Abstract
Caveolin-1, a 21- to 24-kd integral membrane protein, is primarily implicated as a tumor suppressor gene. Transformed cells normally contain reduced or no caveolin-1. Re-expression of caveolin-1 is found in advanced human and mouse prostate adenocarcinomas. To explore its potential role in tumorigenesis and tumor progression of human lung cancers, we used the well-characterized cell line (CL) series of lung adenocarcinoma cells with increasing cellular invasiveness to show that expression of caveolin-1 mRNA and protein was up-regulated with enhanced invasion/metastatic capability of CL cells. Reintroducing the caveolin-1 gene into the less invasive, caveolin-1-negative CL cells enhanced their invasive capability at least by twofold, as revealed by an in vitro chamber invasion assay. Thus, a correlation exists for both constitutive and induced expression of caveolin-1 in CL cells. Immunohistochemical examination of caveolin-1 was performed in 95 specimens obtained retrospectively from patients who had lung adenocarcinoma either with (35 patients) or without (60 patients) ipsilateral hilar/peribronchial tumor-metastasized lymph nodes. Caveolin-1 immunoreactivity was either totally absent or just barely detectable in a few lung adenocarcinoma cells from cases diagnosed as lung adenocarcinoma without regional lymph node metastasis. In contrast, increased caveolin-1 immunoreactivity both in number and intensity was detected in primary lung adenocarcinoma cells as well as in cancer cells that metastasized to regional lymph nodes from the cases diagnosed as advanced lung adenocarcinoma with nodal metastases. Multivariate analysis considering caveolin-1 immunoreactivity in addition to the established prognostic parameters such as pT stage, pN in these patients confirmed that caveolin-1 is an independent functional predictor of poor survival. We further revealed that up-regulated caveolin-1 in CL cells is necessary for mediating filopodia formation, which may enhance the invasive ability of lung adenocarcinoma cells.
Caveolin-1, a 21- to 24-kd protein, is the principal component of caveolae, which are special invaginated microdomains of the plasma membrane present in most mammalian cells. 1 It is well established that caveolin-1 is a tumor suppressor gene. Caveolin-1 mRNA and protein expression are frequently lost in human cancer cell lines. Re-expression of caveolin-1 in oncogenically transformed cell lines inhibits tumor cell growth and reduces tumorigenicity. 2-6 Several mechanisms have been proposed for caveolin-1 to function as a tumor suppressor. Caveolin-1 may exert its tumor-growth inhibition by contact inactivation of signaling molecules such as v-src, Ha-Ras, protein kinase A, PKC, and p42/44 MAP kinase within caveolae. 7-10 In addition, down-regulation of caveolin-1 in colon carcinoma cells has been shown to prevent the degradation of inducible nitric oxide synthase via the proteosome pathway, which, in turn, increases the local nitric oxide concentration to facilitate tumorigenesis. 11
Caveolin-1 can also function as a tumor metastasis-promoting molecule, which is unrelated to its obvious function of cell growth inhibition. 12 Elevated expression of caveolin-1 is found to be associated with progression of prostate, colon, and breast carcinoma. 13,14 Inhibition of c-myc-induced apoptosis by caveolin-1 was recently proposed to promote progression of prostate cancer, 15 and it may serve as a prognostic indicator for these patients. 16 Nonetheless, it still remains unclear whether and how caveolin-1 can potentiate tumor progression in other types of human cancer.
In the present study, we investigated the expression pattern of caveolin-1 both in a series of lung carcinoma cell lines (CLs) with varying invasive/metastatic ability 17 and in 95 paraffin-embedded specimens of clinically well-defined and pathologically proven lung adenocarcinoma. Stably transfected cell lines with the induced or constitutively expressed full length of the caveolin-1 gene were established, which were then used for comparison of caveolin-1-induced cell motility and/or invasiveness. Our results suggest that caveolin-1 expression could enhance the invasive capability of lung adenocarcinoma cells by promoting filopodia formation.
Materials and Methods
Cell Culture
CL series of cell lines were established by selection of increasingly invasive cancer cell populations from a clonal cell line of human lung adenocarcinoma, CL1, via a Transwell invasion chamber assay. The invasive capability is increased in the order CL1-0 < CL1-1 < CL1-5 < CL1-5F4. 17,18 CL1E-9 was a cell line selected from CL1-0 cells with a stably transfected pTet-Off vector (a gift from Dr. Steven R. Roffler, Academia Sinica, Taipei, Taiwan).
Cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% heat-inactivated fetal bovine serum and 2 mmol/L of l-glutamine at 37°C, 5% CO2, in a humidified incubator. In separate studies, charcoal-stripped (delipidated) fetal bovine serum (Sigma, St. Louis, MO) was used in the lipid depletion assay.
An SV-40-transformed, nontumorigenic bronchial epithelial cell line, BEAS-2B (CRL9609; American Type Culture Collection, Rockville, MD), was grown in modified F12 medium admixed with Hepes stock solution (1.5 mol/L, pH ∼7.2 to 7.4) and supplemented with growth factors as previously described. 19
Sodiuim Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis and Western Blotting
The cells (1 × 106) were harvested and prepared by application of 500 μl of boiling 2× concentrated electrophoresis sample buffer (125 mmol/L Tris-HCl, pH 6.8, 2% sodium dodecyl sulfate, 5% glycerol, 0.003% bromophenol blue, and 1% β-mercaptoethanol) to each 10-cm-diameter dish. Protein samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membrane, which was subjected to immunoblotting by anti-human caveolin-1 antibody (catalogue number C37120, a mouse monoclonal antibody, 1:1000 dilution; BD Transduction Laboratories, Lexington, KY), phospho-caveolin-1 (pY14) antibody (catalogue number 611582, a mouse monoclonal antibody, 1:2500 dilution; BD Transduction Laboratories), or anti-human Grb7 antibody (catalogue number sc-607, a rabbit polyclonal antibody, 1:500 dilution; Santa Cruz Biotechnology Inc., Santa Cruz, CA), followed by blotting with horseradish peroxidase-conjugated horse anti-mouse or horse anti-rabbit antibody (1:3500 dilution; Amersham, Buckinghamshire, UK). An enhanced chemiluminescence reaction (Amersham) was applied for signal detection. Immunoblotting with anti-human α-tubulin antibody (1:1000 dilution; Oncogene, Darmstadt, Germany) was used as internal marker for both quantity and quality control.
Southern and Northern Analysis
For Southern blotting, genomic DNA was extracted with a NucleoBond Nucleic Acid purification kit (Clontech Laboratories, Palo Alto, CA). The DNA was digested with EcoRI or HindIII, subjected to gel electrophoresis, transferred to nylon membrane, and blotted with a 32P-radiolabeled human caveolin-1 DNA probe.
