Abstract
Wilms’ tumor (WT) has been considered a prototype for arrested cellular differentiation in cancer, but previous studies have relied on selected markers. We have now performed an unbiased survey of gene expression in WTs using oligonucleotide microarrays. Statistical criteria identified 357 genes as differentially expressed between WTs and fetal kidneys. This set contained 124 matches to genes on a microarray used by Stuart and colleagues (Stuart RO, Bush KT, Nigam SK: Changes in global gene expression patterns during development and maturation of the rat kidney. Proc Natl Acad Sci USA 2001, 98:5649–5654) to establish genes with stage-specific expression in the developing rat kidney. Mapping between the two data sets showed that WTs systematically overexpressed genes corresponding to the earliest stage of metanephric development, and underexpressed genes corresponding to later stages. Automated clustering identified a smaller group of 27 genes that were highly expressed in WTs compared to fetal kidney and heterologous tumor and normal tissues. This signature set was enriched in genes encoding transcription factors. Four of these, PAX2, EYA1, HBF2, and HOXA11, are essential for cell survival and proliferation in early metanephric development, whereas others, including SIX1, MOX1, and SALL2, are predicted to act at this stage. SIX1 and SALL2 proteins were expressed in the condensing mesenchyme in normal human fetal kidneys, but were absent (SIX1) or reduced (SALL2) in cells at other developmental stages. These data imply that the blastema in WTs has progressed to the committed stage in the mesenchymal-epithelial transition, where it is partially arrested in differentiation. The WT-signature set also contained the Wnt receptor FZD7, the tumor antigen PRAME, the imprinted gene NNAT and the metastasis-associated transcription factor E1AF.
Wilms’ tumor (WT) is a common pediatric malignancy that recapitulates the histology of the nephrogenic zone of the growing fetal kidney, and it has long been viewed as a prototype of differentiation failure in human neoplasia. 2,3 Certain developmentally regulated genes have already been examined in these tumors. Notable among these are PAX2, which is essential for kidney development, 4-6 and which is expressed in WTs, 7,8 and the WT1 tumor suppressor, which is also essential for kidney development, 9 and which may function downstream of PAX2. 10-14
To gain a more complete view of gene expression in WTs, we have performed an unbiased survey using oligonucleotide microarrays. We sought to identify first a large set of genes that are differentially expressed between WTs and control fetal kidneys and, second, a smaller and more specific subset of genes that are highly expressed in WTs relative to additional control tissues, including a histologically unrelated type of cancer. The results from the first screen statistically confirm the notion, previously based on histology and analysis of selected genes, that cells in WTs are arrested at an early stage of fetal kidney development. The identities of the smaller subset of WT signature genes emphasize high expression in WTs of genes that are essential for kidney formation and that act at the earliest committed stage in the mesenchymal-epithelial transition.
Materials and Methods
Tissues
WTs were an unselected series of primary nonsyndromic tumors resected at Babies Hospital of Columbia University. None of the WTs were anaplastic. Cryopreserved samples of these tumors, control fetal tissues (16- to 22-week gestations), primary Burkitt lymphomas (BLs), and pulmonary adenocarcinomas were obtained from the Columbia University Cancer Center Tissue Bank. The BL cell lines were from the American Type Culture Collection (Rockville, MD).
Microarrays and Probes
HG-U95A GeneChips (Affymetrix, Santa Clara, CA), which query ∼10,000 genes (12,000 probe sets), were used for all analyses. The cRNA probes were synthesized as recommended by Affymetrix. Briefly, total RNA was isolated in two steps using Trizol (Invitrogen, Carlsbad, CA), followed by RNeasy (Qiagen, Valencia, CA) purification. Double-stranded cDNA was generated from 5 μg of total RNA using a polydT oligonucleotide that contained a T7 RNA polymerase initiation site and the Superscript Choice System Kit (Invitrogen). cDNA was phenol/chloroform extracted. Biotinylated cRNA was generated by in vitro transcription using the Bio Array High Yield RNA Transcript Labeling System (Enzo, Farmingdale, NY). The cRNA was purified using RNeasy. cRNA was fragmented according to the Affymetrix protocol, and 15 μg of biotinylated cRNA were hybridized to U95A microarrays (Affymetrix). After scanning, the expression values for each gene were determined using Affymetrix GeneChip software version 4.0.
Data Analysis
Two software packages, GeneSpring (Silicon Genetics, Redwood City, CA) and Genes at Work, which utilizes the Structural Pattern Localization Analysis by Sequential Histograms, SPLASH, algorithm, 15,16 were used for analyzing the GeneChip data. Simple statistical and expression value cutoffs were done using the filter functions of GeneSpring. The GeneTree function, a hierarchical clustering tool in GeneSpring that creates dendrograms of genes based on the extent of relatedness of their expression patterns, was used for nonsupervised clustering. Supervised clustering of the initial set of differentially expressed genes was done using the pattern discovery algorithm of Genes at Work, 15 as previously described. 17 For the initial screen using GeneSpring (derivation of sets A and B), only those genes that gave consistent Affymetrix presence calls in the GeneChip data for both the WTs and the fetal kidneys (at least five of six samples with presence calls) were analyzed. The Genes at Work algorithm, used for deriving the smaller set of WT signature genes, accepted all data, including Affymetrix absence calls. Among the WT signature genes, tissue samples for which particular probe sets gave absence calls were invariably control tissues with very low absolute signals for these probe sets. However, no significant differences were seen between the overall percentage of absence calls (all genes on the microarray) among the different tissues.
Northern Blotting
Total RNA from WTs and fetal kidneys, prepared using Trizol reagent (Invitrogen), was resolved on formaldehyde-containing agarose gels and transferred to Nytran membranes (Schleicher and Schuell, Keene, NH). Probes for Northern blotting were partial cDNAs prepared by reverse transcriptase-polymerase chain reaction using gene-specific primers. Hybridization was in ULTRAhyb buffer (Ambion, Austin, TX) at 42°C degrees. Probes were stripped by boiling the membranes in 0.1% sodium dodecyl sulfate/0.1× standard saline citrate solution for 10 minutes, or were allowed to decay without stripping.
Immunohistochemistry
Sections of WTs and fetal kidneys were deparaffinized in two changes of xylene for 5 minutes each. Sections were hydrated through graded ethanols. Antigen retrieval was performed in 1 mmol/L of ethylenediaminetetraacetic acid buffer by boiling the slides in a microwave oven for 8 minutes at the maximum power and, sequentially, boiling for 15 minutes at a reduced power. Endogenous biotin was blocked by two steps of incubation for 10 to 20 minutes with egg white and 5% skim milk in 1× Tris-buffered saline (TBS) containing 0.5% bovine serum albumin and 0.1% NaN3, respectively. Between the two blocking step for biotin, slides were treated with 0.3% hydrogen peroxide in 0.1% NaN3 to block endogenous peroxidase activity. Slides were washed three times in 1× TBS buffer containing 0.1% Tween-20 (TBS-T) and then incubated with 5% of the blocking serum for 30 minutes in a humidified chamber. After three TBS-T washes, slides were hybridized with polyclonal antipeptide antibodies against SIX1 (Santa Cruz Biotechnology, Santa Cruz, CA), PAX2/8 (Santa Cruz), or SALL2 (CeMines, Evergreen, CO) at room temperature overnight. After washing in TBS-T, slides were incubated with biotinylated secondary antibodies (Vector Laboratories, Burlingame, CA) for 30 to 40 minutes at room temperature. Antigen-antibody complexes were developed by the Vectastain ABC kits (Vector Laboratories) and a chromogenic substrate, diaminobenzidine (DAKO, Carpinteria, CA). Sections were counterstained with hematoxylin.
