Abstract
Fluorescence in situ hybridization (FISH) is difficult to accomplish using thin-sections of paraffin-embedded lymphoid tissue because of the high cellularity and truncated cells that interfere with accurate scoring of individual nuclei. We modified and tested a new technique to isolate individual nuclei from tissue cores of paraffin-embedded tissue processed with xylene, proteinase K, citric acid, and pepsin. The efficacy of this method to study paraffin-embedded tissue was investigated in six normal lymph nodes or tonsils and 32 malignant lymphomas including five mantle cell, five follicular, five Burkitt, five extranodal marginal zone lymphomas of mucosa-associated lymphoid tissue, five anaplastic large-cell, and seven diffuse large B-cell. Fusion of CCND1 and IgH, BCL2 and IgH, c-myc and IgH, and MALT1 and API2 were detected using probes with a dual-fusion FISH strategy. Anomalies involving ALK and BCL6 were detected using break-apart FISH probes. FISH studies were successful for each of the 38 specimens. Chromosome anomalies were detected in each malignant specimen, but not in the normal lymphoid tissue. The correct chromosome anomaly was detected in 22 of 22 specimens with genetic abnormalities that were established by other genetic techniques. This FISH technique is useful to detect chromosome anomalies with high sensitivity and specificity in paraffin-embedded tissue and may provide important diagnostic and prognostic genetic information.
The World Health Organization recognizes that genetic abnormalities are one of the most reliable criteria for classification of malignant lymphomas. 1 New genetic methods using fluorescent-labeled DNA and in situ hybridization (FISH) are valuable for detection of genetic anomalies. 2 The application of FISH methods to lymphomas has been hindered by the wide use of paraffin-embedded tissue to study and store lymphoid tissue. FISH methods are best applied to fresh cell suspensions that have been fixed with methanol and glacial acetic acid. Most FISH studies of paraffin-embedded tissue have previously been performed on thin-sections. Although the thin-section technique is relatively easy to prepare and can be correlated with hematoxylin and eosin (H&E) slides, there are limitations with this approach. For example, overlapping nuclei and truncated cells inherent in standard thin-sections, interfere with accurate scoring of individual nuclei. Moreover, thin-sections are difficult because the paraffin and standard fixatives often interfere with hybridization of DNA probes to target loci.
New methods are emerging to isolate individual nuclei from paraffin-embedded lymphoid tissue that make application of routine FISH studies feasible in clinical practice. 3-5 The isolation of individual nuclei from thick-sections described in previous investigations 5 have helped circumvent some of the problems associated with thin-sections. However, this method requires large amounts of tissue and is time consuming and laborious. In addition, FISH results cannot be correlated with the area of interest on the H&E slide. In the method described in the present investigation, whole nuclei were extracted from needle core biopsies taken from paraffin blocks. We found this technique to be superior to both the thin-section and isolation of nuclei from thick-section techniques. The method uses minimal tissue, can be directed to the area of interest on the H&E slide, is technically simple to perform, and produces individual nuclei in which FISH signals are bright, planar, and easy to score. The efficacy of this method was tested in six normal lymph nodes or tonsils, and 32 malignant lymphomas including five mantle cell, five follicular, five Burkitt, five extranodal marginal zone lymphomas of mucosa-associated lymphoid tissue (MALT), five anaplastic large-cell, and seven diffuse large B-cell.
Materials and Methods
All lymphomas met the diagnostic morphological and phenotypic criteria of the World Health Organization classification of hematolymphoid neoplasms. Twenty-two of the 32 malignant lymphoma specimens had independent confirmation of a specific genetic abnormality by routine cytogenetic analysis [all follicular lymphomas had t(14;18)(q32;q21) and two Burkitt lymphomas had translocations involving 8q24], polymerase chain reaction studies for CCND1/IgH (all mantle cell lymphomas), and NPM/ALK1 (all anaplastic large-cell lymphomas), and reverse transcriptase-polymerase chain reaction studies for API2/MALT1 translocations (all MALT lymphomas).
Hematopathologists (RM, ER, PK) selected appropriate paraffin-embedded tissue blocks from the Mayo Clinic files and marked the region that contained malignant cells based on comparison with H&E sections. Malignant lymphomas and normal lymphoid tissues were studied together and the diagnosis of each sample was unknown to the technologists (SP, SB) who performed FISH studies. This study was approved by the Mayo Clinic Institutional Review Board.