For Northern blotting, total RNA was extracted from exponentially growing cells by use of Trizol reagent (Life Technologies, Inc., Rockville, MD). Twenty μg of total RNA per lane was denatured, electrophoresed through 1% agarose-formaldehyde gels, transferred to nylon membrane, and hybridized by a 32P-radiolabeled human caveolin-1 DNA probe. After several washes, the blot was exposed to X-ray film (Eastman Kodak Company, Rochester, NY) and developed. Hybridization with a 32P-radiolabeled human GAPDH DNA probe was performed as a control.
Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR) and DNA Sequencing
cDNA was synthesized from total RNA extracted from CL cells by use of a Superscript amplification kit (Life Technologies, Inc.). PCR was performed with primers flanking the amino-terminal and C-terminal sequence of the human caveolin-1 gene (forward primer: 5′-GGAATTCCACGGGCCAGCATGTCT-3′; reverse primer: 5′-GCTCTAGAGCTATTTC TTTCTGCAAGTTGATG-3′). The cDNA clones were sequenced by a dye termination sequencing method (ABI PRISM BigDye terminator cycle sequencing ready reaction kit; Applied Biosystems, Foster City, CA).
Microarray Analysis
A cDNA microarray and membranes were prepared as previously described. 20 Briefly, mRNA was extracted from each CL cell line and reverse-transcribed into cDNA labeled with biotin. The biotin-labeled cDNA was used as probes and hybridized with the membranes that contained 9600 nonredundant expressed sequence tag clones selected from human cDNA libraries. After stringent washing, a color reaction toward biotin was initiated and detected by colorimetric measurement.
Immunohistochemistry
Paraffin-embedded 5-μm-thick sections were deparaffinized, heated in citrate buffer (0.01 mol/L), treated with 0.3% H2O2, and rehydrated. After blocking, sections were incubated with anti-caveolin-1 IgG (1:1000, BD Transduction Laboratories) for 30 minutes at room temperature, washed, and then blotted with biotin-labeled bridge antiserum (1:200; Vector Laboratories, Burlingame, CA). After several washes with phosphate-buffered saline (PBS), sections were incubated with a solution of avidin and biotin-conjugated peroxidase complex (Vector Laboratories) for 30 minutes at room temperature. Diaminobenzidine colorization was applied, and slides were further counterstained with hematoxylin. After serial dehydration, slides were mounted for microscopic examination (Olympus, Tokyo, Japan). For each specimen, the entire population of cancer cells in the lung tissue and regional lymph nodes was scanned. All specimens were evaluated without knowledge of the patients’ clinical data.
Statistical Analysis
The correlation of caveolin-1 immunoreactivity with patients’ clinical variables was analyzed by chi-square and Fisher exact tests for categorical association. McNemar’s test was used for the trend of caveolin-1 expression in primary sites and nodal metastases. The survival-predictive value of possible predictors such as caveolin-1 immunoreactivity was tested univariately by use of a Kaplan-Meier estimate-of-survival curve and was compared by log-rank test. Survival analysis of caveolin-1 immunostaining and of other clinical variables was performed with the Cox proportional-hazards regression model. All analyses were done with statistics software (Statview version 5.0; SAS Institute, Cary, NC).
Plasmids
The full-length cDNA encoding human caveolin-1 was amplified by RT-PCR with caveolin-1-specific primers flanked by restriction sites and RNA isolated from surgically resected human lung tissue serving as template. The resulting cDNA was purified and then subcloned into the expression vector. Restriction mapping and DNA sequencing verified the correct orientation and sequence of the insert.
Plasmid pcDNA3.1/Myc-caveolin-1 was constructed by subcloning into pcDNA3.1/Myc-His B (Invitrogen, Carlsbad, CA) by use of EcoRI and XbaI sites, and the resulting c-Myc epitope tag was fused to the COOH terminus of caveolin-1. Plasmid pTRE2-caveolin-1, which allowed expression of caveolin-1 in the absence of tetracycline in transfected Tet-Off cells (CL1E-9, gift from Dr. Steven R. Roffler), was constructed by subcloning of the cDNA of caveolin-1 into the BamHI and XbaI restriction sites of pTRE-2 (Clontech).
Transfection and Selection of Stable Cell Lines
Parental CL1E-9 cells were co-transfected with pTRE2-caveolin-1 and pTK-Hyg containing hygromycin resistance by use of lipofectamine transfectants (Life Technologies, Inc.). After selection in medium supplemented with hygromycin B (200 μg/ml, Clontech), resistant colonies were isolated via trypsinization with cloning rings. Individual clones were screened for tetracycline-regulated expression of caveolin-1 by Western blot analysis of cells treated with varying concentrations of doxycycline (final concentration, 0.01 μg/ml to 20 μg/ml; Sigma).
In addition, caveolin-1 constitutive expression in CL1-0 cell line was established by transfection of plasmid pcDNA3.1/Myc-caveolin-1, and the stable clones were selected by geneticin, 800 μg/ml (G418; Life Technologies, Inc.). Mock-transfected CL1-0 cells were established by transfection of plasmid pcDNA3.1/Myc-His and were selected by 800 μg/ml of G418.
In Vitro Invasion Assay
We used a membrane invasion chamber system to investigate the invasive capability of CL cells transfected with Tet-Off inducible caveolin-1 constructs or pcDNA3.1/Myc-caveolin-1 plasmid. A polycarbonate membrane containing 10-μm-sized pores (Neuro Probe, Inc., Gaithersburg, MD) was coated with a mixture of laminin (50 μg/ml, Sigma), type IV collagen (50 μg/ml, Sigma), and gelatin solution (2 mg/ml in 10 mmol/L glacial acetic acid, Bio-Rad). The membrane was placed between the upper and lower well plates of a modified Boyden chamber (Neuro Probe, Inc.). Cells were suspended in Dulbecco’s modified Eagle’s medium plus 10% fetal bovine serum culture medium and were seeded into the upper wells at a density of 2 × 104 cells per well. After incubation for 48 hours at 37°C, the membrane was inverted and stained with Trypan blue, and the cell number was counted under a microscope. The cells in the lower chamber were also collected and counted.
Growth Curve
Cells (4 × 104) were evenly plated in 35-mm tissue culture dishes and incubated at 37°C on day 0. From the next day (first day) until the eighth day, the total cell number per dish was counted by trypsinization and measurement by Coulter counting. Three independent counts were made for each cell line.