Results
Genes Differentially Expressed between WTs and Fetal Kidneys
Nephrogenesis is an ongoing process, in which reciprocal inductive interactions of branches of the ureteric bud with overlying metanephric mesenchyme are played out repeatedly as the uretic system ramifies and the kidney grows. 3,14 Mid-gestation fetal kidneys are therefore a composite of fully differentiated, differentiating, and undifferentiated cells, and gene expression in whole fetal kidney is an average over these cells. WTs are more homogeneous, but these neoplasms can also contain a spectrum of cell types, which are classified as blastemal, epithelial, and stromal. Despite this heterogeneity, we reasoned that an initial WT versus whole fetal kidney comparison could be useful in eliminating a large number of genes whose average expression did not differ between these two sources, and identifying sets of differentially expressed genes. Genes found to be overexpressed in WTs by this comparison might reflect either pathological overexpression on a per-cell basis, or stage-appropriate expression in a cell type that is numerically overrepresented in WTs. Both types of net overexpression are potentially interesting, and the latter category might be informative concerning the stage at which the primitive-appearing blastemal cells in WTs are blocked in their ability to differentiate.
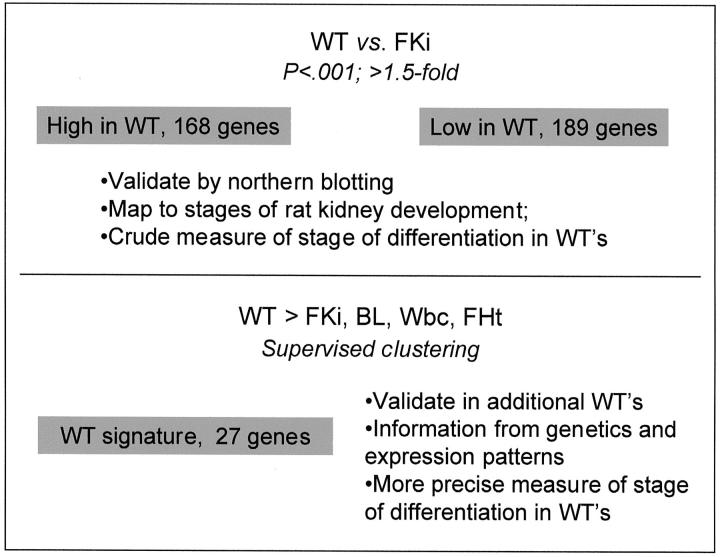
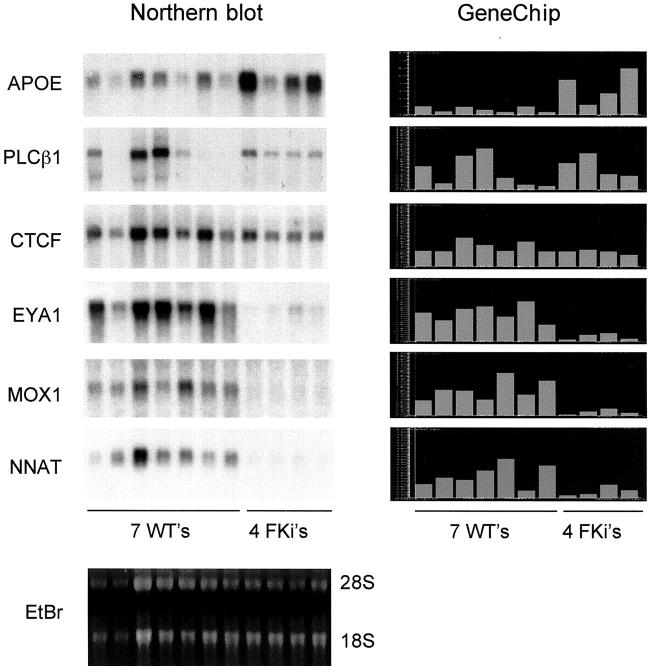
We therefore first performed microarray analysis with cRNA probes from six sporadic (nonsyndromic) WTs, using six whole mid-gestation (16 to 22 weeks) fetal kidneys as the comparison group. We used simple statistical and expression value criteria to select an initial set of differentially expressed genes (Figure 1) ▶ . This produced a set of 357 differentially expressed genes, represented by 396 oligonucleotide probes. Clustering using the GeneTree function of GeneSpring yielded two clades, with 168 genes more highly expressed in WTs than in fetal kidneys (set A), and 189 genes more highly expressed in fetal kidneys than in WTs (set B). To verify the accuracy of the microarray data, we performed Northern blotting of total RNAs from WTs and fetal kidneys and hybridized the blot successively with cDNA probes for six genes that showed various expression patterns. The Northern blotting results for each gene correlated well with the microarray data (Figure 2) ▶ .
Figure 1.
Strategy for analyzing gene expression in WTs. For a gene to be included in set A or set B, we first required a value of P < 0.001 by a chi-square test for differential expression between the WTs and the control fetal kidneys. We additionally required a normalized expression value of >1.5 relative to the experiment mean in at least three samples, either WTs or fetal kidneys.
Figure 2.
Validation of the GeneChip data by comparisons with Northern blotting. Left: A single Northern blot was successively hybridized with cDNA probes for the indicated genes. Right: The GeneChip expression data (normalized to the experiment mean) are displayed with the same order of samples. EtBr, ethidium bromide stain of the Northern gel showing 28S and 18S ribosomal RNAs.
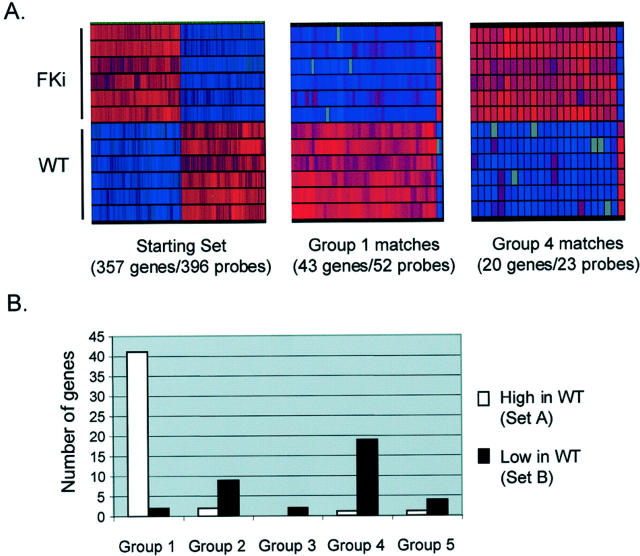
We next mapped our initial set of differentially expressed genes to all genes on the RG-U34A Affymetrix microarray that was previously used by Stuart and colleagues 1 in a study of normal kidney development in the rat. These authors queried gene expression in whole rat kidneys at seven successive stages of development, from 13 days postcoitum (dpc) to adulthood. Although the developing kidney is a heterogeneous mixture of cell types at each of these stages, the less differentiated cellular populations predominate at the early stages. Stuart and colleagues 1 used statistical criteria to establish five clusters of genes (group 1 to group 5) with characteristic temporal patterns of expression. In their classification, group 1 genes are highly expressed at the earliest stage examined, 13 dpc, a stage at which the ureteric bud has first contacted the metanephric mesenchyme, and decline in expression thereafter. Genes in the other groups peak in their expression at progressively later stages: for example, group 2 genes reach their highest expression between 17 dpc and 19 dpc, whereas group 4 genes become highly expressed only at birth (21 dpc) and group 5 genes show high expression only in fully mature adult kidneys. 1 We used the FASTA search algorithm as implemented in the Genetics Computer Group (GCG) package, 18,19 to query a database of all genes represented on the U34A GeneChip with each of the 357 differentially expressed genes in our initial set. Of these 357 genes, 124 had convincing matches by stringent criteria (E score, ≤40; Z-score, >800, with evidence of orthology from inspection of the gene annotations) among the rat genes represented on the U34A GeneChip. Of these, 81 genes fell into groups 1 to 5, whereas the remainder were not detected by Stuart and colleagues 1 as varying significantly during development and so were not assigned to groups. Genes in our set A (high in WTs) fell overwhelmingly into group 1 (42 of 45 genes; 93%; Figure 3A ▶ ), whereas genes in set B (low in WTs) fell predominantly into groups 2 to 5 (34 of 36 genes; 94%; Figure 3A ▶ ). That the expression of these genes was generally consistent among the different WT cases, and among the fetal kidney controls, is shown in Figure 3B ▶ .