Tissue Core Collection
For each specimen two tissue sample cores were collected using a 20-gauge 1.5-inch blunt needle (Sherwood Medical Company, St. Louis, MO) that was pushed through the entire block. A stainless steel wire was then threaded through the needle to force the tissue core into a 0.65-mL microcentrifuge tube.
Extraction of Nuclei
The paraffin was dissolved at room temperature with three 10-minute changes of xylene (100 μl each) in the microcentrifuge tube. For each change, the xylene was removed with a micropipetter (10 to 200 μl) being careful to leave the tissue cores intact at the bottom of the tube. The tissue was then rehydrated with 100 μl of 95%, 75%, and 50% ethanol (EtOH) for 2 minutes each. The 50% EtOH was removed and the tissue was manually disaggregated with the tip of a partially straightened large paper clip. Enzymatic digestion was then performed by adding 100 μl of freshly prepared proteinase K solution (0.005% proteinase K, 30 U/mg protein, in 0.05 mol/L Tris hydroxymethyl aminomethane hydrochloride (pH 7.0), 0.01 mol/L ethylenediaminetetraacetic disodium salt, and 0.01 mol/L sodium chloride) to the microcentrifuge tube. The specimen was incubated at 37°C for 30 minutes. To aid with enzymatic digestion, the sample was vortexed for 3 seconds at 5-minute intervals during this incubation period. Nuclei were pelleted using a mini microcentrifuge (6000 rpm) for 10 minutes. Proteinase K was carefully removed with a micropipetter and the nuclei washed by resuspension with vortexing in 100 μl of phosphate-buffered saline (PBS). The PBS solution was removed and the nuclei fixed by resuspension with vortexing in two changes of freshly prepared fixative (three parts methanol and one part glacial acetic acid). The nuclei were resuspended in 100 μl of fixative. Fixed nuclei suspensions were stored at −70°C until it was convenient to perform FISH studies.
Pretreatment and FISH
Slide preparations were made by placing 8 μl of nuclei suspension within a 13-mm circle on a slide that was positively charged (order number ER-329 from Erie Scientific, Portsmouth, NH) to enhance cell adhesion with the slide surface. The circle was used to define the hybridization site and mark the location where FISH probes were applied. The slide was then dried in a 65°C oven for 15 minutes. A humidified microwave oven was used to minimize evaporation and was prepared as follows: a 2 L beaker filled with water was placed in a microwave oven and the microwave oven was operated for 5 minutes. Slides were then placed in a Coplin jar containing 10 mmol/L of citric acid (pH 6.0) and processed in the humidified microwave oven for 10 minutes. After the citric acid treatment, slide preparations were placed in a Coplin jar containing 2× standard saline citrate (pH 7.0) solution at 37°C for 15 minutes. Slides were then transferred to a Coplin jar containing freshly prepared 0.4% pepsin solution (0.16 g pepsin, 2770 U/mg solid, in 40 ml 0.9% pH 1.5 sodium chloride). The slides were then dehydrated with 70%, 85%, and 100% EtOH for 2 minutes each.
FISH Probes
For purposes of this paper, fluors that produce a red or orange signal are denoted as R, green signals as G, and yellow or adjacent R and G signals as F to indicate an R/G signal fusion. For each probe, the probe name, chromosomal locus, chromosome anomaly detected, and vendor is summarized in Table 1 ▶ .
Table 1.
Summary of DNA FISH Probes Used in this Investigation
| Probes | Hybridization loci | Anomaly detected | Source |
|---|---|---|---|
| CCND1, IgH | 11q13, 14q32 | t(11;14)(q13;q32) | Commercial* |
| c-MYC, IgH | 8q24, 14q32 | t(8;14)(q24;q32) | Commercial* |
| ALK | 2p23 | t(2;var)(p23;var) | Commercial* |
| API2, MALT1 | 11q21, 18q21 | t(11;18)(q21;q21) | Home Brew† |
| BCL2, IgH | 14q32, 18q21 | t(14;18)(q32;q21) | Commercial* |
| BCL6 | 3q27 | t(3;var)(q27;var) | Commercial* |
*Vysis, Inc., Downers Grove, IL.