Indirect Immunofluorescence
Cells were cultured on glass coverslips placed in 12-well culture dishes. After several rinses with PBS, cells were fixed in 4% paraformaldehyde for 10 minutes. Cells were then permeabilized in PBS containing 0.1% Triton X-100 and 0.05% sodium dodecyl sulfate for 4 minutes at 37°C, and blocked in PBS containing 0.2% bovine serum albumin and 0.1% saponin for 30 minutes at room temperature. For subcellular localization of caveolin-1, monoclonal mouse anti-human caveolin-1 antibody (1:1000 dilution, BD Transduction Laboratories) was used at room temperature for 1 hour, washed three times with PBS, and then incubated with fluoro-conjugated secondary antibody (1:1000; Molecular Probes, Eugene, OR) for another 40 minutes. For F-actin staining, 0.185 μmol/L of rhodamine phalloidin (Molecular Probes) was used. The results were observed under a Leica epifluorescence microscope.
Results
Elevated Caveolin-1 Expression in CL Cells Correlated with Enhanced Cell Invasive Ability
To investigate the biological role of caveolin-1 in the progression of lung adenocarcinoma, we analyzed its expression in CLs with varying invasive/metastatic ability. 17,18 As shown in Figure 1A ▶ , expression of caveolin-1 protein in the low-invasive CL1-0 and CL1-1 cells was undetectable. In contrast, it was abundantly expressed in the highly invasive CL1-5 and CL1-5F4 cells. We noted that both the α-isoform and the β-isoform of caveolin-1 were expressed in CL1-5 and CL1-5F4 cells.
Figure 1.
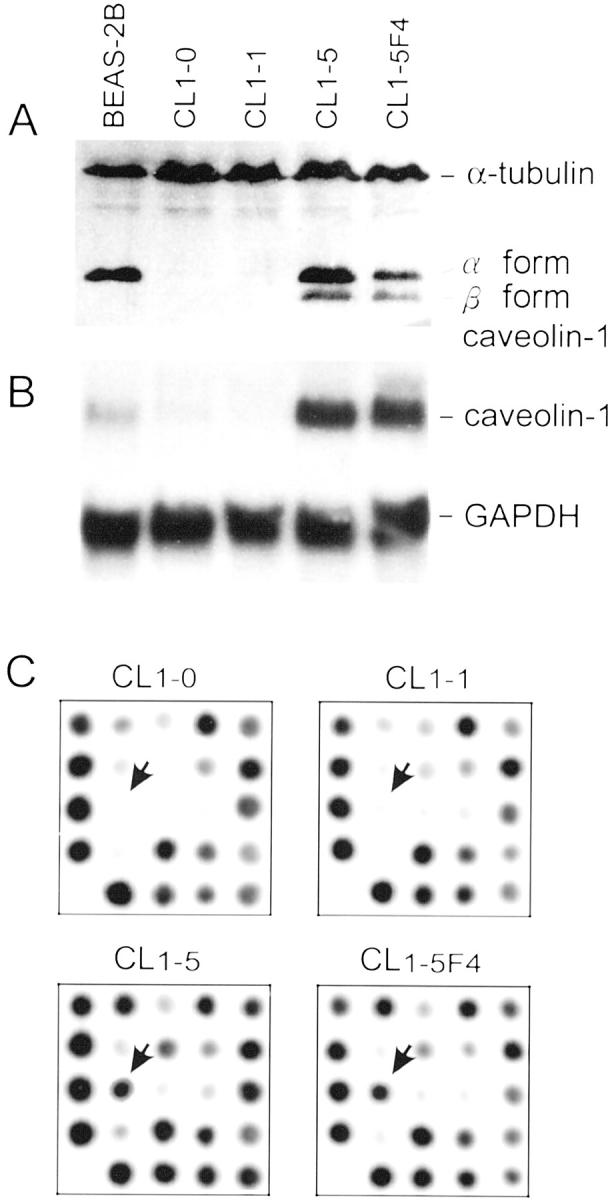
A: Expression of caveolin-1 protein in BEAS-2B cells, CL1-5, and CL1-5F4 cells, but not in CL1-0 and CL1-1 cells. Both the α-isoform and the β-isoform of caveolin-1 were present. Staining for α-tubulin was used as control. B: Northern blotting probed by full length of caveolin-1 cDNA. Moderate transcripts were seen in BEAS-2B cells, absent or low in CL1-0 and CL1-1 cells, and abundant in CL1-5 and CL1-5F4 cells. GAPDH blotting was used as control. C: Close-up view of microarray images. The arrows indicate that caveolin-1 was overexpressed in CL1-5 and CL1-5F4 cells.
Northern blot analysis showed that the expression of a caveolin-1 transcript of 3.0 kb was detected in CL1-5 and CL1-5F4 cells (ie, caveolin-1+ve), but not in CL1-0 and CL1-1 cells (ie, caveolin-1−ve) (Figure 1B) ▶ . A similar result was obtained with a DNA microarray (Figure 1C) ▶ . Taken together, the expression of caveolin-1 RNA and protein was positively associated with CL cells with enhanced invasive ability.
Next, we performed Southern blot analysis of the caveolin-1 gene to exclude the possibility of deletion, translocation, or gene amplification that might cause an increased caveolin-1 transcript. No global difference in the caveolin-1 genomic structure was found among the four CL cell lines. Sequencing analysis of the RT-PCR product of caveolin-1 derived from total RNA of these cell lines did not reveal any point mutation, insertion, or deletion of caveolin-1 transcripts.
Positive Correlation of Caveolin-1 Expression in Pathological Lesions of Human Primary Lung Adenocarcinoma with Hilar/Peribronchial Lymph Node Metastasis
To underscore the importance of the above-mentioned findings in CL cell lines, we examined the status of caveolin-1 expression in specimens obtained from patients with lung adenocarcinoma. In this retrospective study, 95 specimens diagnosed as moderately differentiated lung adenocarcinoma were obtained from the Department of Pathology, National Taiwan University Hospital, during the period of 1998 to 1999. Among the 95 lung cancer specimens, 35 cases (labeled as “P/N+”) already had tumor cells metastasizing to the ipsilateral hilar or peribronchial lymph nodes. The other 60 cases (labeled as “P/N−”) had tumor cells in the lung without evidence of tumor metastasis to regional lymph nodes.
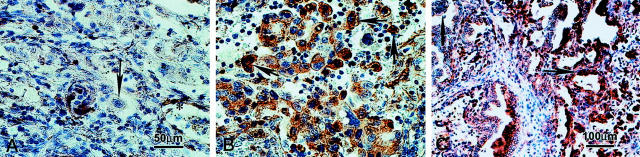
Caveolin-1 immunoreactivity was normally localized to fibroblasts, type I pneumocytes, and endothelial cells of blood vessels in all tissue specimens examined, which could serve as an internal quality control of immunohistochemistry. Caveolin-1 immunoreactivity, however, was not seen in primary lung adenocarcinoma cells in the great majority (66%, 62 of 95) of cases. In approximately 30% (29 of 95) of the cases, regardless of their status of tumor metastasis, caveolin-1 immunoreactivity with weak to moderate intensity was observed in a small percentage (ranging from 0 to 30% with a mean ∼5%) of primary lung adenocarcinoma cells (Figure 2A) ▶ . Only in 4% (4 of 95) of the cases did the percentage of caveolin-1-positive tumor cells at the primary lung lesion sites exceed 30%; all four of these patients happened to have metastatic tumor cells in lymph nodes.