Figure 3.
Correlation of gene expression in WTs with normal kidney development in the rat. A: Genes identified in the current study that mapped to genes identified by Stuart and colleagues. 1 Genes that are more highly expressed in WTs than in whole fetal kidneys map predominantly to the earliest group of Stuart and colleagues, 1 whereas genes that are expressed at lower levels in WTs than in fetal kidneys map to the later groups. B: Graphical representations (GeneTree function, GeneSpring) showing the uniformity of relative overexpression or underexpression across multiple genes and multiple samples. Genes are arrayed along the x axis; WT and fetal kidney samples are shown on the y axis. Gene expression is automatically color-coded with red indicating higher and blue lower expression. Gray indicates Affymetrix absence calls. The group 1 matches are nearly all more highly expressed in WTs, whereas the group 4 matches are nearly all more highly expressed in fetal kidneys. Color range is set at 1 = normal, 6 = maximum.
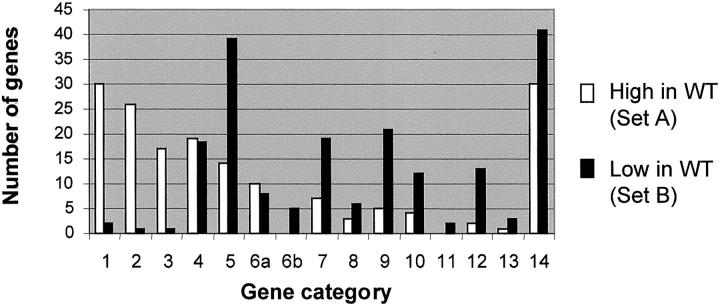
We next classified the differentially expressed genes according to known or predicted function, by reviewing sequence information and the associated scientific literature using public databases (http://www.ncbi.nlm.nih.gov/). We annotated each of the genes in set A and set B, and assigned them to categories similar to those used by Stuart and colleagues. 1 This showed that genes involved in DNA synthesis or metabolism (including repair), regulation of the cell cycle, and RNA metabolism were greatly overrepresented in set A, whereas genes involved in several other functions were overrepresented in set B (Figure 4 ▶ ; gene identities in supplementary data, http://icg.cpmc.columbia.edu/tycko/data/). The results in terms of DNA synthesis or metabolism and regulation of the cell cycle confirm the intuitive expectation that proliferating tumor cells should overexpress genes in these categories, whereas the data for RNA metabolism are less intuitively obvious. But for all three categories, our data closely parallel those of Stuart and colleagues, 1 who found that genes involved in DNA synthesis or metabolism, regulation of the cell cycle, and RNA metabolism, are overexpressed in the earliest stage of metanephric kidney development. Overall, these data from an unbiased screen substantiate the notion, which has previously been based on histology and selected markers, that the cells in WTs are, on average, arrested in their differentiation at a point corresponding to an early stage of development of the metanephric kidney.
Figure 4.
Functional categories of genes differentially expressed between WTs and fetal kidneys. The categories are: 1, DNA synthesis or metabolism (including repair); 2, cell cycle regulation (including mitotic spindle); 3, RNA metabolism (not including transcription factors); 4, transcriptional regulation (including transcription factors); 5, cell signaling; 6a, protein metabolism nonproteolytic; 6b, protein metabolism proteolytic; 7, small molecule metabolism (enzymes); 8, small molecule transport; 9, extracellular matrix, adhesion, and serum proteins; 10, intracellular trafficking; 11, energy metabolism; 12, cytoskeleton (not including mitotic spindle); 13, protection against oxidative stress; 14, unknown function.
Derivation of WT Signature Genes
To derive a smaller set of genes that might give more precise information about the phenotype of WTs, we sought to screen-out those genes that were highly expressed as a nonspecific consequence of cellular proliferation. To do this, we added microarray data from a heterologous (histologically unrelated) highly proliferative tumor type, BL. We obtained data from a series of four primary BLs and four BL cell lines. To eliminate any genes with nonspecific housekeeping functions that may have persisted after the initial WT/fetal kidney screening, we also added heterologous normal control tissues: peripheral blood leukocytes from four healthy individuals, and six mid-gestation fetal hearts. Automated nonsupervised clustering (GeneTree function of GeneSpring) of the initial set of differentially expressed genes (set A and set B) according to their expression in these different tissues yielded patterns that clearly distinguished each type of tumor and normal tissue (Figure 5A) ▶ .
Figure 5.
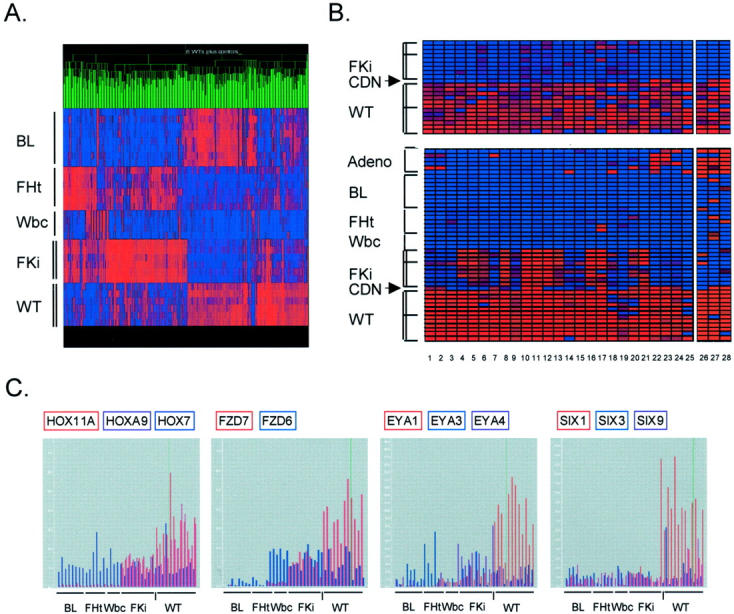
Derivation of WT signature genes. A: Nonsupervised clustering (GeneTree function of GeneSpring) of the 357 genes that were initial identified as differentially expressed between WTs and fetal kidneys. Many of the high in WT/low in fetal kidney genes are also high in the BLs. The color range is normal = 1, maximum = 6. All data are included, with absence calls represented by pure blue. B: WT signature genes. The 27 highest-scoring genes identified by SPLASH are shown here, after clustering using the GeneTree function of GeneSpring (columns 1 and 2 are two probe sets both identifying CRABP2). The identities of these genes are in Table 1 ▶ . Tissues are: AdenoCA, pulmonary adenocarcinoma (six primary tumors); BL, Burkitt lymphoma (four primary tumors and four cell lines); FHt, mid-gestation fetal hearts (six samples); Wbc, normal peripheral blood (four samples); FKi, mid-gestation fetal kidneys (six samples in A and the same six samples below the dividing line in B, with three additional fetal kidneys above the line); CDN, differentiating cystic nephroblastoma (one case); WT, (six WTs in A and the same six WTs below the dividing line in B, with six additional WTs above the line). Tissues used for analysis by Genes at Work are bracketed; the adenocarcinomas were not included in deriving the WT signature genes. The genes in set C were identified by SPLASH using only data from the samples shown in A. The additional WTs and fetal kidneys, above the dividing lines, therefore validate the signature set. The differentiating cystic nephroblastoma shows a distinct pattern of gene expression. Because the data are normalized to all tissues, displaying the WT and fetal kidney samples alone (top) emphasizes the differences between them, while displaying these samples together with the heterologous tissues highlights their close affinity. Color ranges are set at 1 = normal, 6 = maximum (top) and 2 = normal, 10 = maximum (bottom). C: Differential expression of genes in the HOX-, FZD-, EYA-, and SIX-families. Microarray data are normalized to the experiment mean. Each bar represents the value for a single sample. BL, Burkitt lymphoma (eight samples); FHt, fetal heart (six samples); Wbc, peripheral blood leukocytes (four samples); FKi, fetal kidney (nine samples); WT, Wilms’ tumor (12 samples). The vertical dash indicates the single case of differentiating cystic nephroblastoma.
