase-polymerase chain reaction (RT-PCR), as previously described. 19 The samples were freshly frozen in liquid nitrogen and stored at −70°C until use. Nine additional sarcoma samples were obtained from the Cooperative Human Tissue Network, all originally diagnosed as synovial sarcoma or MFH. A histological review of all of the synovial sarcomas was performed (CRA), and the additional tumors from the Cooperative Human Tissue Network were also subject to RT-PCR analysis of their SYT-SSX status. The histological review of one synovial sarcoma sample from the Cooperative Human Tissue Network classified this tumor as a fibrosarcoma and the RT-PCR analysis showed that this tumor was lacking the SYT-SSX fusion transcript. The histological review and RT-PCR analysis of this case were performed in a mutually blinded manner. This fibrosarcoma tumor sample was retained in the study as a part of the control group. The final study group therefore consisted of 14 synovial sarcomas, 4 MFH, and 1 fibrosarcoma.
Total cellular RNA was isolated from frozen tumor specimens using two rounds of extraction with Trizol reagent (Invitrogen, Carlsbad, CA). RNA from an osteosarcoma cell line, OsA-CL, 20 was used as the constant reference RNA and included in each hybridization to allow normalization of the expression of each clone relative to the reference for each sample. Total RNA was isolated with the RNeasy kit (Qiagen, Valencia, CA) and further purified by Trizol reagent according to the manufacturer’s recommendations.
cDNA Microarrays and Image Analysis
The 6548 sequence-verified human cDNAs used in this study were obtained under a Cooperative Research and Development Agreement with Research Genetics (ResGen, Huntsville, AL). Gene names are according to Build 138 of the UniGene human sequence collection (http://www.ncbi.nlm.nih.gov/UniGene/). PCR products generated from these clones were printed onto glass slides as described. 21 Hybridization, scanning, and image analysis were performed as previously described (www.nhgri.nih.gov/DIR/microarray). 22,23 Briefly, fluorescently labeled cDNA was synthesized from ∼100 μg of tumor RNA or ∼200 μg of cell line RNA by oligo(dT)-primed polymerization in the presence of Cy3 or Cy5 dUTP (Amersham Pharmacia Biotech, Piscataway, NJ). Image analyses were performed using DeArray software (Scanalytics, Fairfax, VA). 24 The two fluorescent images (red and green channel) obtained constitute the raw data from which differential gene expression ratio values were calculated. The ratios of the red intensity to the green intensity (R/G) for all targets were determined, and the ratio normalization was performed based on 88 preselected internal control genes that are stable for most experiments.
Statistical Analyses
Multidimensional scaling plots and weighted gene lists were generated as previously described (www.nhgri.nih.gov/DIR/microarray). 25,26 For each tumor group partition, a discriminative weight was first assigned to each gene, and a random permutation test was then performed to assess the significance of the discriminative weight based on its ability to differentiate the two groups tested. Genes were filtered before participating in the selection process and random permutation by requiring the average measurement quality score across each tumor group, assigned by the DeArray software, to be greater than 0.4. On average 3000 of 6500+ genes survived this measurement quality criterion. The gene selection algorithm along with the random permutation test (with 1000 permutations) was applied to: all samples, monophasic versus biphasic synovial sarcomas, and SYT-SSX1-positive versus SYT-SSX2-positive synovial sarcomas. For the final lists of genes differentially expressed between two tumor groups studied, a minimum expression level was set by requiring the average natural logarithm (ln) of the relative red intensity (RRI) to be ≥1.5 for all synovial sarcoma samples. 23
TMAs
A TMA was constructed containing 37 synovial sarcoma samples, all previously confirmed by RT-PCR to have either the SYT-SSX1 (n = 21) or SYT-SSX2 (n = 16) gene fusion. TMAs of 1-mm cores of the tumors were made using a TMAer (Beecher Instruments, Silver Spring, MD). Donor paraffin blocks for all arrayed tumors were verified histologically by a surgical pathologist (CRA). The TMA contained 32 monophasic and 5 biphasic cases. All cores of biphasic cases scored in this study were confirmed to contain glandular elements. Of the 14 cases studied by cDNA microarray analysis, 5 were represented on the TMA (all monophasic, two SYT-SSX1 and three SYT-SSX2). To evaluate immunoreactivity of MFH and fibrosarcoma cases, we used sections of a high-grade soft tissue sarcoma TMA (gift of Carlos Cordon-Cardo, Memorial Sloan-Kettering Cancer Center, New York, NY) previously described elsewhere in detail. 27
Immunohistochemistry (IHC)
IHC stains were performed in the Memorial Sloan-Kettering Cancer Center Core Research IHC Laboratory. Five-μm formalin-fixed paraffin-embedded sections were cut and placed on positively charged slides and IHC was performed using the avidin-biotin complex method, with heat-induced epitope retrieval in citrate buffer. The antigenicity of all 37 arrayed synovial sarcomas was confirmed by the presence of positive immunostaining for vimentin (DAKO, Carpinteria, CA) in synovial sarcoma spindle cells. IHC for ERBB2 (Herceptest, DAKO) was performed and scored according to the manufacturer’s instructions. Briefly, this scoring is as follows: 0, no membrane staining, 1+, faint partial membrane staining, 2+, weak complete membrane staining in >10% of cells, 3+, intense complete membrane staining in >10% of cells. In breast carcinomas, scores of 0 and 1+ are considered negative and scores of 2+ and 3+ are considered positive and indicate eligibility for Herceptin therapy.
For localization of IGFBP2 expression, we used IGFBP2 polyclonal antibody (C-18 goat antibody; Santa Cruz Biotechnology, Santa Cruz, CA), diluted 1:1000 in citrate buffer at pH 6.0. BCL2 antibody was obtained from DAKO. Antibody to IGF2 (H-103 rabbit polyclonal antibody, Santa Cruz Biotechnology) was evaluated but performed poorly on control materials in our hands.
Fluorescence in Situ Hybridization (FISH)
FISH for ERBB2 was performed using a Vysis paraffin pretreatment kit and following the manufacturer’s instruction for the PathVysion probe kit for ERBB2 (Vysis, Downers Grove, IL). At least 50 nuclei were scored for the probes for chromosome 17 (centromeric, Spectrum Green) and ERBB2 (locus-specific, Spectrum Orange). FISH images were obtained using an Olympus BX40 epifluorescent microscope and captured on the Applied Image Analysis System (Applied Imaging, Pittsburgh, PA).