Figure 2.
Caveolin-1 immunoreactivity in lung adenocarcinoma and ipsilateral hilar/peribronchial lymph nodes with tumor metastases. A: Negative or faint staining of caveolin-1 in the primary lung adenocarcinoma (arrow). B: Positive caveolin-1 staining was detected in cancer cells in a lymph node with metastatic lung adenocarcinoma. The caveolin-1-positive staining was in the cytoplasm, some with a granular pattern. C: Differential staining intensity of caveolin-1 in metastatic carcinoma cells with varying degrees of differentiation.
In contrast, immunohistochemical examination of ipsilateral hilar/peribronchial lymph nodes (35 cases) with metastatic lung adenocarcinoma (P/N+) revealed moderate to intense caveolin-1 immunoreactivity in the cytoplasm of varying percentages of cancer cells (ranging from 10 to 95%) in all lymph nodes except one (Figure 2B) ▶ . In 28.6% (10 of 35) of these cases (P/N+), the number of caveolin-1+ve tumor cells in lymph nodes exceeded 30% (ranging from 30 to 90%). The staining intensity in the tumor cells varied slightly, being weaker in the more differentiated glandular part of the tumor cells, but stronger in the poorly differentiated tumor cells (Figure 2C) ▶ .
Altogether, if we artificially chose 30% caveolin immunoreactivity as a cutoff value for the assignment of positivity (or negativity) of caveolin-1 staining, four cases had caveolin-1 positivity both at the primary lesion site and at the regional nodal metastasis. Six cases had caveolin-1 positivity in the nodal metastasis site, but not at the primary site. Twenty-five cases were caveolin-1-negative (ie, <30%) both at the primary lung lesion site and at nodal metastases. No cases had caveolin-1 positivity at the primary lung lesion site, but were caveolin-1-negative at the nodal metastasis site. McNemar’s test for the caveolin-1 expression in the primary site and nodal metastases analyzed in this way was statistically significant (P = 0.0313). Therefore, there was a trend toward caveolin-1 expression in regional nodal metastases rather than in the primary lung lesion site.
Caveolin-1 as a Predictor of Poor Survival of Patients with Lung Adenocarcinoma
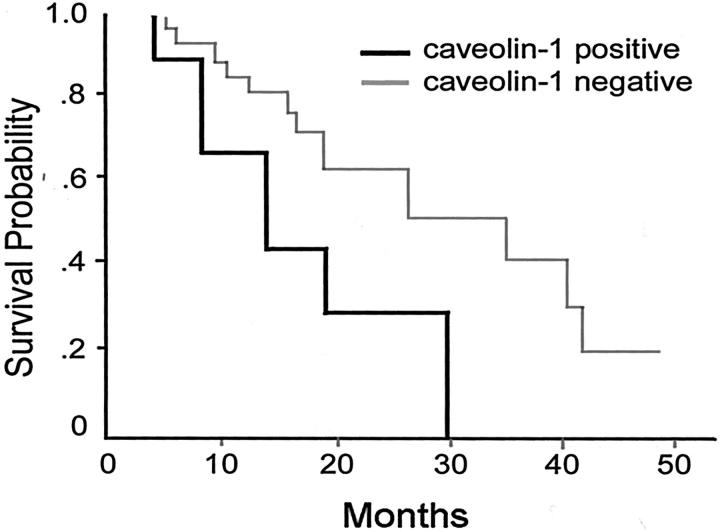
Analysis of the association of caveolin-1 expression with the patient’s survival rate was performed on the 35 patients of lung adenocarcinoma with regional nodal metastases (P/N+). The patients included 22 men and 13 women (mean age, 58.4 ± 14 years). The clinical stage of the disease was IIa in 1 patient, IIb in 1 patient, IIIa in 24 patients, IIIb in 4 patients, and stage IV in 5 patients according to the criteria of the International System for Staging Lung Cancer. The survival time after surgery for these 4 caveolin-1+ve and 31 caveolin-1−ve (at the primary lung lesion site) cases was calculated by the Kaplan-Meier method (Figure 3) ▶ . The survival time was not significantly different (P = 0.0704) between the two groups, but a trend toward a poor prognosis for caveolin-1+ve patients was noted. In contrast, by analyzing caveolin-1 positivity in regional hilar/peribronchial lymph nodes, the survival time for 10 caveolin-1+ve and 25 caveolin-1−ve (nodal metastases) cases was significantly different (P = 0.0153) (data not shown).
Figure 3.
Statistically positive correlation between caveolin-1 expression and poor survival rate. The survival time after surgery for these 4 caveolin-1+ve and 31 caveolin-1−ve cases was calculated by the Kaplan-Meier method. The survival time showed a slight trend toward a poor prognosis for caveolin-1+ve patients.
Multivariate analysis was performed for caveolin-1 immunoreactivity and other factors commonly used as prognosis predictors such as stage, smoking, age, sex, and chemotherapy in these 4 caveolin-1+ve and 31 caveolin-1−ve patients (at the primary lung lesion site). Performance status was excluded from analysis because all of the patients had received the same surgical procedure (ie, pulmonary lobectomy with local lymph node dissection). The univariate log-rank test for each variable was tested, and the result is listed in Table 1 ▶ . When all variables were evaluated, the multiple cog regression model suggested that the expression of caveolin-1 was an independent factor for prediction of poor survival in patients with pulmonary adenocarcinoma (hazard ratio 7.198; P = 0.0041) (Table 1) ▶ .
Table 1.