More importantly, it was evident from the GeneTree display that a substantial proportion of genes in set A (high in WTs) were in fact highly expressed in the primary BLs and in the BL cell lines (Figure 5A) ▶ . In fact, examination of the composition of clusters containing the genes that were simultaneously high in WT and in BL showed that they contained all of the genes that fell into the DNA synthesis/repair and cell cycle categories, as well as other genes that are known to be markers of cellular proliferation. A few examples are Ki67 antigen, CDK2, cyclin B, Ku80 DNA-binding protein, and several kinesin genes. This confirmed the expectation that most of these genes would be nonspecific markers of cellular proliferation. A smaller percentage of the genes in set A were highly expressed in fetal heart and/or in peripheral blood leukocytes, but as expected these control tissues tended instead to express genes from set B (Figure 5A) ▶ .
Given these observations, we were confident that we had chosen a useful combination of control tissues. We therefore used a supervised learning algorithm in the Genes at Work package, 16 to rank all genes based on the difference in their expression between WTs and these control tissues. The 30 probe sets with the highest Z-scores (measure of probability) included 29 genes, with 1 gene (CRABP2) identified by two different probe sets. When this set was examined in an expanded series of WTs and fetal kidneys (six additional WTs and three additional fetal kidneys), all but 2 of the 29 genes showed significantly higher expression in the additional WTs compared to the control tissues. The resulting 27 WT signature genes, which we refer to as set C, are shown in Figure 5B ▶ and listed in Table 1 ▶ . In addition to the positive validation of these genes in additional WTs, as a further test of specificity we examined their expression in microarray data from one case of a related but distinct type of pediatric kidney tumor, cystic differentiating nephroblastoma. This case showed a distinct pattern of gene expression, with nine of the set C genes silent, and several others detected only at low levels (Figure 5B) ▶ .
Table 1.
Genes in Set C
| Figure 5b ▶ | Probe set | Gene product | Symbol | GenBank |
|---|---|---|---|---|
| 1 | 1057_at | Retinoic acid-binding protein II | CRABP2 | M97815 |
| 2 | 41783_at | Retinoic acid-binding protein II | CRABP2 | M97815 |
| 3 | 36010_at | Mox1 homeodomain protein | MOX1 | U10492 |
| 4 | 39051_at | Neuronatin alpha and neuronatin beta | NNAT | U31767 |
| 5 | 37605_at | Pro-alpha1 type II collagen | COL2A1 | L10347 |
| 6 | 1058_at | Brush-1; Was-family protein | WASF3 | S69790 |
| 7 | 40292_at | DBCCR1; IB3089A; retinoic acid inducible | DBCCR1 | AF027734 |
| 8 | 33222_at | Frizzled-7 | FZD7 | AB017365 |
| 9 | 37567_at | Zinc finger protein, Hsal2; KIAA0360 | SALL2 | X98834 |
| 10 | 37809_at | Homeobox gene; HOXA9 | HOXA9 | U41813 |
| 11 | 33248_at | EST matching HOXA11 | HOXA11 | H94842 |
| 12 | 38184_at | Pax2 paired box protein | PAX2 | M89470 |
| 13 | 38952_s_at | Alpha-2 type IV collagen | COL4A2 | M33653 |
| 14 | 35396_at | Hyaluronan synthase | HAS2 | U54804 |
| 15 | 37073_at | EYA1C: eyes absent homolog 1 | EYA1 | AJ000098 |
| 16 | 38749_at | G-protein coupled receptor GPR39 | GPR39 | AI936826 |
| 17 | 40358_at | DNA-binding protein GLI3 | GLI3 | M57609 |
| 18 | 35200_at | HMGI-C high-mobility group factor | HMGIC | X92518 |
| 19 | 37283_at | MGCR gene, protein of unknown function | MGCR1, MN1 | X82209 |
| 20 | 37630_at | Chordin-like gene; Unigene Hs.82223 | (not assigned) | AL049176 |
| 21 | 38853_at | HE6; seven transmembrane-domain receptor | HE6 | X81892 |
| 22 | 38545_at | Inhibin beta, B-subunit | INHBB | M31682 |
| 23 | 40004_at | SIX1 protein; homeobox protein | SIX1 | X91868 |
| 24 | 41395_at | Keratan sulfate Gal-6-sulfotransferase | CHST1 | AB003791 |
| 25 | 31946_s_at | HBF-2 forkhead-family transcription factor | HBF2 | X74143 |
| 26 | 2084_s_at | E1A-F; Ets-family transcription factor | E1AF | D12765 |
| 27 | 157_at | PRAME-1 tumor antigen | PRAME | U65011 |
| 28 | 41764_at | EST similar to APOCI | APOCI | AA976838 |
Identities of WT Signature Genes
Set C was depleted for genes that are involved directly in the general functions of DNA replication and repair and cell-cycle control, but it was enriched for genes encoding transcription factors. The fraction of human-coding sequences devoted to transcription factors has been estimated at 6% based on genomic sequencing. 20 Of the genes in set A, 12% encoded known transcription factors, but 41% of genes in set C (11 of 27) were in this category (Figure 5B ▶ and Table 1 ▶ ). As discussed in detail below, many of the 11 transcription factor genes in set C are informative as developmental markers. Four of these genes, PAX2, EYA1, HBF2, and HOXA11 have been previously shown to be essential for metanephric development in mice and/or humans. 5,6,21-25 A fifth gene in set C, SALL2, has not been previously implicated in kidney development, but it is homologous to SALL1, encoding a Spalt-family transcription factor that is essential for normal kidney development in mice and humans. 26,27 Some of the other transcription factor genes in set C are also likely to function in kidney development and, as discussed below, two of them, MOX1 and SIX1 are known to act in the PAX/EYA pathway.
Additional genes in set C, not encoding transcription factors, are also likely to participate in the formation of the metanephric kidney. Signaling by Wnt-family proteins, including Wnt-4, is essential for the epithelial conversion of the metanephric mesenchyme. 28 This occurs via interaction of Wnt ligands with one or more Frizzled-family receptors, and the FZD7 (Frizzled-7) gene, in set C, is a candidate for this function. Interestingly, Wnt signaling is essential for appropriate expression of a chicken Spalt-family protein, cSal1. 29 Other genes in set C have unknown functions. One of these, neuronatin (NNAT), is subject to genomic imprinting, such that it is expressed only from the paternal allele. 30 This gene, which encodes a proteolipid, produces abundant mRNA in various fetal tissues, but is silenced on differentiation of neurons and other cell types. 30,31
The FZD7 gene, like HOXA11, SIX1, and EYA1, belongs to a family with multiple members, so we asked whether the microarray data could give some indication of specificity within these gene families. Using selected genes on the Hu95A array that gave consistent presence calls in one or more tissue type, it was possible to compare the expression patterns of HOXA11 and HOXA9 versus HOX7, SIX1 versus SIX3 and SIX9, EYA1 versus EYA3 and EYA4, and FZD6 versus FZD7 (Figure 5C) ▶ . These comparisons confirmed the specificity of the overexpression of HOXA11, HOXA9, SIX1, EYA1, and FZD7 in the WTs.
Many of the set C genes are transcribed at detectable levels in fetal kidneys, whereas few are detected in the heterologous controls (Figure 5B) ▶ . This confirms the intuition that there should be a close relationship between the fetal kidney and WT. However, three genes in set C, E1AF, PRAME, and APOCI, differed from the others in that they were expressed at high levels in WTs, but were undetectable in the normal fetal kidneys. These genes are less likely to have roles in kidney development, and more likely to be overexpressed because of neoplastic transformation per se. E1AF encodes an Ets-family transcription factor that is overexpressed in several types of metastatic tumor cell lines, 32,33 and PRAME encodes a cell surface antigen that is highly expressed in melanomas and acute leukemias. 34,35 APOCI produces a serum apolipoprotein. Although not expressed in BL, these genes are frequently expressed in other tumor types, as confirmed by high signals seen in our microarray data from a series of pulmonary adenocarcinomas (Figure 5B) ▶ , and by queries of the Unigene database (http://www.ncbi.nlm.nih.gov/unigene). Several of the adenocarcinomas also expressed SIX1 (see Discussion).