†Remstein, E., et al. 7
FISH and Posthybridization Wash
The following method was used for each probe set. A total of 3 μl of probe solution prepared by the manufacturer’s recommendation was applied to the hybridization site and covered with a 12-mm circle coverslip. The coverslip edges were sealed with a continuous bead of rubber cement. Specimens were processed with a HYBrite instrument (Vysis Inc., Downers Grove, IL) programmed for melt temperature 80°C, melt time 8 minutes, hybridization temperature 37°C, and hybridization time 4 to 20 hours. After hybridization, coverslips were gently removed and the slides were washed in a Coplin jar filled with 0.4× standard saline citrate (pH 7.0) at 72°C for 2 minutes. The slides were transferred to a Coplin jar filled with 2× standard saline citrate/Nonidet P-40 solution (pH 7.0) at room temperature for 1 minute. Nuclei were counterstained with a mixture of 10 μl of 1000 ng/ml of 4′,6′-diamidino-2-phenylindole dihydrochloride and Vectashield antifade (Vector Laboratories, Inc., Burlingame, CA) at a ratio of 1:10. A 24 × 50-mm coverslip was then placed over the hybridization sites.
Microscopy
Analysis was done using a fluorescence microscope equipped with a 100-W mercury lamp. To view signals a dual-pass SpectrumOrange/SpectrumGreen filter set or a dual pass fluorescein isothiocyanate/Texas Red filter set was used. Individual fluors were observed using a single-pass SpectrumGreen, single pass SpectrumOrange, single-pass fluorescein isothiocyanate, or a single pass Texas Red filter.
Statistical Studies
A total of 200 consecutive qualifying interphase nuclei were scored from each specimen. The normal range for 200 nuclei was calculated using data from lymph nodes, tonsils, or bone marrow from separate unpublished studies. The upper bound of the normal range was determined using a one-sided 95% confidence interval for observing the maximum number of nuclei for each false-positive signal pattern seen in 200 scoreable nuclei using the binomial distribution. The normal cutoff was 2.5% for CCND1/IgH fusion, c-myc/IgH fusion, BCL2/IgH fusion, MALT1/API2 fusion, and 4.5% for break-apart of ALK and BCL6.
Results
Success of Nuclei Extraction and FISH
Individual nuclei were successfully extracted from each of the 38 specimens on the first attempt with this method. FISH was successful on the first hybridization attempt for 31 of 38 specimens. Two MALT lymphomas required a repeat of the FISH and posthybridization wash to obtain successful FISH results. Two diffuse large B-cell lymphomas and one MALT produced successful results after the nuclear extraction process was repeated. The remaining two diffuse large B-cell lymphomas were initially unsuccessful for all probes and required pretreatment with a commercial steamer instead of a humidified microwave oven to achieve successful FISH studies.
Probe Performance
The probe sets and theoretical signal patterns for ALK, CCND1/IgH, and MALT1/API2 were established in previous studies. 5-7 The performance of BCL2/IgH, c-myc/IgH, and BCL6 were established in separate but unpublished studies. Briefly, the analytical sensitivity for each of these probes was 100% based on analysis of 100 consecutive metaphases from a normal phytohemagglutinin-stimulated blood specimen. The expected signal pattern and hybridization locus without evidence of cross-hybridization was observed on 100% of 200 targets for BCL2/IgH, c-myc, and BCL6. Based on pilot studies of bone marrow specimens with known translocations, strict scoring criteria were developed for each probe set to distinguish between normal and abnormal cells. Representative nuclei from lymph nodes or tonsils with expected signal patterns for each probe set are shown in Figure 1 ▶ . For each probe set, 100% of nuclei from a normal paraffin-embedded lymph node or tonsil specimen had a normal signal pattern. These results were considered acceptable probe performance for interphase analysis using each of these probe sets.
Figure 1.