Results
cDNA Microarray Analyses
Fourteen synovial sarcoma samples, 10 monophasic (five SYT-SSX1 and five SYT-SSX2) and 4 biphasic (three SYT-SSX1 and one SYT-SSX2) tumors, were subjected to cDNA microarray analysis. The comparison group was a set of five spindle cell sarcomas of soft tissue, including four MFH and one fibrosarcoma. Expression profiles were generated based on calculated intensity ratios relative to OsA-CL. The data from these hybridizations can be found at www.nhgri.nih.gov/DIR/Microarray/main.html.
Using the weighted discriminator method with a random permutation test, we found a significant overabundance of discriminating genes relative to the random assignments. There were 153 genes distinguishing synovial sarcoma from MFH/fibrosarcoma (weight value >1.8 and P ≤ 0.004) as compared to three genes for the random assignment. Using 50 of these genes expressed above a minimum level in all synovial sarcomas, a strong separation between synovial sarcoma and MFH could be displayed by hierarchical clustering (Figure 1a) ▶ and multidimensional scaling (Figure 1b) ▶ . We found notable overexpression in synovial sarcoma, relative to the MFH and fibrosarcoma samples, of the genes encoding insulin-like growth factor binding protein 2 (IGFBP2), ERBB2, insulin-like growth factor II (IGF2), fibroblast growth factor receptor 3 (FGFR3), OLFM1 (olfactomedin, also known as noelin-1 or NOE1), 28 TLE2 (transducin-like enhancer of split 2), 29 CNTNAP1 (contactin-associated protein, also known as CASPR) 30 , DRPLA (dentatorubral-pallidoluysian atrophy gene, also known as atrophin-1), 31 CRABP1 (cellular retinoic acid-binding protein 1), and the PRAME (preferentially expressed antigen in melanoma) tumor antigen gene, 32 among others. We should note that SSX1 and SSX2 clones were not included on the arrays used and therefore possible differential signals because of their overexpression in the context of the SYT-SSX fusion transcripts of synovial sarcomas could not be assessed.
Figure 1.
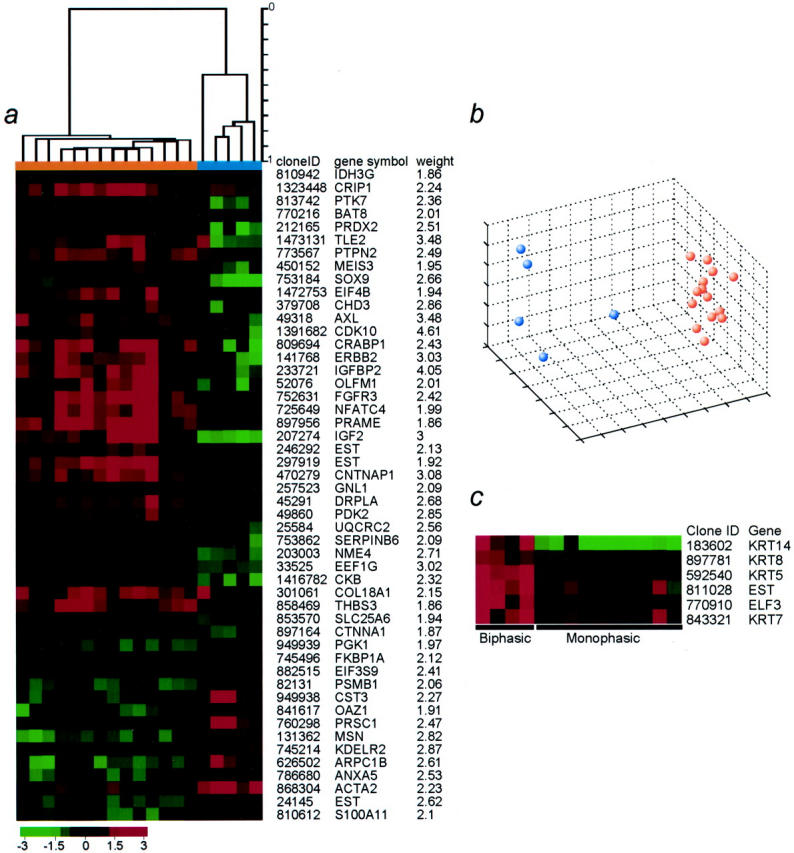
a: Hierarchical clustering dendrogram separating synovial sarcomas (orange) and MFH/fibrosarcoma (blue). The dendrogram shown on top was generated using a list of 50 genes with the most power to separate the groups based on weight value and a significant expression level in synovial sarcoma, [average ln(RRI) for the synovial sarcomas ≥1.5]. The weight value is listed to the right. A pseudocolored representation of gene expression ratios is shown according to the scale below. b: Multidimensional scaling plot for all 19 tumor samples studied. The similarity of gene expression profiles between any pair of tumor samples was assessed by Pearson correlation coefficients based on expression levels for the 50 genes presented in a. Orange dots denote synovial sarcoma and blue dots denote MFH or fibrosarcoma. c: Genes significantly expressed in biphasic synovial sarcoma as compared to monophasic synovial sarcoma. The four columns to the left represent the expression levels for the biphasic tumors, and the 10 columns to the right represent expression levels for the monophasic tumors. For inclusion, each gene met the following criteria: average ln(RRI) ≥2 for the biphasic and ≤1 for the monophasic tumors, red intensity >500, and ratio ≥1.2 for all biphasic tumors. The color scale is as in b.
Statistical analysis by a random permutation test also showed a separation between biphasic and monophasic synovial sarcomas with 21 genes distinguishing these two classes (weight >2.1 and P ≤ 0.04) as compared to two genes for the random assignment. The genes most significantly and specifically expressed in biphasic tumors were the genes encoding keratins 14, 8, 5, and 7, ELF3, and an anonymous expressed sequence tag (Figure 1c) ▶ .
We also compared expression profiles between SYT-SSX1- and SYT-SSX2-positive synovial sarcomas in two different analyses. The first included all synovial sarcoma samples (eight SYT-SSX1 versus six SYT-SSX2), and the second analysis examined monophasic tumors only (five SYT-SSX1 versus five SYT-SSX2). No significant differences between our actual data and the random assignments were found (data not shown). This negative finding may be unexpected given the known correlations of SYT-SSX fusion type with survival and areas of glandular epithelial differentiation, 33,34 but the study group and the technical platform may have been inadequate to detect the likely much smaller differences in gene expression between subsets of the same tumor type.