Distribution of SIX1 and SALL2 Proteins in Fetal Kidneys and WTs
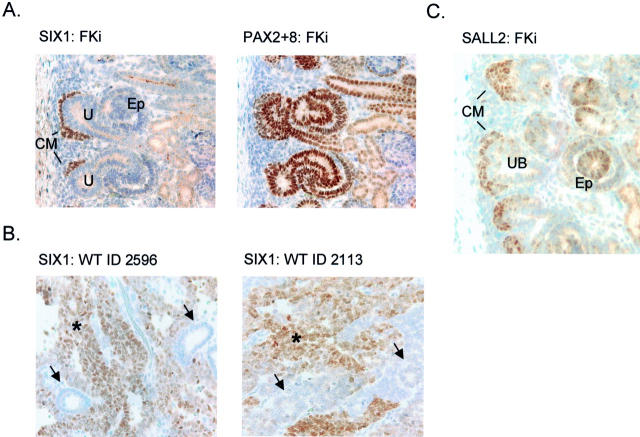
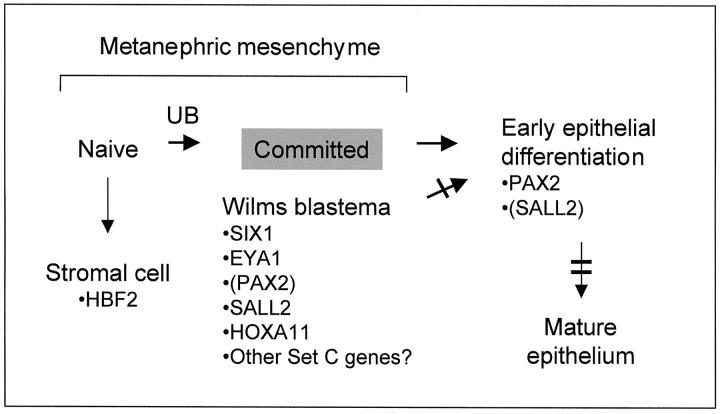
The relative overexpression of particular genes in WTs compared to whole fetal kidneys might reflect either a uniform difference in expression on a per-cell basis, or a greater numerical abundance of a particular type of cell in the WTs that might be present as minor cell population in fetal kidneys. Because we suspected that most of the genes in set C would fall into the latter category, we assessed the tissue distribution of SIX1 and SALL2 proteins in sections of WTs and normal fetal kidneys. For SIX1, the expression in fetal kidney was highly specific for the condensing mesenchyme, which was seen closely apposed to the tips of the ureteric bud branches (Figure 6A) ▶ . These cells have received and responded to an inductive signal from the ureteric bud epithelium, but they do not yet show morphological transformation into epithelial structures. As expected based on previous studies, 8,36 a control antibody recognizing both PAX2 and PAX8 also stained these cells, but reacted more strongly with ureteric bud epithelium and with differentiating epithelial structures deeper in the developing renal cortex (Figure 6A) ▶ . In two WTs examined by immunohistochemistry, SIX1 protein was detected only in the blastemal elements (Figure 7B) ▶ . Like SIX1, SALL2 protein strongly marked the condensing mesenchyme, but it was also detectable at lower intensity and in fewer cells in the differentiating epithelial structures (Figure 6C) ▶ . These findings, together with published developmental information for several of the other genes in set C, indicate that for many of these genes their overexpression in WTs reflects the numerical predominance of neoplastic cells arrested at a specific developmental stage. The composition of set C can therefore be used to establish a working model for failure of cellular differentiation in WTs (Figure 7) ▶ .
Figure 6.
Immunohistochemical detection of SIX1 and SALL2 proteins in human fetal kidney and in WTs. A: Serial sections of a 20-week gestation human fetal kidney were developed with anti-SIX1 and anti-PAX2 + 8 polyclonal antisera. SIX1 is expressed only in the earliest condensing mesenchyme (metanephric blastema), which is seen directly apposed to the uretic bud-derived epithelium. PAX2 + 8 detects expression of these two proteins in the condensing mesenchyme, and there is a strong signal in differentiating epithelial structures (S-shaped and comma-shaped bodies) deeper in the kidney, 8,50 as well as in ureteric bud. 36 Weak cytoplasmic staining in differentiated epithelial cells is nonspecific (seen in control with secondary antibody alone). B: SIX1 is expressed in the blastemal component of WTs (asterisks), but not in the epithelial areas (arrows). This pattern was seen in three of three cases of WT examined. In these same cases the epithelial component stained strongly with anti-PAX2 + 8 (not shown). C: Anti-SALL2 polyclonal antiserum gives a strong and uniform signal in the condensing mesenchyme, but cells with nuclear SALL2 are also seen at lower frequencies and with somewhat reduced staining in differentiating epithelial structures. Ureteric bud derivatives are negative for SALL2. Weak cytoplasmic staining in differentiated epithelial cells is nonspecific. CM, condensing mesenchyme; U, ureteric bud branches; Ep, differentiating epithelial structures; Gl, primitive glomeruli; Tu, differentiated renal tubules.
Figure 7.
Differentiation arrest in WTs inferred from gene expression profiling. A schematic of the normal metanephric differentiation program is shown. The neoplastic blastema, which comprises the bulk of many WTs, is arrested at the position indicated by the gray box, and relevant WT signature genes are indicated. The single bar indicates a partial differentiation arrest in WTs, and the double bar indicates a full block of differentiation. Parentheses indicate low but detectable expression. UB indicates the inductive interaction with the ureteric bud.
Discussion
WT has long been considered a prototype for arrested cellular differentiation in cancer, but previous assessments of the WT phenotype have relied on histology and selected markers. The current data, from an unbiased survey of ∼10,000 genes, support the general statement that cells in WTs are arrested in their differentiation at an early stage of metanephric development. Access to microarray data from a previous study of normal kidney development in an animal model facilitated this interpretation. 1 This emphasizes the benefits of using standardized methods for microarray-based studies, and making the primary data available through the Internet. A second lesson of experimental design that may be pertinent for other studies is that the direct comparison of tumor biopsies with whole-organ control samples, although it relies on averaging across heterogeneous cell populations, can still produce informative data.
Our set C was derived by an unbiased approach that identified those genes with higher expression in WTs than in the homologous tissue fetal kidney and the histologically unrelated tissues fetal heart, peripheral blood, and BL. Empirically, this set of genes was strikingly enriched in developmentally essential genes and stage-specific markers of the metanephros, and depleted in genes with more general functions. We have therefore used the operational term “WT signature” to describe this set. But few genes are expected to be completely tissue-specific, and the expression of the set C genes is not restricted to WTs. The SIX1 gene is highly expressed in some human breast cancers and in rhabdomyosarcomas, 37,38 and we found high expression of SIX1 mRNA, as well as several other genes from set C, in several pulmonary adenocarcinomas. Nor are these genes specific for normal metanephros: Six1 mRNA is expressed in the developing somites, 39 and Eya1 is essential both for kidney and for inner ear and craniofacial bone development. 21 By Northern blotting, human FZD7 mRNA is most abundant in normal fetal kidney and in adult skeletal muscle, but it is also detectable in fetal lung, adult heart, and placenta. 40 However, in these multiple sites, these developmental regulators play homologous functions. Eya1, for example, is necessary for the survival of condensing mesenchyme, both in the metanephros and in the otic vesicle. 21
Disproportionately represented in set C were genes in the PAX pathway, which included 4 of the 27 genes in this set. Genes in the PAX-, SIX-, MOX-, and EYA-families act together in a shared pathway for tissue growth and differentiation. EYA-family proteins bind directly to members of the SIX family of homeodomain proteins, and they provide a transcriptional co-activator function. 41 The expression of Six1 and Eya2 is induced by Pax3 expression in murine myogenic stem cells, 42 paralleling genetic interactions that were originally described in Drosophila eye development, with Pax (eyeless) acting upstream of Eya (eyes absent), which in turn acts upstream of Six (sine oculis). 43,44 Pax3 acts synergistically with Eya2 to induce muscle differentiation, 39 and Eya2 also shows synergy with Six1 in this system. 39 Eya1 is required for expression of Six2 (and probably Six1) in condensing metanephric mesenchyme of the murine kidney, and for expression of Six1 in the otic vesicle, suggesting positive feedback of Eya/Six heterodimers on Six-family genes. 21 There is also evidence for positive feedback of Eya/Six on Pax genes. 45 The Mox1 and Mox2 proteins bind directly to Pax1 and Pax3 proteins, respectively. 46 Mox2 is essential for myogenesis in mice, 47 but it is not yet known whether Mox1 is essential for kidney development.