Analysis of Parameters Including Caveolin-1 Immunoreactivity and Other Clinical Prognostic Factors in 35 Patients with Nodal Metastases (P/N+)
| Prognostic factors | No. | No. dead % (n) | Log rank test P value | Cog regression | |
|---|---|---|---|---|---|
| Hazard ratio* (95% CI†) | P | ||||
| Caveolin-1‡ | |||||
| Yes | 4 | 100% (4) | 0.0704 | 7.198 (1.872–27.671) | 0.0041¶ |
| No | 31 | 58% (18) | |||
| Age, years | |||||
| <65 | 21 | 67% (14) | 0.6364 | 1.361 (0.508–3.650) | 0.5402 |
| ≥65 | 14 | 57% (8) | |||
| Sex | |||||
| Male | 22 | 64% (14) | 0.5364 | 0.542 (0.122–2.411) | 0.4211 |
| Female | 13 | 62% (8) | |||
| Smoking | |||||
| Yes | 16 | 63% (10) | 0.6713 | 0.696 (0.129–3.745) | 0.6727 |
| No | 19 | 63% (12) | |||
| Chemotherapy | |||||
| Yes | 14 | 64% (9) | 0.4806 | 2.774 (0.887–8.678) | 0.0796 |
| No | 21 | 67% (14) | |||
| Stage§ | |||||
| Early | 26 | 62% (16) | 0.3082 | 1.706 (0.520–5.604) | 0.3784 |
| Late | 9 | 67% (6) | |||
*Ratio is calculated by comparing the first category to the second category: e.g., yes versus no for caveolin-1.
†CI, confidence interval.
‡Cutoff value, 30%.
§Early stage: stages II and IIIa; late stage: stages IIIb and IV.
¶Significant.
Reintroduction of Caveolin-1 Expression into Less Invasive CL1-0 Cells Causes Increases in Cell Invasive Ability
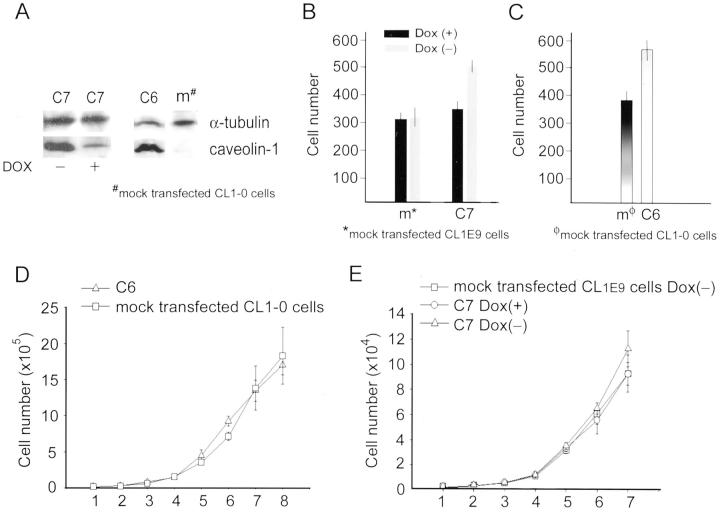
To assess whether the presence of caveolin-1 in CL cells may potentiate cellular invasive capability, we transfected caveolin-1−ve CL1E-9 cells with plasmid harboring a full-length human caveolin-1-encoding cDNA under the control of a tetracycline-responsive element-pTRE2-cav-1 and pTK-Hyg. Several clones were isolated and checked for the status of caveolin-1 expression via Western blotting and immunofluorescence studies. Basal caveolin-1 expression levels and levels regulated by doxycycline (Dox) were not identical in these clones, with clone 7 (C7) expressing the highest amount. As shown in Figure 4A ▶ , withdrawal of Dox increased the expression of caveolin-1 in the C7 clone 15-fold, but had no effect on mock-transfected CL1E-9 cells. When we used varying concentrations of Dox, a dose response for caveolin-1 expression in C7 clone cells was noticeably present.
Figure 4.
The caveolin-1−ve CL1E-9 cells were transfected with plasmid harboring a full-length human caveolin-1 encoding cDNA under the control of a tetracycline-responsive element. A: A clone (C7) was selected and examined for its caveolin-1 expression, which was reduced nearly 15-fold on addition of 20 μg/ml of doxycycline (Dox) into the culture medium (left). Intense caveolin-1 expression was also confirmed in a constitutively expressing cell line, C6, but not in the mock-transfected CL1-0 cells (right). B: The invasive capability was correlated with the ability of cells to express caveolin-1. For C7 cells, addition of Dox to the culture medium resulted in a decrease in both caveolin-1 expression and invasion ability. Dox treatment or no treatment did not affect the invasion ability of CL1E-9 cells (B). C: Increased invasion ability was noted in C6 cells with constitutive caveolin-1 expression, as compared with that of the mock-transfected CL1-0 cells. D and E: The growth rates of caveolin-1+ve C6 and C7 cells (with or without tetracycline treatment) compared with those of mock-transfected caveolin-1−ve CL1-0 cells or CL1E-9 cells were similar, if not identical.
We used an in vitro reconstituted basement membrane invasion assay to measure cell invasive ability. A 48-hour incubation period was chosen based on the fact that the time point for the maximal caveolin-1 protein expression was 24 to 36 hours after withdrawal of Dox from the culture medium. As shown in Figure 4B ▶ , re-expression of caveolin-1 in C7 cells caused approximately one and a half-fold increases in cellular chamber invasive capability.
We also established a constitutively expressing cell line clone 6 (C6) transfected with pcDNA3.1/Myc-caveolin-1 plasmid containing the full length of caveolin-1 cDNA (Figure 4C) ▶ . The invasive capability was also increased in clone C6 cells in comparison with mock-transfected, caveolin-1−ve CL1-0 cells.
Considering that the cell proliferation rate may bias the interpretation of the results of the in vitro invasion assay, we also examined the growth rates of C6 and C7 cells compared with those of mock-transfected or parental CL cells. The growth curves for C6 (Figure 4D) ▶ and C7 cells (with or without tetracycline treatment) (Figure 4E) ▶ and caveolin-1−ve CL1-0 cells or CL1E-9 cells were similar, if not identical.
Constitutive or Induced Expression of Caveolin-1 Induced Filopodia Formation
In tumor cells, high levels of actin polymerization and rearrangement of actin filaments are required for the formation of pseudopodia, which, in turn, are needed for the invasion of malignant cells into tissues. Confocal laser scan microscopy of caveolin-1+ve-CL1-5, -C6, and -C7 Dox(−) cells (without Dox treatment) revealed intense F-actin staining in the periphery of the cells and a redistribution of F-actin toward the leading edge of filopodia (Figure 5) ▶ , which was in strong contrast to the cortex staining of the parental caveolin-1−ve-CL1-0, and caveolin-1−ve-C7 Dox(+) cells (with Dox treatment) where only few filopodia were detectable (Figure 5) ▶ .
Figure 5.