IHC and FISH
The ERBB2 expression detected by cDNA array analysis was further evaluated by IHC and FISH analysis on TMA slides containing 37 synovial sarcomas, including 32 monophasic and 5 biphasic cases, as summarized in Table 1 ▶ . In all biphasic tumors, strong (2+ to 3+) cell membrane immunoreactivity for ERBB2 was observed in the glandular epithelial component (cases SS-51 and SS-52 in Figure 2 ▶ ). Also, three of nine monophasic synovial sarcomas with solid epithelioid areas showed moderate-to-strong (2+) cell membrane ERBB2 reactivity localized to these areas (case SS-67 in Figure 2 ▶ ). Focal weak positivity (1+) was also observed in the spindle cell component in seven other monophasic synovial sarcomas (not illustrated). By FISH analysis there was no ERBB2 gene amplification in any of the immunoreactive cases (data not shown). To evaluate ERBB2 immunoreactivity in MFH and fibrosarcoma, we performed IHC on a high-grade soft tissue sarcoma TMA described elsewhere 27 which included triplicate 0.6-mm cores from 25 cases classified as MFH or myxofibrosarcoma and 6 cases classified as fibrosarcoma. All of these 31 cases were completely negative for ERBB2 by IHC (not illustrated).
Table 1.
Results of ERBB2 IHC on 37 Synovial Sarcomas
| Histological type | 0 to 1+ | 2+ to 3+ |
|---|---|---|
| Monophasic without epithelioid areas (n = 23) | 23 | 0 |
| Monophasic with epithelioid areas (n = 9) | 6 | 3 |
| Biphasic (n = 5) | 0 | 5 |
Chi-square test; P < 0.0001.
Results scored according to standard Dako Herceptest criteria (see Materials and Methods).
Figure 2.
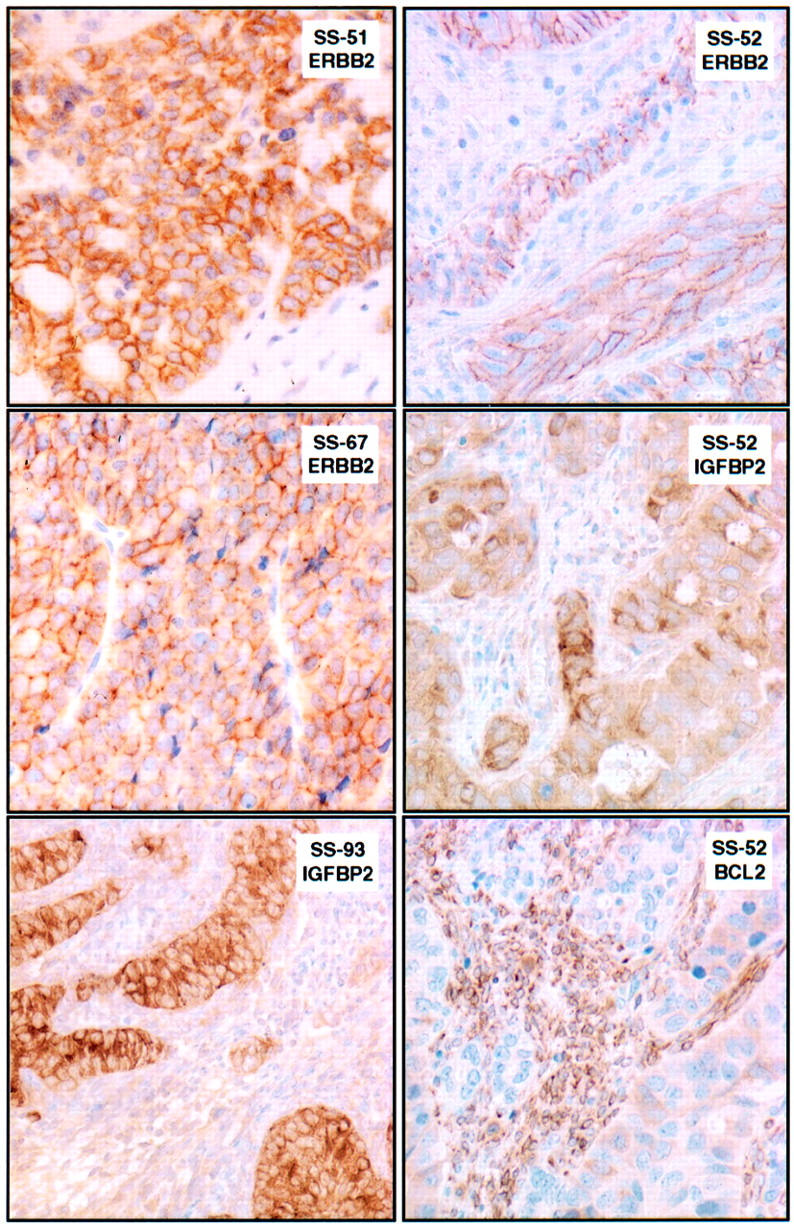
Composite figure showing patterns of ERBB2 and IGFBP2 immunoreactivity in synovial sarcomas from a TMA. Biphasic synovial sarcoma SS-51 shows 3+ ERBB2 immunoreactivity in the glandular epithelial component. Monophasic synovial sarcoma SS-67 shows 2+ ERBB2 immunoreactivity in a solid epithelioid area. Biphasic synovial sarcoma SS-93 shows intense immunoreactivity for IGFBP2 in its epithelial component (lumen formation seen in other fields). Biphasic synovial sarcoma SS-52 shows 2+ ERBB2 immunoreactivity and moderate-to-strong immunoreactivity for IGFBP2 in the glandular epithelial component. In contrast, BCL2 immunostaining, a known marker of the spindle cell component in synovial sarcomas, outlines the nests of primarily BCL2-negative epithelial cells in SS-52.
IGFBP2 also showed prominent membranous and cytoplasmic expression in the epithelial component of almost all biphasic synovial sarcomas (cases SS-52 and SS-93 in Figure 2 ▶ ), and focal to diffuse weaker cytoplasmic expression in the spindle cells and solid epithelioid areas of biphasic and monophasic tumors.
Discussion
Expression-profiling analysis based on biochip technologies has revolutionized the efficiency of analyzing tumors. Several reports have demonstrated the ability of tumor profiling to determine differentially expressed genes in subsets of tumors according to histological, hormonal, or prognostic status for a certain tumor type. 23,35-37 Synovial sarcoma provides a good model for gene expression analysis, because transcriptional deregulation may be central to its pathogenesis and the specific SYT-SSX gene fusion provides an objective marker for supervised analyses. Synovial sarcoma has two well-defined histological subgroups and two subgroups based on the two major types of SYT-SSX fusions. There is also a correlation between the histological subtype and the SSX gene involved in the fusion transcript, the majority of biphasic tumors contain the SYT-SSX1 fusion transcript, whereas the monophasic tumors have both fusion transcripts more or less equally represented. 19,34 We used a statistical approach to allow determination of subsets of tumors and to identify genes with the most power to discriminate between the patterns of gene expression in SYT-SSX-positive synovial sarcomas and other soft tissue sarcomas, to find genes significantly expressed in synovial sarcoma. The use of a relatively small comparison group of five other sarcomas allowed the identification of a discriminatory gene list that represents a broader gene expression profile than if many other sarcomas had been included. We found variably high expression of IGFBP2, ERBB2, IGF2, OLFM1, 28 TLE2 (transducin-like enhancer of split 2), 29 CNTNAP1 (contactin-associated protein, also known as CASPR) 30 , andDRPLA, 31 CRABP1, and PRAME 32 in synovial sarcoma, as compared to MFH and fibrosarcoma (Figure 1a) ▶ . In addition to these named genes, our cDNA microarrays also contained many unnamed transcripts in the form of expressed sequence tags, some of which were differentially expressed and may present new opportunities for gene discovery.