Mice mutant for Pax2 fail to develop normal kidneys 6 and provide an exaggerated model for the human haploinsufficiency disorder renal-coloboma syndrome. Similarly, deletion or mutation of Eya1 in the mouse germline caused defective morphogenesis in the kidney. 21,23 These mice are severe models for another human disorder, the brachio-oto-renal (BOR) syndrome, which is because of EYA1 haploinsufficiency and which is associated with renal dysgenesis, inner ear defects, and craniofacial abnormalities. 22
Although there is no evidence that it acts in the PAX pathway, the Sall1 gene, closely related to Sall2, is also essential for kidney development. 27 Mouse embryos lacking this gene are an exaggerated model for a human genetic syndrome, Townes-Brocks syndrome, that is associated with renal hypoplasia and other developmental abnormalities because of heterozygosity for mutations in SALL1. 26 SALL2 was included in set C, but our analysis did not identify SALL1 in either set A or set C, even though this gene has been reported as overexpressed in WTs. 48 A probe set querying SALL1 is present on the HG-U95A microarray, and examination of the data showed that this gene is indeed highly expressed in some, but not all, WTs (not shown). This intertumor variability explains the lack of inclusion of this gene in set C.
Optimally, conclusions about the stage of differentiation in WTs should be based on large panels of markers. Our analysis has identified a well-studied marker, PAX2, and several new ones with informative expression patterns in the normal fetal kidney. Eya1 is expressed only at the earliest stage of commitment of the metanephric mesenchyme to the epithelial-mesenchymal transition, as indicated by in situ hybridization in developing mouse kidneys. 21,49 Pax2 is also most highly expressed in this early condensing mesenchyme, but in both mouse and human Pax2 mRNA is also detected at lower levels in later mesenchyme-derived structures (comma- and S-shaped-bodies) and in ureteric bud. 8,50 Like Eya1 and, as we have shown, SIX1, the Hoxa11 gene is a specific marker for the earliest committed metanephric mesenchyme. 25 The murine Bf2 gene (human orthologue HBF2) is expressed in the primitive stromal cells of the metanephric kidney, 24 so this gene does not fit the pattern of Pax2, Eya1, Hoxa11, and Six1. In this regard, it may be relevant that of the genes in set C, HBF2 showed the greatest variability in expression among the different cases of WT (Figure 5B) ▶ . This may be because WTs can contain varying proportions of stromal-lineage cells, but two technical problems, namely variable composition in different areas of the same tumor and lack of an effective anti-HBF2 antibody, have prevented us from testing this.
Although we have focused on transcription factors, a number of the other genes in set C are likely to affect the mesenchymal-epithelial transition. The Wnt pathway and frizzled-family receptors, such as FZD7, were mentioned above. Another crucial signaling pathway in metanephric development is mediated by ligands in the bone morphogenetic protein (Bmp) family and requires sulfated glycosaminoglycans in the extracellular matrix. 51,52 So, the presence of a carbohydrate sulfotransferase gene, CHST1, in set C is not surprising.
A working model for differentiation arrest in WTs, shown in Figure 7 ▶ , is consistent with the available data. In this model, WTs arise from a primitive mesenchymal cell that is capable of differentiating in the stromal and epithelial directions. A differentiation block, which is partial, is present at the transition from committed mesenchyme to early epithelium. This is consistent with the accumulation in most WTs of an abundant blastemal component that expresses PAX2, SIX1, EYA1, SALL2, and HOXA11. Primitive epithelial structures can form in WTs from the cells that succeed in passing this block, but these are often a minority of cells. Cadherin genes are useful markers for the successive stages of epithelial differentiation, with cadherin-11 being widely expressed in both naïve and committed metanephric mesenchyme, and cadherin-6 and E-cadherin expressed in successively more mature epithelial structures. 53 E-cadherin appeared in our set B (supplementary data), and consistent with our model, the microarray data ranks the expression of these genes in WTs in the order cadherin-11 > cadherin-6 > E-cadherin (data not shown).
What might be the functional importance of the signature genes that we have identified? There is no evidence that genes in set C play a primary role as proto-oncogenes in initiating Wilms’ tumorigenesis, but they may have essential downstream functions. In particular, because mutations of several of these genes cause renal hypoplasia or aplasia because of failed proliferation of nonneoplastic metanephric mesenchyme, and because additional set C genes act coordinately in these same pathways, these genes may also be required to maintain cell survival and proliferation in the blastema of WTs.
Acknowledgments
We thank J. Licht and M. Goldberg for insightful discussions, A. Saxena for advice on immunohistochemistry, G. Cattoretti for immunohistochemistry protocols (http://icg.cpmc.columbia.edu/cattoretti/protocol/), and V. Miljkovic for microarray hybridization and advice on data management.
Footnotes
Address reprint requests to B. Tycko, Berrie Research Pavillion, 1150 St. Nicholas Ave., New York, NY 10032. E-mail: bt12@columbia.edu.
Supported by grants from the National Institutes of Health (to B. T.) and from the Human Frontiers Science Project (to U. K.).