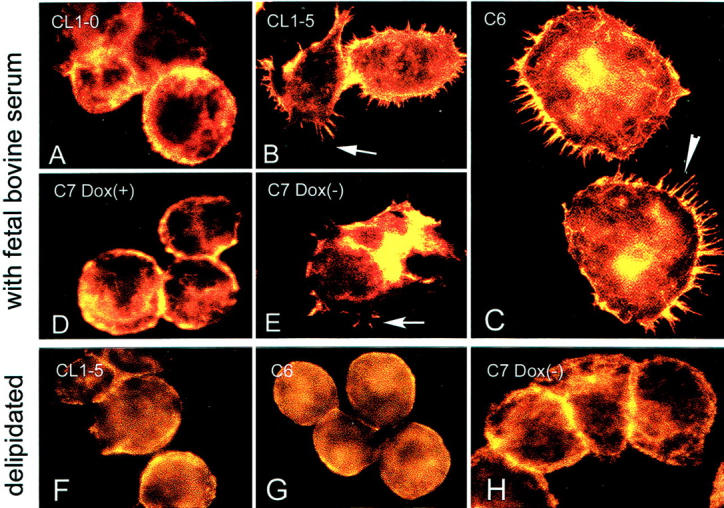
Caveolin-1+ve CL1-5 cells (B), C6 cells (C), and C7-Dox(−) cells (D) showed abundant filopodia formation. The caveolin-1−ve CL1-0 cells (the parental cells for C6, A) and C7-Dox(+) cells (E) did not reveal filopodia formation in cultures. However, the ability to form filopodia in CL1-5, C6, and C7-Dox(−) cells was abolished when cells were cultured in delipidated medium (F to H). A rhodamine phalloidin staining was used to highlight the presence of F-actin in filopodia and at the peripheral cytoplasmic area.
Cells Cultured under Delipidated Condition Lost the Ability to Form Filopodia
Cells cultured under delipidated condition lost the ability to form filopodia despite the presence of caveolin-1 expression (Figure 5) ▶ . Delipidated culture condition seemed not to affect the steady-state expression of actin or the process of actin polymerization because there was little difference in the fluorescence intensity of rhodamine in the cytoplasmic periphery of cells. The findings suggest that not only the caveolin-1 protein itself, but also its proper subcellular localization to lipid component play an important role in mediating filopodia formation.
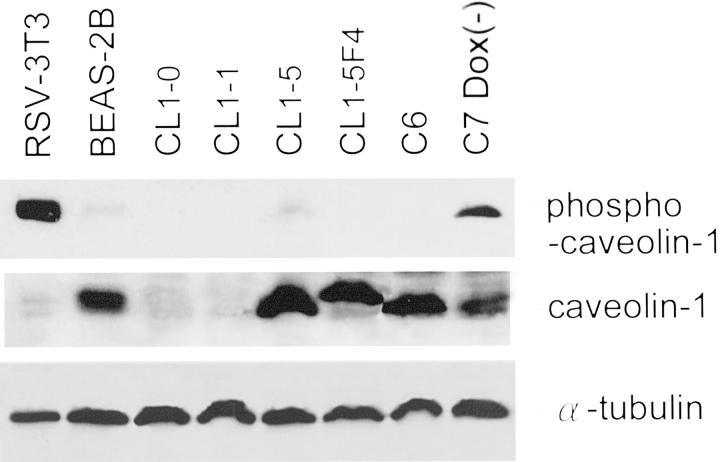
Caveolin-1-Positive CL Cells with Enhanced Invasive Capability Had Little Detectable Amount of Tyrosine-14 Phosphorylated Caveolin-1
Previously, Lee and colleagues 21 have shown that binding of Grb7 to tyrosine 14-phosphorylated caveolin-1 functionally stimulates cell migration. To evaluate whether such a mechanism operated in the differential invasive ability of lung adenocarcinoma cell lines, we performed Western blotting of phospho-caveolin-1 protein and caveolin-1 protein. As shown in Figure 6 ▶ , Y14-phosphorylated-caveolin-1 was either totally absent or barely detectable in CL1-5, CL1-5F4 cells, and caveolin-1 constitutively expressing cell line clone 6 (C6) despite the presence of caveolin-1. Furthermore, Grb7 was not detectable by RT-PCR and Western blotting in the caveolin-1+ve CL1-5, and CL1-5F4 cells (data not shown). Although C7 cells without Dox treatment did express Y14-phosphorylated caveolin-1 as well as caveolin-1, Grb7 expression was not as abundant as the noncancerous lung epithelial BEAS-2B cells. Therefore, tyrosine-14 phosphorylated caveolin-1 complex with Grb7 plays only a small role in mediating cell invasiveness of advanced lung adenocarcinoma cells.
Figure 6.
Y14-phosphorylated caveolin-1 was not always detectable in caveolin-1+ve lung adenocarcinoma cells. Western blotting revealed expression of phospho-caveolin-1 protein in RSV-3T3 cell lysate (a positive control for phospho-caveolin-1 protein) and C7-Dox(−) cells, but totally absent or just barely detectable in CL1-0, CL1-1, CL1-5, CL1-5F4, and C6 cells. Staining for caveolin-1 and α-tubulin is also shown for comparison.
Discussion
Our results are consistent with the dual functions of caveolin-1 both as a tumor suppressor gene and as a metastasis-promoting gene. In the low-invasive lung cancer cell line CL1-0, caveolin-1 expression is absent or extremely low, which is consistent with the fact that down-regulated caveolin-1 expression facilitates cell transformation. However, in the other two cell lines, CL1-5 and CL1-5F4, which were selected for higher cell invasiveness, both caveolin-1 mRNA and protein expression are substantially up-regulated, which is consistent with the proposed metastasis-promoting function of caveolin-1 in advanced adenocarcinoma. 13,14 When we introduced caveolin-1 expression into the less invasive CL1-0 cells, the ability of cell invasiveness was enhanced by at least 150%, as revealed by invasion chamber assay.
The same observation was also obtained from our immunohistochemical study of caveolin-1 expression in the formalin-fixed, paraffin-embedded specimens of human lung adenocarcinoma and of the ipsilateral hilar/peribronchial lymph nodes. In our collected 95 cases of lung adenocarcinoma, 35 patients already had regional lymph node metastases, of which virtually all (except one) showed caveolin-1 positivity in 10 to 95% of the nodal metastasized tumor cells. This is in contrast to a small percentage of caveolin-1 immunoreactivity (generally 0 to 30% with a mean ∼5%) in primary lung adenocarcinoma cells in 29 among 95 cases tested. The morphology of cancer cells with caveolin-1 expression is more pleomorphic and anaplastic compared with those without caveolin-1 expression. The trend of caveolin-1 expression positively associated with nodal metastases is statistically significant via analysis of our specimens. Furthermore, the expression of caveolin-1 in lung adenocarcinoma is an independent factor predicting survival time in stage IIa and higher stages. Our result is consistent with those of other reports that caveolin-1 is a prognostic factor for disease progression in prostate cancer. 16
Several mechanisms have been elucidated to explain how down-regulated caveolin-1 helps cell transformation during tumorigenesis. The reciprocal negative transcriptional regulation between caveolin-1 and other growth factor or signal transducers such as α-folate receptor, p42/44 MAP kinase, and the neu oncogene has been proposed to account for cell transformation in various cell types. 2-4,6-8,22 In addition, transcriptional silencing by methylation of CpG islands in the 5′ promoter region of the caveolin-1 gene was once found in two human breast cancer cell lines that failed to express the caveolin-1 protein. 23 Our previous microarray data from the comparison of genes differentially expressed in cell lines with varying invasive capability suggested a possible reciprocal transcriptional regulation between caveolin-1 and HEK2, TNF-R, or protein kinase C during lung cell transformation, 20 although further experimentation is needed to confirm this guess.