IGF2 is thought to enhance tumor growth by acting in an autocrine manner via the IGF1 receptor (IGF1R), and increasing evidence supports the crucial role of IGF1R in malignant transformation and tumor growth. 38 IGF2 has also been shown to be induced by the PAX3-FKHR fusion transcript suggesting a role in the oncogenesis of rhabdomyosarcoma. 39,40 IGF signaling has also been implicated in the biology of synovial sarcoma 41 and other primitive tumors, such as Ewing’s sarcoma. 42,43 IGFBP2, as an IGF1-binding protein, is known to regulate IGF1 signaling primarily in a negative manner, by sequestering IGF1 from its receptor. 38 IGFBP2 has also been implicated in cell adhesion. 44 IGFBP2 is expressed in a developmental stage-specific manner, with predominant expression in highly proliferative fetal tissues, notably in the limbs. In mouse limb morphogenesis, IGFBP2 is prominent in the apical ectodermal ridge whereas IGF1 is present in the limb mesenchymal cells. 45 In synovial sarcomas, IGF1 expression appears restricted to the spindle cell component 46 and thus seems to recapitulate the reciprocal expression of IGF1 and IGFBP2 in the developing limb. This raises the possibility that the IGFBP2 expression observed in synovial sarcoma represents a recapitulation of stage-specific expression in an IGFBP2-positive progenitor cell, possibly to modulate the proliferative effects of IGF signaling in favor of epithelial differentiation.
IGFBP2 has been identified as a differentially overexpressed transcript in several other expression-profiling studies, including androgen-independent prostate cancer 47 and high-grade gliomas. 48,49 Its association with several different cancers suggests that it may contribute to tumorigenic potential, an observation with some experimental support. 50 Indeed, there is a positive correlation between tumor grade and IGFBP2 expression. 44 Although some studies have proposed a role for IGFBP2 in the nucleus, 44 we observed only cytoplasmic immunostaining in the positive synovial sarcomas studied in the present series. Another interesting aspect of the IGF system is the emerging information on serum IGF/IGFBP levels as clinical markers in tumors expressing these proteins. 51,52 This raises the possibility that IGFBP2 overexpression in synovial sarcoma could be associated with elevated serum IGFBP2, providing a potential marker for monitoring of tumor burden.
There are to our knowledge no previous articles describing the systematic analysis of ERBB2 status in synovial sarcoma in the literature, but data on a few cases are found within some early studies of ERBB2. One study including four synovial sarcomas examined ERBB2 levels by Northern blotting and found low levels in two cases and moderate levels in two cases. 53 Another study included six cases and found them to be negative by IHC. 54 In neither study were the histological types of the synovial sarcomas specified.
ERBB2 normally forms part of a heterodimeric receptor tyrosine kinase by dimerizing with ERBB1 (EGFR) or ERBB3. 55 Its tumorigenic effects are thought to operate primarily through enhancement of ligand-dependent heterodimerization that activates the MAPK and PI3K/AKT downstream signaling cascades. 55 We find that strong ERBB2 protein expression is seen in the epithelial component of all biphasic synovial sarcomas, and approximately one third of monophasic tumors, notably those with solid epithelioid areas. FISH analysis demonstrated that this overexpression was not because of gene amplification. The association of fusion type and histological type may result in an indirect association of fusion type and ERBB2 positivity; however, our data suggest that ERBB2 positivity is associated primarily with histological type, not fusion type.
The interrelationship of the ERBB2 and HGF/MET signaling systems is potentially interesting, given the pattern of expression of HGF and MET in synovial sarcoma. MET is a receptor tyrosine kinase normally expressed mainly in epithelial cells. Its ligand, HGF (hepatocyte growth factor/scatter factor), a glycoprotein secreted mainly by peri-epithelial mesenchymal cells, is capable of paracrine induction of tubular or glandular differentiation in epithelial cells. Co-expression of MET and HGF in the same cells is unusual and probably nonphysiological. By in situ hybridization, Kuhnen and colleagues 56 found MET and HGF co-expressed in the epithelial cells of all biphasic synovial sarcomas and in the spindle cells of six of nine monophasic cases. We have also confirmed the prominent expression of MET in the glandular areas of biphasic synovial sarcomas by IHC (CRA and ML, unpublished results). In the breast, MET is required for tubulogenesis, whereas ERBB2 is required for lobuloalveolar differentiation. 57 HGF and MET (HGF receptor) can induce heregulin, a ligand of ERBB2 heterodimers. 58
In breast cancer, aberrant ERBB2 overexpression is associated with cell cycle deregulation because of, at least in part, up-regulation and posttranslational stabilization of CCND1, 55 as well as chemoresistance mediated through the anti-apoptotic effects of increased p53 degradation 59 and p21 up-regulation. 55 In contrast, studies of synovial sarcomas indicate that the epithelial component, found in the present study to specifically express ERBB2, has a notably lower mitotic rate than the spindle cell component. 33 In synovial sarcoma, BCL2 is expressed predominantly in the spindle cells. 33 The reciprocal relationship of BCL2 and ERBB2 expression in synovial sarcoma is also intriguing because it suggests the hypothesis that ERBB2 expression may compensate for the absence of BCL2 in the epithelial component by the anti-apoptotic effects of the p53 and p21 deregulation outlined above. KIT has recently been reported as another potentially anti-apoptotic gene specifically expressed in the epithelial cells of synovial sarcoma. 60 Aside from its fundamental interest, the finding of strong ERBB2 in a subset of synovial sarcomas also provides a biological basis for evaluating Herceptin (trastuzumab) as a potential therapeutic agent, at least in model systems (xenografts, cell lines).