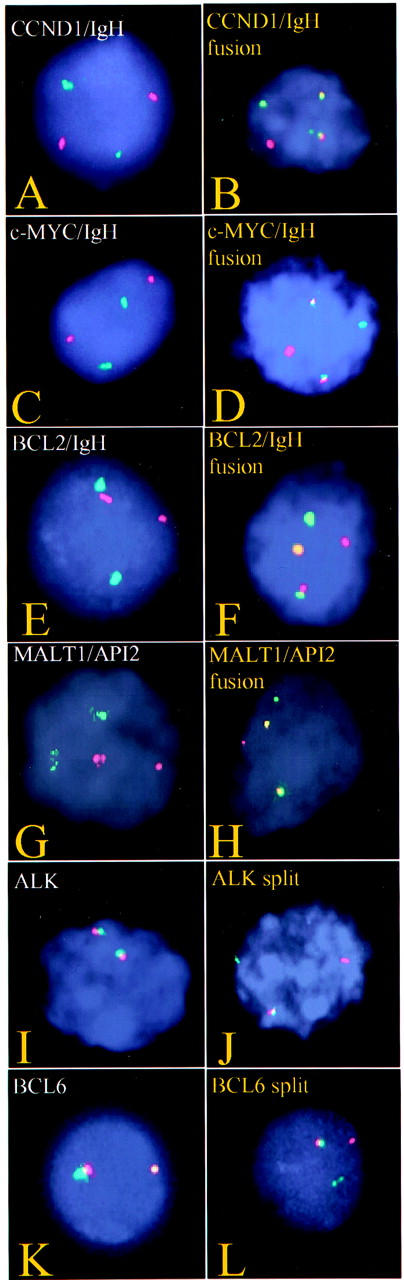
Representative nuclei extracted from paraffin-embedded tissue showing normal (left) and abnormal (right) signal patterns for CCND1 and IgH (A and B), c-myc and IgH (C and D), BCL2 and IgH (E and F), MALT1 and API2 (G and H), ALK (I and J), and BCL6 (K and L).
Mantle Cell Lymphoma
Six specimens were studied with probes for CCND1 and IgH: five were known to have an abnormal CCND1/IgH FISH pattern from previous studies 5 and one was a normal lymph node. Results were abnormal in 81 to 98% of nuclei among the five mantle cell lymphomas and 0% of nuclei in the normal lymph node (Figure 2) ▶ . The signal pattern was 1R1G2F in each of four mantle cell lymphomas. This result is consistent with the expected signal pattern for cells with a t(11;14)(q13;q32) and reciprocal fusion of portions of CCND1 and IgH loci. The signal pattern in one mantle cell lymphoma was 1R1G1F. This result most likely suggests fusion of IgH/CCND1 loci and loss of a partial signal for IgH and CCND1. The signal pattern for the normal lymph node was normal, 2R2G.
Figure 2.
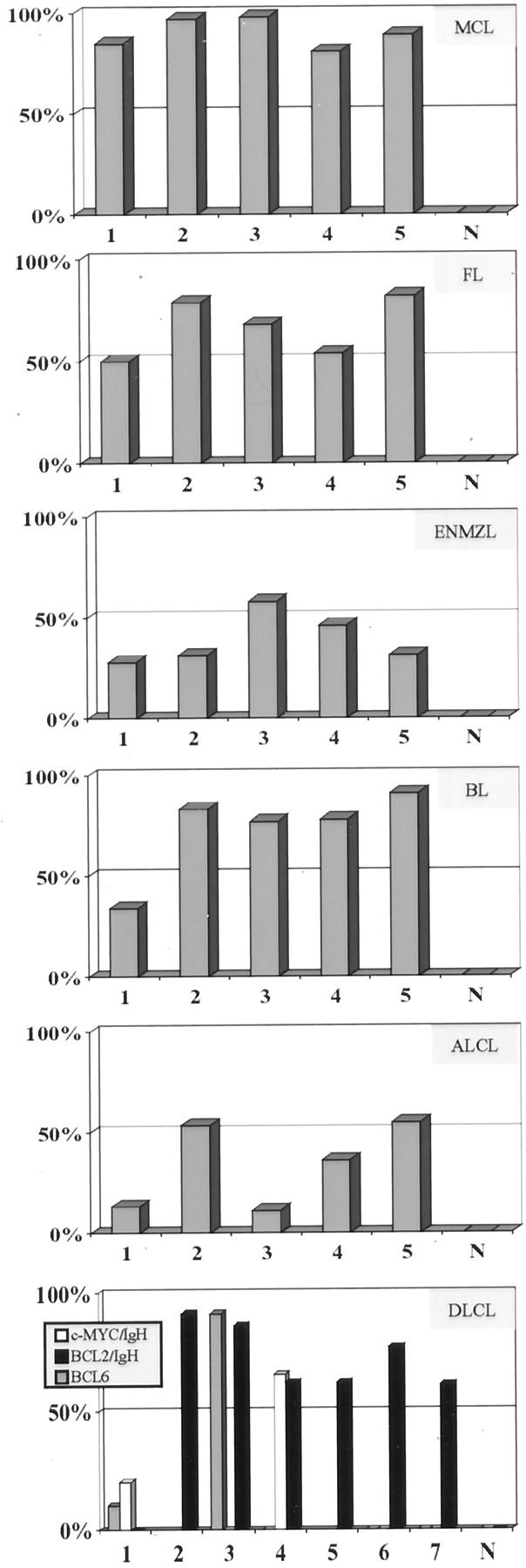
For each lymphoma type tested, the five cases and normal lymphoid tissue (N) are shown on the x axis. The percent nuclei with abnormal signal patterns in each case are shown on the y axis. The following lymphomas and probe sets are shown: MCL (mantle cell lymphoma, CCND1/IgH), FL (follicular lymphoma, BCL2/IgH), ENMZL (extranodal marginal zone lymphoma, MALT1/API2), BL (Burkitt lymphoma, c-myc/IgH), ALCL (anaplastic large-cell lymphoma, ALK), DLCL (diffuse large B-cell lymphoma, IgH, c-myc, BCL2, and BCL6).