Why a tumor suppressor gene whose inactivation is necessary for tumor transformation can be re-expressed to facilitate tumor progression remains a puzzle. Such a paradoxical phenomenon is not limited to caveolin-1; other molecules such as CD44 and granulocyte/macrophage-colony stimulation factor (GM-CSF) have also been reported to exert the seemingly opposite functions in tumorigenesis. 24,25 GM-CSF can promote tumor cell growth and survival, but it also can stimulate tumor cell rejection. A recent study of transgenic mice epidermally overexpressing either GM-CSF or GM-CSF antagonist revealed that both up- and down-regulation of GM-CSF activity in the murine skin increase susceptibility to skin cancer by independent mechanisms. 24 Up-regulated expression of CD44, an 80-kd transmembrane glycoprotein, has been implicated in cell transformation as well as in metastasis formation in breast, bladder, and colon cancers. By contrast, loss of CD44 expression accompanies cell transformation in Burkitt’s lymphoma and neuroblastoma. It seems that the type of action of CD44 depends on the type of cells expressing it.
As for caveolin-1, it was recently reported that a point mutation in caveolin-1 at codon 132 (P132L) was found in invasive human scirrhous breast cancers, and that mutant caveolin-1 expression could induce cellular transformation and promote invasion ability. 26 Our CL cells did not reveal any point mutation or any deletion or insertion of the caveolin-1 gene. Therefore, another mechanism must exist for the invasion-promoting capability of caveolin-1 in lung adenocarcinoma. It also has been proposed that phosphorylated caveolin-1 complexed with Grb7 augments EGF-stimulated cell migration, and such effect is blocked if the phosphorylation site on caveolin-1 is mutated (Y14A mutation). 21 In our CL cells, Y14-phosphorylated caveolin-1 is not obvious. Furthermore, Grb7 is generally not detected in CL cells with higher invasive ability. Thus, Y14-phosphorylated caveolin-1 may not be the major mechanism exploited by caveolin-1+ve CL cells for enhancing cell invasiveness.
In summary, our study clearly revealed that up-regulated caveolin-1 in CL cells is necessary for mediating filopodia formation, which may enhance cell migration and, in part, the invasive ability of lung adenocarcinoma cells. Caveolin-1, the principal component of caveolae, binds cholesterol and sphingolipids to form platforms and serves to support membrane traffic and signal transduction. 27 Thus, it is not totally unexpected that the formation of filopodia mediated by up-regulated caveolin-1 in cells requires the presence of lipid in culture medium. Recently, Saltiel and colleagues reported that cholesterol depletion effectively disrupted the co-localization of F-actin with caveolae. 28 Using a yeast two-hybrid screen, the actin-binding filamin is identified as a ligand for caveolin-1. 29 Possibly, the interaction between caveolin-1 and actin or filamin 29 provides a physical link for caveolae and cytoskeleton, and, thus participating in filopodia formation. How caveolin-1 interacts with the intracellular cytoskeleton (especially that in filopodia) in cells with various transformation or differentiation states will be another interesting issue for understanding the role of caveolin-1 as a tumor metastasis-promoting molecule.
Acknowledgments
We thank Dr. T. S. Jou and Dr. J. Y. Shih for technical consultation; and C. J. Chang, K. M. Liao, and T. W. Chen for assistance with statistical analysis.
Footnotes
Address reprint requests to Su-Ming Hsu, M.D., Department of Pathology, National Taiwan University Hospital, 1-1 Jen-Ai Rd., Taipei, Taiwan 100. E-mail: smhsu@ha.mc.ntu.edu.tw.
Supported by the National Science Council (Taiwan) grants 89-2320-B-002-177, 90-2314-B-002-186 to P.-H. H. and National Health Research Institutes (Taiwan) grant EX-91-8704SL, Ministry of Education (Taiwan) grant 89-B-FA01-1-4 to S.-M. H.
C.-C. H. and P.-H. H. contributed equally to this work.
References
- 1.Rothberg K, Heuser JE, Donzell WC, Ying YS, Glenney JR, Anderson RGW: Caveolin, a protein component of caveolae membrane coats. Cell 1992, 68:673-682 [DOI] [PubMed] [Google Scholar]
- 2.Koleske AJ, Baltimore D, Lisanti MP: Reduction of caveolin and caveolae in oncogenically transformed cells. Proc Natl Acad Sci USA 1995, 92:1381-1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Engelman JA, Wykoff CC, Yasuhara S, Song KS, Okamoto T, Lisanti MP: Recombinant expression of caveolin-1 in oncogenically transformed cells abrogates anchorage-independent growth. J Biol Chem 1997, 272:16374-16381 [DOI] [PubMed] [Google Scholar]
- 4.Lee SW, Reimer CL, Oh P, Campbell DB, Schnitzer JE: Tumor cell growth inhibition by caveolin re-expression in human breast cancer cells. Oncogene 1998, 16:1391-1397 [DOI] [PubMed] [Google Scholar]
- 5.Racine C, Belanger M, Hirabayashi H, Boucher M, Chakir J, Couet J: Reduction of caveolin 1 gene expression in lung carcinoma cell lines. Biochem Biophys Res Commun 1999, 255:580-586 [DOI] [PubMed] [Google Scholar]
- 6.Bender FC, Reymond MA, Bron C, Quest AF: Caveolin-1 levels are down-regulated in human colon tumors, and ectopic expression of caveolin-1 in colon carcinoma cell lines reduces cell tumorigenicity. Cancer Res 2000, 60:5870-5878 [PubMed] [Google Scholar]