Our cDNA array analysis also detected a limited set of differentially expressed genes between mono- and biphasic synovial sarcomas (Figure 1c) ▶ . Among the most significantly expressed genes in biphasic tumors are the genes encoding keratins 14, 8, 5, and 7. According to our cDNA microarray data, these keratins are more abundant in biphasic tumors, which is explained by the glandular epithelial component in these tumors. Our results are in total agreement with a recent report showing extensive IHC reactivity for keratins 14, 8, and 7 in biphasic synovial sarcomas. 61 We also detected keratin 5 expression in biphasic tumors, which has previously not been specifically reported.
ELF3, E74-like factor 3 (also known as ESE-1 or ESX), was also found to be overexpressed in biphasic tumors (Figure 1c) ▶ . Interestingly, ELF3 is an ets-family transcription factor identified as a critical regulator of epithelial differentiation. 62 Indeed, it is one of only a few transcription factors known to be expressed specifically in epithelial cells of different types, but not in hematopoietic cells. In situ hybridization has confirmed its expression in the epithelium of ductules and lobules in breast tissue. 63 Its expression in biphasic synovial sarcomas would be consistent with their prominent epithelial differentiation. Notably, the genes for keratins 8 and 18 contain ETS-binding sites in their promoters and have been proposed as direct or indirect targets of ELF3. 62 Furthermore, ELF3 is up-regulated by ERBB2 signaling and may in turn bind and transactivate the ERBB2 promoter. 64,65 Thus, our results, taken together with previous data on MET and HGF expression in synovial sarcoma, suggest the presence of a coordinated glandular epithelial differentiation program in these tumors, possibly involving cooperative interactions between ELF3, the MET/HGF pathway, and ERBB2 signaling. Recent data also suggest a role for down-regulation or mutational inactivation of E-cadherin in the reversion from epithelial to spindle cell phenotype in at least some cases of synovial sarcoma. 66
Finally, we note the differential expression of several genes preferentially expressed in neural or neuroectodermal cells including OLFM1 28 , TLE2, 29 CNTNAP1, 30 and DRPLA 31 in synovial sarcomas relative to the comparison sarcoma group. Although their expression in synovial sarcoma tumor tissue remains to be localized, the observation is intriguing given a previous report of immunohistochemical evidence of neuroectodermal differentiation in synovial sarcoma. 18 OLFM1 (also known as NOE1) was also found to be significantly expressed in Ewing’s sarcoma and neuroblastoma in a previous expression profiling study from our group. 23
Our high-throughput studies of gene expression patterns, complemented by TMA studies, confirm the distinctive expression profile of synovial sarcoma, provide leads for the study of glandular morphogenesis in this tumor, reveal candidate targets for transcriptional deregulation by the SYT-SSX fusion proteins, and identify a possible novel therapeutic target, ERBB2, in a subset of cases. More broadly, the present results further strengthen the notion that specific translocation-derived chimeric transcription factors are associated with unique gene expression profiles, a concept first established in alveolar rhabdomyosarcoma (which contains the PAX3-FKHR fusion), 22,39 and more recently confirmed in other cancers with chromosomal translocations. 23,67
Finally, it is interesting to compare our results with those of Nielsen and colleagues, 68 published after the present study was completed. They studied eight synovial sarcomas and compared them to gastrointestinal stromal tumors, schwannomas, leiomyosarcomas, malignant fibrous histiocytomas, and liposarcomas. Synovial sarcoma-associated genes shared by their study and the present include CRABP1, PRAME, and OLFM1 (also known as NOE1) (see Figure 3 and web Table 3 in Nielsen et al 68 ), suggesting that these may be especially robust features of the synovial sarcoma expression profile. However, most of the gene lists were nonoverlapping. For instance, some key findings of the two studies appear discordant, but the differences can be at least partly explained. Thus, Nielsen and colleagues 68 observed prominent EGFR expression in synovial sarcoma. Although not as notable in our study, EGFR did indeed show higher expression among synovial sarcomas in our cDNA microarray data, but did not rank among the 50 most discriminating genes. Regarding the apparent discordance in ERBB2 data between the two studies, it should be noted that the study of Nielsen and colleagues 68 included only monophasic synovial sarcomas, thereby explaining why this component of the gene expression profile (and IGFBP2 and ELF3) did not figure prominently in their results. These differences in specific expression profiles highlight the strong impact of differences in study design and study groups. It is also notable that aside from OLFM1, Nielsen and colleagues 68 also found differential expression of several other potential neural-associated genes in synovial sarcoma (see Figure 3 and web Table 3 in Nielsen et al 68 ), namely ZIC2 (zinc finger protein of cerebellum 2), ATSV (axonal transporter of synaptic vesicles), NSP (neuroendocrine-specific protein), and NOGOR (nogo receptor). These data along with similar data from our study discussed above suggest that the potential neuroectodermal phenotype of synovial sarcoma may warrant closer examination.
Acknowledgments
We thank Kimberly A. Gayton, John Leuders, and Robert L. Walker (National Human Genome Research Institute) and Irina Linkov (Memorial Sloan-Kettering Cancer Center) for excellent technical assistance; and Carlos Cordon-Cardo (Memorial Sloan-Kettering Cancer Center) for providing sections of a high-grade soft tissue sarcoma TMA.
Footnotes
Address reprint requests to Marc Ladanyi, M.D., Memorial Sloan-Kettering Cancer Center, Department of Pathology, 1275 York Ave., New York, NY 10021. E-mail: ladanyim@mskcc.org.
Supported by a fellowship grant from the Swedish Medical Research Council (to S. V. A.), and by grant PO1 CA47179 from the National Institutes of Health (to M. L.).
Present address of S. V. A.: Wyeth Nordic CR&D, Dalvägen 12, SE-171 36 Solna, Sweden. E-mail: allands@war.wyeth.com.