References
- 1.Stuart RO, Bush KT, Nigam SK: Changes in global gene expression patterns during development and maturation of the rat kidney. Proc Natl Acad Sci USA 2001, 98:5649-5654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miyagawa K, Kent J, Schedl A, van Heyningen V, Hastie ND: Wilms’ tumour—a case of disrupted development. J Cell Sci 1994, 18:S1-S5 [DOI] [PubMed] [Google Scholar]
- 3.Davies JA, Perera AD, Walker CL: Mechanisms of epithelial development and neoplasia in the metanephric kidney. Int J Dev Biol 1999, 43:473-478 [PubMed] [Google Scholar]
- 4.Torres M, Gomez-Pardo E, Dressler GR, Gruss P: Pax-2 controls multiple steps of urogenital development. Development 1995, 121:4057-4065 [DOI] [PubMed] [Google Scholar]
- 5.Sanyanusin P, McNoe LA, Sullivan MJ, Weaver RG, Eccles MR: Mutation of PAX2 in two siblings with renal-coloboma syndrome. Hum Mol Genet 1995, 4:2183-2184 [DOI] [PubMed] [Google Scholar]
- 6.Favor J, Sandulache R, Neuhauser-Klaus A, Pretsch W, Chatterjee B, Senft E, Wurst W, Blanquet V, Grimes P, Sporle R, Schughart K: The mouse Pax2(1Neu) mutation is identical to a human PAX2 mutation in a family with renal-coloboma syndrome and results in developmental defects of the brain, ear, eye, and kidney. Proc Natl Acad Sci USA 1996, 93:13870-13875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dressler GR, Douglass EC: Pax-2 is a DNA-binding protein expressed in embryonic kidney and Wilms tumor. Proc Natl Acad Sci USA 1992, 89:1179-1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eccles MR, Yun K, Reeve AE, Fidler AE: Comparative in situ hybridization analysis of PAX2, PAX8, and WT1 gene transcription in human fetal kidney and Wilms’ tumors. Am J Pathol 1995, 146:40-45 [PMC free article] [PubMed] [Google Scholar]
- 9.Kreidberg JA, Sariola H, Loring JM, Maeda M, Pelletier J, Housman D, Jaenisch R: WT-1 is required for early kidney development. Cell 1993, 74:679-691 [DOI] [PubMed] [Google Scholar]
- 10.Torban E, Goodyer PR: Effects of PAX2 expression in a human fetal kidney (HEK293) cell line. Biochim Biophys Acta 1998, 1401:53-62 [DOI] [PubMed] [Google Scholar]
- 11.Dehbi M, Ghahremani M, Lechner M, Dressler G, Pelletier J: The paired-box transcription factor, PAX2, positively modulates expression of the Wilms’ tumor suppressor gene (WT1). Oncogene 1996, 13:447-453 [PubMed] [Google Scholar]
- 12.McConnell MJ, Cunliffe HE, Chua LJ, Ward TA, Eccles MR: Differential regulation of the human Wilms tumour suppressor gene (WT1) promoter by two isoforms of PAX2. Oncogene 1997, 14:2689-2700 [DOI] [PubMed] [Google Scholar]
- 13.Donovan MJ, Natoli TA, Sainio K, Amstutz A, Jaenisch R, Sariola H, Kreidberg JA: Initial differentiation of the metanephric mesenchyme is independent of WT1 and the ureteric bud. Dev Genet 1999, 24:252-262 [DOI] [PubMed] [Google Scholar]
- 14.Schedl A, Hastie ND: Cross-talk in kidney development. Curr Opin Genet Dev 2000, 10:543-549 [DOI] [PubMed] [Google Scholar]
- 15.Califano A: SPLASH: structural pattern localization analysis by sequential histograms. Bioinformatics 2000, 16:341-357 [DOI] [PubMed] [Google Scholar]
- 16.Califano A, Stolovitzky G, Tu Y: Analysis of gene expression microarrays for phenotype classification. Proc Int Conf Intell Syst Mol Biol 2000, 8:75-85 [PubMed] [Google Scholar]
- 17.Klein U, Tu Y, Stolovitzky G, Mattioli M, Cattoretti G, Husson H, Freedman A, Inghirami G, Cro L, Baldini L, Neri A, Califano A, Dalla-Favera R: Gene expression profiling of B cell chronic lymphocytic leukemia reveals a homogeneous phenotype related to memory B cells. J Exp Med 2001, 194:1625-1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pearson WR: Rapid and sensitive sequence comparison with FASTP and FASTA. 1990:pp 63-98 Academic Press San Diego [DOI] [PubMed]
- 19.: Genetics Computer Group: Wisconsin Package, version 10.0. 1999. Madison
- 20.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Hudson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Miklos GLG, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, Levine AJ, Roberts RJ, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu-Threideh J, Beasley E, Biddick K, Bonazzi V, Brandon R, Cargill M, Chandramouliswaran I, Charlab R, Chaturvedi K, Deng Z, Di Francesco V, Dunn P, Eilbeck K, Evangelista C, Gabrielian AE, Gan W, Ge W, Gong F, Gu Z, Guan P, Heiman TJ, Higgins ME, Ji RR, Ke Z, Ketchum KA, Lai Z, Lei Y, Li Z, Li J, Liang Y, Lin X, Lu F, Merkulov GV, Milshina N, Moore HM, Naik AK, Narayan VA, Neelam B, Nusskern D, Rusch DB, Salzberg S, Shao W, Shue B, Sun J, Wang ZY, Wang A, Wang X, Wang J, Wei MH, Wides R, Xiao C, Yan C, Yao A, Ye J, Zhan M, Zhang W, Zhang H, Zhao Q, Zheng L, Zhong F, Zhong W, Zhu SC, Zhao S, Gilbert D, Baumhueter S, Spier G, Carter C, Cravchik A, Woodage T, Ali F, An H, Awe A, Baldwin D, Baden H, Barnstead M, Barrow I, Beeson K, Busam D, Carver A, Center A, Cheng ML, Curry L, Danaher S, Davenport L, Desilets R, Dietz S, Dodson K, Doup L, Ferriera S, Garg N, Gluecksmann A, Hart B, Haynes J, Haynes C, Heiner C, Hladun S, Hostin D, Houck J, Howland T, Ibegwam C, Johnson J, Kalush F, Kline L, Koduru S, Love A, Mann F, May D, McCawley S, McIntosh T, McMullen I, Moy M, Moy L, Murphy B, Nelson K, Pfannkoch C, Pratts E, Puri V, Qureshi H, Reardon M, Rodriguez R, Rogers YH, Romblad D, Ruhfel B, Scott R, Sitter C, Smallwood M, Stewart E, Strong R, Suh E, Thomas R, Tint NN, Tse S, Vech C, Wang G, Wetter J, Williams S, Williams M, Windsor S, Winn-Deen E, Wolfe K, Zaveri K, Abril JF, Guigó R, Campbell MJ, Sjolander KV, Karlak B, Kejariwal A, Mi H, Lazareva B, Hatton T, Narechania A, Diemer K, Muruganujan A, Guo N, Sato S, Bafna V, Istrail S, Lippert R, Schwartz R, Walenz B, Yooseph S, Allen D, Basu A, Baxendale J, Blick L, Caminha M, Carnes-Stine J, Caulk P, Chiang YH, Coyne M, Dahlke C, Mays AD, Dombroski M, Donnelly M, Ely D, Esparham S, Fosler C, Gire H, Glanowski S, Glasser K, Glodek A, Gorokhov M, Graham K, Gropman B, Harris M, Heil J, Henderson S, Hoover J, Jennings D, Jordan C, Jordan J, Kasha J, Kagan L, Kraft C, Levitsky A, Lewis M, Liu X, Lopez J, Ma D, Majoros W, McDaniel J, Murphy S, Newman M, Nguyen T, Nguyen N, Nodell M, Pan S, Peck J, Peterson M, Rowe W, Sanders R, Scott J, Simpson M, Smith T, Sprague A, Stockwell T, Turner R, Venter E, Wang M, Wen M, Wu D, Wu M, Xia A, Zandieh A, Zhu X: The sequence of the human genome. Science 2001, 291:1304-1351 [DOI] [PubMed] [Google Scholar]
- 21.Xu PX, Adams J, Peters H, Brown MC, Heaney S, Maas R: Eya1-deficient mice lack ears and kidneys and show abnormal apoptosis of organ primordia. Nat Genet 1999, 23:113-117 [DOI] [PubMed] [Google Scholar]
- 22.Abdelhak S, Kalatzis V, Heilig R, Compain S, Samson D, Vincent C, Weil D, Cruaud C, Sahly I, Leibovici M, Bitner-Glindzicz M, Francis M, Lacombe D, Vigneron J, Charachon R, Boven K, Bedbeder P, Van Regemorter N, Weissenbach J, Petit C: A human homologue of the Drosophila eyes absent gene underlies branchio-oto-renal (BOR) syndrome and identifies a novel gene family. Nat Genet 1997, 15:157-164 [DOI] [PubMed] [Google Scholar]