Burkitt Lymphoma
Six specimens were studied with probes for c-myc and IgH: five were known to have Burkitt lymphoma and one was a normal lymph node. Among the Burkitt lymphomas, two had previous conventional cytogenetic studies on fresh samples: one was known to have a t(8;14)(q24;q32), and one was known to have a t(2;8)(p12;q24). The other three Burkitt lymphomas had no previous genetic tests performed. Four Burkitt lymphomas had an abnormal signal pattern (1R1G2F) in 77 to 97% of nuclei (Figure 2) ▶ . This result is consistent with cells that have a t(8;14)(q24;q32) and reciprocal forms of fusion for portions of c-myc and IgH loci. The signal pattern in one Burkitt lymphoma was 3R2G in 34% of nuclei. This patient was known to have a t(2;8)(p12;q24) and the results are consistent with separation of the red c-myc signal. The signal pattern for the normal lymph node was normal, 2R2G.
Follicular Lymphoma
Six specimens were studied with probes for BCL2 and IgH: five were known to have a t(14;18)(q32;q21) by conventional cytogenetic studies of bone marrow from these patients and one was a normal lymph node. The results were abnormal in 50 to 82% of nuclei among the five follicular lymphomas and 0% of nuclei in the normal lymph node (Figure 2) ▶ . The signal pattern was 1R1G2F in four follicular lymphomas and 1R1G3F in one follicular lymphoma. These results are consistent with the expected signal pattern for cells that had a t(14;18)(q32;q21) associated with reciprocal fusion of portions of BCL2 and IgH loci. The 1R1G3F signal pattern is consistent with t(14;18)(q32;q21) with an extra copy of either BCL2/IgH fusion or IgH/BCL2 fusion. The signal pattern for the normal lymph node was normal, 2R2G.
MALT
Six specimens were studied with probes for MALT1 and API2: five were known to have MALT1 and API2 fusion from previous studies 7 and one was a hyperplastic tonsil. The results were abnormal in 28 to 58% of nuclei among the five MALT lymphomas and 0% of nuclei in the tonsil (Figure 2) ▶ . The signal pattern was 1R1G2F in each of the five MALT lymphomas. This result is consistent with the expected signal pattern for cells that had a t(11;18)(q21;q21) and reciprocal fusion of portions of the MALT1 and API2 loci. The signal pattern for the hyperplastic tonsil was normal, 2R2G.
Anaplastic Large-Cell Lymphoma
Six specimens were studied with probes for ALK: five were known to have an abnormal ALK locus by previous FISH and immunohistochemical studies 6 and one was a hyperplastic tonsil. The results were abnormal in 11 to 53% of nuclei among the five anaplastic large-cell lymphomas and 0% of nuclei in the tonsil (Figure 2) ▶ . The signal pattern was 1R1G1F in four anaplastic large-cell lymphomas and 2R1G1F in one other anaplastic large-cell lymphoma. This result is consistent with the expected signal pattern for cells with a translocation involving the ALK locus (2p23) and separation of the ALK hybridization site. The signal pattern for the hyperplastic tonsil was normal, 2F.