References
- 1.Lewis JJ, Antonescu CR, Leung DH, Blumberg D, Healey JH, Woodruff JM, Brennan MF: Synovial sarcoma: a multivariate analysis of prognostic factors in 112 patients with primary localized tumors of the extremity. J Clin Oncol 2000, 18:2087-2094 [DOI] [PubMed] [Google Scholar]
- 2.Spillane AJ, A’Hern R, Judson IR, Fisher C, Thomas JM: Synovial sarcoma: a clinicopathologic, staging, and prognostic assessment. J Clin Oncol 2000, 18:3794-3803 [DOI] [PubMed] [Google Scholar]
- 3.Trassard M, Le DV, Hacene K, Terrier P, Ranchere D, Guillou L, Fiche M, Collin F, Vilain MO, Bertrand G, Jacquemier J, Sastre-Garau X, Bui NB, Bonichon F, Coindre JM: Prognostic factors in localized primary synovial sarcoma: a multicenter study of 128 adult patients. J Clin Oncol 2001, 19:525-534 [DOI] [PubMed] [Google Scholar]
- 4.Rosen G, Forscher C, Lowenbraun S, Eilber F, Eckardt J, Holmes C, Fu YS: Synovial sarcoma: uniform response of metastases to high dose ifosfamide. Cancer 1994, 73:2506-2511 [DOI] [PubMed] [Google Scholar]
- 5.Clark J, Rocques PJ, Crew AJ, Gill S, Shipley J, Chan AML, Gusterson BA, Cooper CS: Identification of novel genes, SYT and SSX, involved in t(X;18)(p112;q112) translocation found in human synovial sarcoma. Nat Genet 1994, 7:502-508 [DOI] [PubMed] [Google Scholar]
- 6.De Leeuw B, Balemans M, Olde Weghuis D, Geurts van Kessel A: Identification of two alternative fusion genes, SYT-SSX1 and SYT-SSX2, in t(X;18)(p112;q112)-positive synovial sarcomas. Hum Mol Genet 1995, 4:1097-1099 [DOI] [PubMed] [Google Scholar]
- 7.Crew AJ, Clark J, Fisher C, Gill S, Grimer R, Chand A, Shipley J, Gusterson BA, Cooper CS: Fusion of SYT to two genes, SSX1 and SSX2, encoding proteins with homology to the Kruppel-associated box in human synovial sarcoma. EMBO J 1995, 14:2333-2340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Skytting B, Nilsson G, Brodin B, Xie YT, Lundeberg J, Uhlen M, Larsson O: A novel fusion gene, SYT-SSX4, in synovial sarcoma. J Natl Cancer Inst 1999, 91:974-975 [DOI] [PubMed] [Google Scholar]
- 9.Ladanyi M, Bridge JA: Contribution of molecular genetic data to the classification of sarcomas. Hum Pathol 2000, 31:532-538 [DOI] [PubMed] [Google Scholar]
- 10.dos Santos NR, de Bruijn DR, Geurts van Kessel A: Molecular mechanisms underlying human synovial sarcoma development. Genes Chromosom Cancer 2001, 30:1-14 [DOI] [PubMed] [Google Scholar]
- 11.Ladanyi M: Fusions of the SYT and SSX genes in synovial sarcoma. Oncogene 2001, 20:5755-5762 [DOI] [PubMed] [Google Scholar]
- 12.Kato H, Tjernberg A, Zhang W, Krutchinsky A, An W, Takeuchi T, Ohtsuki Y, Sugano S, Chait B, Roeder RG: SYT associates with human SNF/SWI complexes and the C-terminal region of its fusion partner SSX1 targets histones. J Biol Chem 2002, 277:5498-5505 [DOI] [PubMed] [Google Scholar]
- 13.Hiraga H, Nojima T, Abe S, Sawa H, Yamashiro K, Yamawaki S, Kaneda K, Nagashima K: Diagnosis of synovial sarcoma with reverse transcriptase-polymerase chain reaction: analyses of 84 soft tissue and bone tumors. Diagn Mol Pathol 1998, 7:102-110 [DOI] [PubMed] [Google Scholar]
- 14.Birdsall S, Osin P, Lu YJ, Fisher C, Shipley J: Synovial sarcoma specific translocation is associated with both epithelial and spindle cell components. Int J Cancer 1999, 82:605-608 [DOI] [PubMed] [Google Scholar]
- 15.Kasai T, Shimajiri S, Hashimoto H: Detection of SYT-SSX fusion transcripts in both epithelial and spindle cell areas of biphasic synovial sarcoma using laser capture microdissection. Mol Pathol 2000, 53:107-110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noguchi SI, Ueki T, Kawauchi S, Fukuda T, Matsuura H, Sonoda T, Tsuneyoshi M: Establishment and characterization of a new synovial sarcoma cell line, SN-SY-1: special reference to bcl-2 protein and SYT-SSX1 hybrid transcripts. Int J Cancer 1997, 72:995-1002 [DOI] [PubMed] [Google Scholar]
- 17.Yakushiji T, Yonemura K, Tsuruta J, Nishida K, Kato T, Takagi K: Capacity for epithelial differentiation in synovial sarcoma: analysis of a new human cell line. J Clin Pathol 2000, 53:525-531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noguera R, Navarro S, Cremades A, Rosello-Sastre E, Pellin A, Peydro-Olaya A, Llombart-Bosch A: Translocation (X;18) in a biphasic synovial sarcoma with morphologic features of neural differentiation. Diagn Mol Pathol 1998, 7:16-23 [DOI] [PubMed] [Google Scholar]
- 19.Kawai A, Woodruff J, Healey JH, Brennan MF, Antonescu CR, Ladanyi M: SYT-SSX gene fusion as a determinant of morphology and prognosis in synovial sarcoma. N Engl J Med 1998, 338:153-160 [DOI] [PubMed] [Google Scholar]
- 20.Roberts WM, Douglass EC, Peiper SC, Houghton PJ, Look AT: Amplification of the gli gene in childhood sarcomas. Cancer Res 1989, 49:5407-5413 [PubMed] [Google Scholar]
- 21.De Risi J, Penland L, Brown PO, Bittner ML, Meltzer PS, Ray M, Chen Y, Su YA, Trent JM: Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nat Genet 1996, 14:457-460 [DOI] [PubMed] [Google Scholar]
- 22.Khan J, Simon R, Bittner M, Chen Y, Leighton SB, Pohida T, Smith PD, Jiang Y, Gooden GC, Trent JM, Meltzer PS: Gene expression profiling of alveolar rhabdomyosarcoma with cDNA microarrays. Cancer Res 1998, 58:5009-5013 [PubMed] [Google Scholar]
- 23.Khan J, Wei J, Ringnér M, Saal LH, Ladanyi M, Westermann F, Berthold F, Schwab M, Antonescu CR, Peterson C, Meltzer PS: Classification and diagnostic prediction of cancers using gene expression profiling and artificial neural networks. Nat Med 2001, 7:673-679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Y, Dougherty ER, Bittner M: Ratio-based decisions and the quantitative analysis of cDNA microarray images. Biomed Opt 1997, 2:364-374 [DOI] [PubMed] [Google Scholar]