- 7.Galbiati F, Volonte D, Engelman JA, Watanabe G, Burk R, Pestell RG, Lisanti MP: Targeted downregulation of caveolin-1 is sufficient to drive cell transformation and hyperactivate the p42/44 MAP kinase cascade. EMBO J 1998, 17:6633-6648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engelman JA, Chu C, Lin A, Jo H, Ikezu T, Okamoto T, Kohtz DS, Lisanti MP: Caveolin-mediated regulation of signaling along the p42/44 MAP kinase cascade in vivo. A role for the caveolin-scaffolding domain. FEBS Lett 1998, 428:205-211 [DOI] [PubMed] [Google Scholar]
- 9.Carman CV, Lisanti MP, Benovic JL: Regulation of G protein-coupled receptor kinases by caveolin. J Biol Chem 1999, 274:8858-8864 [DOI] [PubMed] [Google Scholar]
- 10.Razani B, Rubin CS, Lisanti MP: Regulation of cAMP-mediated signal transduction via interaction of caveolins with the catalytic subunit of protein. J Biol Chem 1999, 274:26353-26360 [DOI] [PubMed] [Google Scholar]
- 11.Felley-Bosco E, Bender FC, Courjault-Gautier F, Bron C, Quest AF: Caveolin-1 down-regulates inducible nitric oxide synthase via the proteosome pathway in human colon carcinoma cells. Proc Natl Acad Sci USA 2000, 97:14334-14339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hulit J, Bash T, Fu M, Galbiati F, Albanese C, Sage DR, Schlegel A, Zhurinsky J, Shtutman M, Ben-Ze’ev A, Lisanti MP, Pestell RG: The cyclin D1 gene is transcriptionally repressed by caveolin-1. J Biol Chem 2000, 275:21203-21209 [DOI] [PubMed] [Google Scholar]
- 13.Yang G, Truong LD, Timme TL, Ren C, Wheeler TM, Park SH, Nasu Y, Bangma CH, Kattan MW, Scardino PT, Thompson TC: Elevated expression of caveolin is associated with prostate and breast cancer. Clin Cancer Res 1998, 4:1873-1880 [PubMed] [Google Scholar]
- 14.Thompson TC: Metastasis-related genes in prostate cancer: the role of caveolin-1. Cancer Metastasis Rev 1999, 17:439-442 [DOI] [PubMed] [Google Scholar]
- 15.Timme TL, Goltsov A, Tahir S, Li L, Wang J, Ren C, Johnston RN, Thompson TC: Caveolin-1 is regulated by c-myc and suppresses c-myc-induced apoptosis. Oncogene 2000, 19:3256-3265 [DOI] [PubMed] [Google Scholar]
- 16.Yang G, Truong LD, Wheeler TM, Thompson TC: Caveolin-1 expression in clinically confined human prostate cancer: a novel prognostic marker. Cancer Res 1999, 59:5719-5723 [PubMed] [Google Scholar]
- 17.Chu YW, Yang PC, Yang SC, Shyu YC, Hendrix MJ, Wu R, Wu CW: Selection of invasion and metastatic subpopulations from a human lung adenocarcinoma cell line. Am J Respir Cell Mol Biol 1997, 17:353-360 [DOI] [PubMed] [Google Scholar]
- 18.Shih JY, Yang SC, Hong TM, Yuan A, Chen JJ, Yu CJ, Chang YL, Lee YC, Peck K, Wu CW, Yang PC: Collapsin response mediator protein-1 and the invasion and metastasis of cancer cells. J Natl Cancer Inst 2001, 93:1392-1400 [DOI] [PubMed] [Google Scholar]
- 19.Chang MM, Harper R, Hyde DM, Wu R: A novel mechanism of retinoic acid-enhanced interleukin-8 gene expression in airway epithelium. Am J Respir Cell Mol Biol 2000, 22:502-510 [DOI] [PubMed] [Google Scholar]
- 20.Chen JJ, Wu R, Yang PC, Huang JY, Shen YP, Han MH, Kao WC, Lee PJ, Chiu TF, Chang F, Chu YW, Wu CW, Peck K: Profiling expression patterns and isolating differentially expressed genes by cDNA microarray system with colorimetry detection. Genomics 1998, 51:313-324 [DOI] [PubMed] [Google Scholar]
- 21.Lee H, Volonte D, Galbiati F, Iyengar P, Lublin DM, Bregman DB, Wilson MT, Campos-Gonzales R, Bouzahzah B, Pestell RG, Scherer PE, Lisanti MP: Constitutive and growth factor-regulated phosphorylation of caveolin-1 occurs at the same site (Tyr-14) in vivo: identification of a c-Src/Cav-1/Grb7 signaling cassette. Mol Endocrinol 2000, 14:1750-1775 [DOI] [PubMed] [Google Scholar]
- 22.Bagnoli M, Tomassetti AT, Figini M, Flati S, Dolo V, Canevari S, Miotti S: Downregulation of caveolin-1 expression in human ovarian carcinoma is directly related to α-folate receptor overexpression. Oncogene 2000, 19:4754-4763 [DOI] [PubMed] [Google Scholar]
- 23.Engelman JA, Zhang XL, Lisanti MP: Sequence and detailed organization of the human caveolin-1 and 2 genes located near the D7S522locus (7q31.1). Methylation of a CpG island in the 5′ promoter region of the caveolin-1 gene in human breast cancer cell lines. FEBS Lett 1999, 448:221-230 [DOI] [PubMed] [Google Scholar]
- 24.Mann A, Breuhahn K, Schirmacher P, Wilhelmi A, Beyer C, Rosenau A, Özbek S, Rose-John S, Blessing M: Up- and down-regulation of granulocyte/macrophage-colony stimulating factor activity in murine skin increase susceptibility to skin carcinogenesis by independent mechanisms. Cancer Res 2001, 61:2311-2319 [PubMed] [Google Scholar]
- 25.Herrlich P, Morrison H, Sleeman J, Orain-Rousseau V, König H, Weg-Remers S, Ponta H: Cd44 acts as a growth- and invasiveness-promoting molecule and as a tumor-suppressing cofactor. Ann NY Acad Sci 2000, 910:106-118 [DOI] [PubMed] [Google Scholar]
- 26.Hayashi K, Matsuda S, Machida K, Yamamoto T, Fukuda Y, Nimura Y, Hayakawa T, Hamaguchi M: Invasion activating caveolin-1 mutation in human scirrhous breast cancers. Cancer Res 2001, 61:2361-2364 [PubMed] [Google Scholar]
- 27.Anderson RG, Jacobson K: A role for lipid shells in targeting proteins to caveolae, rafts and other lipid domains. Science 2002, 296:1821-1825 [DOI] [PubMed] [Google Scholar]
- 28.Kanzaki M, Pessin JE: Caveolin-associated filamentous actin (Cav-actin) defines a novel F-actin structure in adipocyte. J Biol Chem 2002, 277:25867-25869 [DOI] [PubMed] [Google Scholar]
- 29.Stahlhut M, Deurs BV: Identification of filamin as a novel ligand for caveolin-1: evidence for the organization of caveolin-1-associated membrane domains by the actin cytoskeleton. Mol Biol Cell 2000, 11:325-337 [DOI] [PMC free article] [PubMed] [Google Scholar]