Diffuse Large B-Cell Lymphoma
Eight specimens were studied with probes for IgH, CCND1, MALT1, API2, ALK, c-myc, BCL2, and BCL6. Seven of these specimens were known to have diffuse large B-cell lymphoma that evolved from follicular lymphoma and one was a normal lymph node. None of these specimens had previous genetic studies. The results were normal for CCND1/IgH, MALT1/API2, and ALK. The results were abnormal for BCL2/IgH fusion in 61 to 91% of nuclei among six lymphomas (Figure 2) ▶ . One of these specimens also had an abnormality of BCL6 in 91% of nuclei. Another one of these specimens had c-myc/IgH fusion in 66% of nuclei. The other diffuse large B-cell lymphoma had fusion of c-myc and IgH in 20% of nuclei and a BCL6 abnormality in 10% of nuclei. The signal pattern for each probe for the normal lymph node was normal.
Discussion
The results of this investigation demonstrate the efficacy of extracting nuclei from paraffin-embedded lymphoid tissues to perform successful interphase FISH studies. This method worked on a variety of lymphomas including mantle cell, follicular, Burkitt, MALT, anaplastic large-cell, and diffuse large B-cell. Moreover, this method allowed for successful hybridization of a variety of different FISH probes including BCL6, BCL2, c-myc, CCND1, ALK, IgH, MALT1, and API2.
Several methods have been used to extract individual nuclei from paraffin-embedded lymphoma tissue to perform various immunohistochemical stains. 3 Modifications of these methods have led to techniques that can be applied to FISH testing. 4,5 The method used in the present investigation combines the best aspects of two different published methods. 4,5
We previously investigated a method to extract nuclei from 50-μm sections of paraffin-embedded tissue to study mantle cell lymphoma 5 and MALT lymphoma. 7 That method required a significant quantity of tissue, six hours of processing time to extract individual nuclei, and did not consistently produce successful FISH studies. The present method requires a minute quantity of tissue (Figure 3) ▶ , 2 hours to extract nuclei, and consistently produces successful FISH studies. The timesavings associated with the current method results from elimination of a number of centrifugation steps, the replacement of pepsin with proteinase K enzymatic treatment, and processing tissue in a single tube eliminating the need for multiple tube transfers. Moreover, these changes reduce loss of cells and tissue during the extraction procedure.
Figure 3.
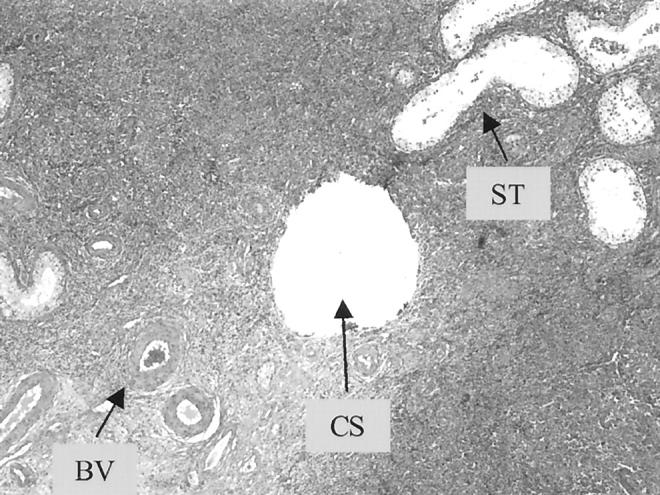
A routine H&E section of a large B-cell lymphoma in a testis after sampling by the paraffin-core technique used in this study. The relative size of the core is <0.55 mm (CS) and is comparable to adjacent small blood vessels (BV) and seminiferous tubules (ST).
The use of needle cores proved to be more efficacious than thick-sections to collect samples from paraffin-embedded tissue. In addition to requiring far less tissue, this procedure allowed sampling from exact areas of interest and was less destructive to the specimen because the cores are small (<0.55 mm in diameter) (Figure 3) ▶ . The sample was also easy to collect in the cytogenetics laboratory using common instruments.
The method used in this study was problematic for two diffuse large B-cell lymphoma specimens. The reason is not clear, but we considered several possible causes. Each of these specimens was fixed with B5. Because formalin was used to fix 28 of these specimens and B5 to fix 10 specimens, we believe this method works equally well on either of these forms of fixation. One specimen was 13 years old and other was 10 years old. However, the age of specimens used in this investigation ranged from 14 years to a few months. Although we were successful with even older specimens than either of the problematic specimens, it may be important that these two specimens were among the oldest in the investigation. Another factor that may affect hybridization efficiency of fluorescent-labeled DNA in paraffin-embedded tissue is the time from specimen collection to fixation. Although we do not have information regarding this variable for our specimens, it is reasonable to speculate that specimens that are allowed to deteriorate before fixation may be challenging for testing.