- 25.Bittner M, Meltzer P, Trent J: Data analysis and integration: of steps and arrows. Nat Genet 1999, 22:213-215 [DOI] [PubMed] [Google Scholar]
- 26.Bittner M, Meltzer P, Chen Y, Jiang Y, Seftor E, Hendrix M, Radmacher M, Simon R, Yakhini Z, Ben-Dor A, Sampas N, Dougherty E, Wang E, Marincola F, Gooden C, Lueders J, Glatfelter A, Pollock P, Carpten J, Gillanders E, Leja D, Dietrich K, Beaudry C, Berens M, Alberts D, Sondak V, Hayward N, Trent J: Molecular classification of cutaneous malignant melanoma by gene expression profiling. Nature 2000, 406:536-540 [DOI] [PubMed] [Google Scholar]
- 27.Hoos A, Stojadinovic A, Mastorides S, Urist MJ, Polsky D, Di Como CJ, Brennan MF, Cordon-Cardo C: High Ki-67 proliferative index predicts disease specific survival in patients with high-risk soft tissue sarcomas. Cancer 2001, 92:869-874 [DOI] [PubMed] [Google Scholar]
- 28.Barembaum M, Moreno TA, LaBonne C, Sechrist J, Bronner-Fraser M: Noelin-1 is a secreted glycoprotein involved in generation of the neural crest. Nat Cell Biol 2000, 2:219-225 [DOI] [PubMed] [Google Scholar]
- 29.Grbavec D, Lo R, Liu Y, Stifani S: Transducin-like Enhancer of split 2, a mammalian homologue of Drosophila Groucho, acts as a transcriptional repressor, interacts with Hairy/Enhancer of split proteins, and is expressed during neuronal development. Eur J Biochem 1998, 258:339-349 [DOI] [PubMed] [Google Scholar]
- 30.Rios JC, Melendez-Vasquez CV, Einheber S, Lustig M, Grumet M, Hemperly J, Peles E, Salzer JL: Contactin-associated protein (Caspr) and contactin form a complex that is targeted to the paranodal junctions during myelination. J Neurosci 2000, 20:8354-8364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knight SP, Richardson MM, Osmand AP, Stakkestad A, Potter NT: Expression and distribution of the dentatorubral-pallidoluysian atrophy gene product (atrophin-1/drplap) in neuronal and non-neuronal tissues. J Neurol Sci 1997, 146:19-26 [DOI] [PubMed] [Google Scholar]
- 32.van Baren N, Chambost H, Ferrant A, Michaux L, Ikeda H, Millard I, Olive D, Boon T, Coulie PG: PRAME, a gene encoding an antigen recognized on a human melanoma by cytolytic T cells, is expressed in acute leukaemia cells. Br J Haematol 1998, 102:1376-1379 [DOI] [PubMed] [Google Scholar]
- 33.Antonescu CR, Kawai A, Leung DH, Lonardo F, Woodruff JM, Healey JH, Ladanyi M: Strong association of SYT-SSX fusion type and morphologic epithelial differentiation in synovial sarcoma. Diagn Mol Pathol 2000, 9:1-8 [DOI] [PubMed] [Google Scholar]
- 34.Ladanyi M, Antonescu CR, Leung DH, Woodruff JM, Kawai A, Healey JH, Brennan MF, Bridge JA, Neff JR, Barr FG, Goldsmith JD, Brooks JSJ, Goldblum JR, Ali SZ, Shipley JM, Cooper CS, Fisher C, Skytting B, Larsson O: Impact of SYT-SSX fusion type on the clinical behavior of synovial sarcoma. A multi-institutional retrospective study of 243 patients. Cancer Res 2002, 62:135-140 [PubMed] [Google Scholar]
- 35.Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, Bloomfield CD, Lander ES: Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 1999, 286:531-537 [DOI] [PubMed] [Google Scholar]
- 36.Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, Powell JI, Yang L, Marti GE, Moore T, Hudson JJ, Lu L, Lewis DB, Tibshirani R, Sherlock G, Chan WC, Greiner TC, Weisenburger DD, Armitage JO, Warnke R, Staudt LM: Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 2000, 403:503-511 [DOI] [PubMed] [Google Scholar]
- 37.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D: Molecular portraits of human breast tumours. Nature 2000, 406:747-752 [DOI] [PubMed] [Google Scholar]
- 38.Werner H, Le Roith D: New concepts in regulation and function of the insulin-like growth factors: implications for understanding normal growth and neoplasia. Cell Mol Life Sci 2000, 57:932-942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khan J, Bittner ML, Saal LH, Teichmann U, Azorsa DO, Gooden GC, Pavan WJ, Trent JM, Meltzer PS: cDNA microarrays detect activation of a myogenic transcription program by the PAX3-FKHR fusion oncogene. Proc Natl Acad Sci USA 1999, 96:13264-13269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang W, Kumar P, Wang W, Epstein J, Helman L, Moore JV, Kumar S: Insulin-like growth factor II and PAX3-FKHR cooperate in the oncogenesis of rhabdomyosarcoma. Cancer Res 1998, 58:4426-4433 [PubMed] [Google Scholar]
- 41.Xie Y, Skytting B, Nilsson G, Brodin B, Larsson O: Expression of insulin-like growth factor-1 receptor in synovial sarcoma: association with an aggressive phenotype. Cancer Res 1999, 59:3588-3591 [PubMed] [Google Scholar]
- 42.Scotlandi K, Benini S, Sarti M, Serra M, Lollini PL, Maurici D, Picci P, Manara MC, Baldini N: Insulin-like growth factor I receptor-mediated circuit in Ewing’s sarcoma/peripheral neuroectodermal tumor: a possible therapeutic target. Cancer Res 1996, 56:4570-4574 [PubMed] [Google Scholar]
- 43.Toretsky JA, Kalebic T, Blakesley V, LeRoith D, Helman LJ: The insulin-like growth factor-I receptor is required for EWS/FLI-1 transformation of fibroblasts. J Biol Chem 1997, 272:30822-30827 [DOI] [PubMed] [Google Scholar]
- 44.Hoeflich A, Reisinger R, Lahm H, Kiess W, Blum WF, Kolb HJ, Weber MM, Wolf E: Insulin-like growth factor-binding protein 2 in tumorigenesis: protector or promoter? Cancer Res 2001, 61:8601-8610 [PubMed] [Google Scholar]
- 45.Streck RD, Wood TL, Hsu MS, Pintar JE: Insulin-like growth factor I and II and insulin-like growth factor binding protein-2 RNAs are expressed in adjacent tissues within rat embryonic and fetal limbs. Dev Biol 1992, 151:586-596 [DOI] [PubMed] [Google Scholar]
- 46.Roholl PJ, Skottner A, Prinsen I, Lips CJ, Den Otter W, Van Unnik JA: Expression of insulin-like growth factor 1 in sarcomas. Histopathology 1990, 16:455-460 [DOI] [PubMed] [Google Scholar]
- 47.Bubendorf L, Kolmer M, Kononen J, Koivisto P, Mousses S, Chen Y, Mahlamaki E, Schraml P, Moch H, Willi N, Elkahloun AG, Pretlow TG, Gasser TC, Mihatsch MJ, Sauter G, Kallioniemi OP: Hormone therapy failure in human prostate cancer: analysis by complementary DNA and tissue microarrays. J Natl Cancer Inst 1999, 91:1758-1764 [DOI] [PubMed] [Google Scholar]
- 48.Fuller GN, Rhee CH, Hess KR, Caskey LS, Wang R, Bruner JM, Yung WK, Zhang W: Reactivation of insulin-like growth factor binding protein 2 expression in glioblastoma multiforme: a revelation by parallel gene expression profiling. Cancer Res 1999, 59:4228-4232 [PubMed] [Google Scholar]
- 49.Sallinen SL, Sallinen PK, Haapasalo HK, Helin HJ, Helen PT, Schraml P, Kallioniemi OP, Kononen J: Identification of differentially expressed genes in human gliomas by DNA microarray and tissue chip techniques. Cancer Res 2000, 60:6617-6622 [PubMed] [Google Scholar]
- 50.Menouny M, Binoux M, Babajko S: IGFBP-2 expression in a human cell line is associated with increased IGFBP-3 proteolysis, decreased IGFBP-1 expression and increased tumorigenicity. Int J Cancer 1998, 77:874-879 [DOI] [PubMed] [Google Scholar]
- 51.Renehan AG, Painter JE, O’Halloran D, Atkin WS, Potten CS, O’Dwyer ST, Shalet SM: Circulating insulin-like growth factor II and colorectal adenomas. J Clin Endocrinol Metab 2000, 85:3402-3408 [DOI] [PubMed] [Google Scholar]
- 52.Wex H, Vorwerk P, Mohnike K, Bretschneider D, Kluba U, Aumann V, Blum WF, Mittler U: Elevated serum levels of IGFBP-2 found in children suffering from acute leukaemia is accompanied by the occurrence of IGFBP-2 mRNA in the tumour clone. Br J Cancer 1998, 78:515-520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Duda RB, Cundiff D, August CZ, Wagman LD, Bauer KD: Growth factor receptor and related oncogene determination in mesenchymal tumors. Cancer 1993, 71:3526-3530 [DOI] [PubMed] [Google Scholar]
- 54.George E, Niehans GA, Swanson PE, Strickler JG, Singleton TP: Overexpression of the c-erbB-2 oncogene in sarcomas and small round-cell tumors of childhood. An immunohistochemical investigation. Arch Pathol Lab Med 1992, 116:1033-1035 [PubMed] [Google Scholar]
- 55.Harari D, Yarden Y: Molecular mechanisms underlying ErbB2/HER2 action in breast cancer. Oncogene 2000, 19:6102-6114 [DOI] [PubMed] [Google Scholar]
- 56.Kuhnen C, Tolnay E, Steinau HU, Voss B, Muller KM: Expression of c-Met receptor and hepatocyte growth factor/scatter factor in synovial sarcoma and epithelioid sarcoma. Virchows Arch 1998, 432:337-342 [DOI] [PubMed] [Google Scholar]
- 57.Yang Y, Spitzer E, Meyer D, Sachs M, Niemann C, Hartmann G, Weidner KM, Birchmeier C, Birchmeier W: Sequential requirement of hepatocyte growth factor and neuregulin in the morphogenesis and differentiation of the mammary gland. J Cell Biol 1995, 131:215-226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Castagnino P, Lorenzi MV, Yeh J, Breckenridge D, Sakata H, Munz B, Werner S, Bottaro DP: Neu differentiation factor/heregulin induction by hepatocyte and keratinocyte growth factors. Oncogene 2000, 19:640-648 [DOI] [PubMed] [Google Scholar]
- 59.Zhou BP, Liao Y, Xia W, Zou Y, Spohn B, Hung MC: HER-2/neu induces p53 ubiquitination via Akt-mediated MDM2 phosphorylation. Nat Cell Biol 2001, 3:973-982 [DOI] [PubMed] [Google Scholar]
- 60.Tamborini E, Papini D, Mezzelani A, Riva C, Azzarelli A, Sozzi G, Pierotti MA, Pilotti S: c-KIT and c-KIT ligand (SCF) in synovial sarcoma (SS): an mRNA expression analysis in 23 cases. Br J Cancer 2001, 85:405-411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miettinen M, Limon J, Niezabitowski A, Lasota J: Patterns of keratin polypeptides in 110 biphasic, monophasic, and poorly differentiated synovial sarcomas. Virchows Arch 2000, 437:275-283 [DOI] [PubMed] [Google Scholar]
- 62.Oettgen P, Alani RM, Barcinski MA, Brown L, Akbarali Y, Boltax J, Kunsch C, Munger K, Libermann TA: Isolation and characterization of a novel epithelium-specific transcription factor, ESE-1, a member of the ets family Mol Cell Biol 1997, 17:4419-4433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thomas RS, Ng AN, Zhou J, Tymms MJ, Doppler W, Kola I: The Elf group of Ets-related transcription factors. ELF3 and ELF5. Adv Exp Med Biol 2000, 480:123-128 [DOI] [PubMed] [Google Scholar]
- 64.Chang CH, Scott GK, Kuo WL, Xiong X, Suzdaltseva Y, Park JW, Sayre P, Erny K, Collins C, Gray JW, Benz CC: ESX: a structurally unique Ets overexpressed early during human breast tumorigenesis. Oncogene 1997, 14:1617-1622 [DOI] [PubMed] [Google Scholar]
- 65.Neve RM, Ylstra B, Chang CH, Albertson DG, Benz CC: ErbB2 activation of ESX gene expression. Oncogene 2002, 21:3934-3938 [DOI] [PubMed] [Google Scholar]
- 66.Saito T, Oda Y, Sugimachi K, Kawaguchi KK, Tamiya S, Tanaka K, Matsuda S, Sakamoto A, Iwamoto Y, Tsuneyoshi M: E-cadherin gene mutations frequently occur in synovial sarcoma as a determinant of histological features. Am J Pathol 2001, 159:2117-2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Armstrong SA, Staunton JE, Silverman LB, Pieters R, den Boer ML, Minden MD, Sallan SE, Lander ES, Golub TR, Korsmeyer SJ: MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet 2002, 30:41-47 [DOI] [PubMed] [Google Scholar]
- 68.Nielsen TO, West RB, Linn SC, Alter O, Knowling MA, O’Connell JX, Zhu S, Fero M, Sherlock G, Pollack JR, Brown PO, Botstein D, van de Rijn M: Molecular characterisation of soft tissue tumours: a gene expression study. Lancet 2002, 359:1301-1307 [DOI] [PubMed] [Google Scholar]