We investigated possible method modifications to perform successful FISH hybridization for the two diffuse large B-cell lymphoma specimens that failed by the technique used in this study. We substituted the microwave/citric acid step with a commercial steam machine such as those used to steam vegetables. We tried the steamer method using either citric acid (pH 6.0) or ethylenediaminetetraacetic acid (pH 8.0). The ethylenediaminetetraacetic acid-steamer method destroyed cellular structure and was not helpful. The citric acid-steamer method was successful and provided useful results for each of the FISH probes in this investigation. Both specimens proved to be abnormal for BCL2/IgH fusion and normal for all other probes tested. Preliminary experiments suggest that a commercial steamer may be substituted for the humidified microwave oven with improved results in some cases.
The results of FISH studies for chromosome abnormalities involving BCL6, BCL2, c-myc, CCND1, ALK, IgH, MALT1, and API2 were consistent with the type of lymphoma investigated. Each of the mantle cell, follicular, MALT, anaplastic large-cell lymphomas, and two Burkitt lymphoma specimens in this investigation were known to have abnormalities by previous genetic and FISH studies. 5-7 For these cases, the sensitivity of the method was 100%. Because each of the six normal tissues studied had a normal FISH pattern with each probe indicates this method has a high specificity as well. In addition, a variant FISH pattern associated with c-myc break-apart was detected in one patient with Burkitt lymphoma because of a t(2;8)(p12;q24) and was confirmed by cytogenetic studies. Thus, FISH has potential to detect variant chromosome anomalies associated with malignant lymphomas.
The results of this study suggest that using this method to extract individual nuclei from paraffin-embedded specimens and studied with the appropriate FISH probes, a chromosome abnormality can be detected with high specificity and high sensitivity in many lymphomas. Thus, this method should be helpful to hematopathologists to diagnose and classify lymphomas using FISH to detect common gene and chromosome abnormalities.
Footnotes
Address reprint requests to Gordon Dewald, Ph.D., Cytogenetics Laboratory, Hilton 970, Mayo Clinic, 200 First St. SW, Rochester, MN 55905. E-mail: gdewald@mayo.edu.
Supported in part by a grant from Vysis, Inc., Downers Grove, IL.
References
- 1.Jaffe ES, Harris NL, Stein H, Vardiman J: Pathology and genetics of tumours of haematopoietic and lymphoid tissues. World Health Organization of Tumours. 2001, International Agency for Research on Cancer Lyon
- 2.Andreeff M, Pinkel D: Introduction to Fluorescence in Situ Hybridization—Principles and Clinical Applications. 1999:p 455 John Wiley & Sons, Inc., New York
- 3.Hedley DW, Friedlander ML, Taylor IW, Rugg CA, Musgrove EA: Method for analysis of cellular DNA content of paraffin-embedded pathological material using flow cytometry. J Histochem Cytochem 1983, 31:1333-1335 [DOI] [PubMed] [Google Scholar]
- 4.Pickering D, Nelson M, Chan W, Huang J, Dave B, Sanger W: Paraffin tissue core sectioning: an improved technique for whole nuclear extraction and interphase FISH. J Assoc Genet Technol 2001, 27:38-39 [Google Scholar]
- 5.Remstein ED, Kurtin PJ, Buno I, Bailey RJ, Proffitt J, Wyatt WA, Hanson CA, Dewald GW: Diagnostic utility of fluorescence in situ hybridization in mantle-cell lymphoma. Br J Haematol 2000, 110:856-862 [DOI] [PubMed] [Google Scholar]
- 6.Cataldo KA, Jalal SM, Law ME, Ansell SM, Inwards DJ, Fine M, Arber DA, Pulford KA, Strickler JG: Detection of t(2;5) in anaplastic large cell lymphoma: comparison of immunohistochemical studies, FISH, and RT-PCR in paraffin-embedded tissue. Am J Surg Pathol 1999, 23:1386-1392 [DOI] [PubMed] [Google Scholar]
- 7.Remstein E, Kurtin P, James C, Wang X, Meyer R, Dewald G: MALT lymphomas with t(11;18)(q21;q21) and MALT lymphomas with aneuploidy develop along different pathogenetic pathways. Am J Pathol, in press. [DOI] [PMC free article] [PubMed]