- 23.Johnson KR, Cook SA, Erway LC, Matthews AN, Sanford LP, Paradies NE, Friedman RA: Inner ear and kidney anomalies caused by IAP insertion in an intron of the Eya1 gene in a mouse model of BOR syndrome. Hum Mol Genet 1999, 8:645-653 [DOI] [PubMed] [Google Scholar]
- 24.Hatini V, Huh SO, Herzlinger D, Soares VC, Lai E: Essential role of stromal mesenchyme in kidney morphogenesis revealed by targeted disruption of Winged helix transcription factor BF-2. Genes Dev 1996, 10:1467-1478 [DOI] [PubMed] [Google Scholar]
- 25.Patterson LT, Pembaur M, Potter SS: Hoxa11 and Hoxd11 regulate branching morphogenesis of the ureteric bud in the developing kidney. Development 2001, 128:2153-2161 [DOI] [PubMed] [Google Scholar]
- 26.Kohlhase J, Wischermann A, Reichenbach H, Froster U, Engel W: Mutations in the SALL1 putative transcription factor gene cause Townes-Brocks syndrome. Nat Genet 1998, 18:81-83 [DOI] [PubMed] [Google Scholar]
- 27.Nishinakamura R, Matsumoto Y, Nakao K, Nakamura K, Sato A, Copeland NG, Gilbert DJ, Jenkins NA, Scully S, Lacey DL, Katsuki M, Asashima M, Yokota T: Murine homolog of SALL1 is essential for ureteric bud invasion in kidney development. Development 2001, 128:3105-3115 [DOI] [PubMed] [Google Scholar]
- 28.Kispert A, Vainio S, McMahon AP: Wnt-4 is a mesenchymal signal for epithelial transformation of metanephric mesenchyme in the developing kidney. Development 1998, 125:4225-4234 [DOI] [PubMed] [Google Scholar]
- 29.Farrell ER, Munsterberg AE: csal1 is controlled by a combination of FGF and Wnt signals in developing limb buds. Dev Biol 2000, 225:447-458 [DOI] [PubMed] [Google Scholar]
- 30.John RM, Aparicio SA, Ainscough JF, Arney KL, Khosla S, Hawker K, Hilton KJ, Barton SC, Surani MA: Imprinted expression of neuronatin from modified BAC transgenes reveals regulation by distinct and distant enhancers. Dev Biol 2001, 236:387-399 [DOI] [PubMed] [Google Scholar]
- 31.Joseph R, Tsang W, Dou D, Nelson K, Edvardsen K: Neuronatin mRNA in PC12 cells: downregulation by nerve growth factor. Brain Res 1996, 738:32-38 [DOI] [PubMed] [Google Scholar]
- 32.Hida K, Shindoh M, Yoshida K, Kudoh A, Furaoka K, Kohgo T, Fujinaga K, Totsuka Y: Expression of E1AF, an ets-family transcription factor, is correlated with the invasive phenotype of oral squamous cell carcinoma. Oral Oncol 1997, 33:426-430 [DOI] [PubMed] [Google Scholar]
- 33.Habelhah H, Okada F, Kobayashi M, Nakai K, Choi S, Hamada J, Moriuchi T, Kaya M, Yoshida K, Fujinaga K, Hosokawa M: Increased E1AF expression in mouse fibrosarcoma promotes metastasis through induction of MT1-MMP expression. Oncogene 1999, 18:1771-1776 [DOI] [PubMed] [Google Scholar]
- 34.Ikeda H, Lethe B, Lehmann F, van Baren N, Baurain JF, de Smet C, Chambost H, Vitale M, Moretta A, Boon T, Coulie PG: Characterization of an antigen that is recognized on a melanoma showing partial HLA loss by CTL expressing an NK inhibitory receptor. Immunity 1997, 6:199-208 [DOI] [PubMed] [Google Scholar]
- 35.van Baren N, Chambost H, Ferrant A, Michaux L, Ikeda H, Millard I, Olive D, Boon T, Coulie PG: PRAME, a gene encoding an antigen recognized on a human melanoma by cytolytic T cells, is expressed in acute leukaemia cells. Br J Haematol 1998, 102:1376-1379 [DOI] [PubMed] [Google Scholar]
- 36.Poleev A, Fickenscher H, Mundlos S, Winterpacht A, Zabel B, Fidler A, Gruss P, Plachov D: PAX8, a human paired box gene: isolation and expression in developing thyroid, kidney and Wilms’ tumors. Development 1992, 116:611-623 [DOI] [PubMed] [Google Scholar]
- 37.Ford HL, Landesman-Bollag E, Dacwag CS, Stukenberg PT, Pardee AB, Seldin DC: Cell cycle-regulated phosphorylation of the human SIX1 homeodomain protein. J Biol Chem 2000, 275:22245-22254 [DOI] [PubMed] [Google Scholar]
- 38.Khan J, Bittner ML, Saal LH, Teichmann U, Azorsa DO, Gooden GC, Pavan WJ, Trent JM, Meltzer PS: cDNA microarrays detect activation of a myogenic transcription program by the PAX3-FKHR fusion oncogene. Proc Natl Acad Sci USA 1999, 96:13264-13269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heanue TA, Reshef R, Davis RJ, Mardon G, Oliver G, Tomarev S, Lassar AB, Tabin CJ: Synergistic regulation of vertebrate muscle development by Dach2, Eya2, and Six1, homologs of genes required for Drosophila eye formation. Genes Dev 1999, 13:3231-3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sagara N, Toda G, Hirai M, Terada M, Katoh M: Molecular cloning, differential expression, and chromosomal localization of human frizzled-1, frizzled-2, and frizzled-7. Biochem Biophys Res Commun 1998, 252:117-122 [DOI] [PubMed] [Google Scholar]
- 41.Ohto H, Kamada S, Tago K, Tominaga SI, Ozaki H, Sato S, Kawakami K: Cooperation of six and eya in activation of their target genes through nuclear translocation of Eya. Mol Cell Biol 1999, 19:6815-6824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ridgeway AG, Skerjanc IS: Pax3 is essential for skeletal myogenesis and the expression of Six1 and Eya2. J Biol Chem 2001, 276:19033-19039 [DOI] [PubMed] [Google Scholar]
- 43.Halder G, Callaerts P, Flister S, Walldorf U, Kloter U, Gehring WJ: Eyeless initiates the expression of both sine oculis and eyes absent during Drosophila compound eye development. Development 1998, 125:2181-2191 [DOI] [PubMed] [Google Scholar]
- 44.Niimi T, Seimiya M, Kloter U, Flister S, Gehring WJ: Direct regulatory interaction of the eyeless protein with an eye-specific enhancer in the sine oculis gene during eye induction in Drosophila. Development 1999, 126:2253-2260 [DOI] [PubMed] [Google Scholar]
- 45.Pignoni F, Hu B, Zavitz KH, Xiao J, Garrity PA, Zipursky SL: The eye-specification proteins So and Eya form a complex and regulate multiple steps in Drosophila eye development. Cell 1997, 91:881-891 [DOI] [PubMed] [Google Scholar]
- 46.Stamataki D, Kastrinaki M, Mankoo BS, Pachnis V, Karagogeos D: Homeodomain proteins Mox1 and Mox2 associate with Pax1 and Pax3 transcription factors. FEBS Lett 2001, 499:274-278 [DOI] [PubMed] [Google Scholar]
- 47.Mankoo BS, Collins NS, Ashby P, Grigorieva E, Pevny LH, Candia A, Wright CV, Rigby PW, Pachnis V: Mox2 is a component of the genetic hierarchy controlling limb muscle development. Nature 1999, 400:69-73 [DOI] [PubMed] [Google Scholar]
- 48.Ma Y, Singer DB, Gozman A, Ford D, Chai L, Steinhoff MM, Hansen K, Maizel AL: Hsal 1 is related to kidney and gonad development and is expressed in Wilms tumor. Pediatr Nephrol 2001, 16:701-709 [DOI] [PubMed] [Google Scholar]
- 49.Kalatzis V, Sahly I, El-Amraoui A, Petit C: Eya1 expression in the developing ear and kidney: towards the understanding of the pathogenesis of branchio-oto-renal (BOR) syndrome. Dev Dyn 1998, 213:486-499 [DOI] [PubMed] [Google Scholar]
- 50.Winyard PJ, Risdon RA, Sams VR, Dressler GR, Woolf AS: The PAX2 transcription factor is expressed in cystic and hyperproliferative dysplastic epithelia in human kidney malformations. J Clin Invest 1996, 98:451-459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bullock SL, Fletcher JM, Beddington RS, Wilson VA: Renal agenesis in mice homozygous for a gene trap mutation in the gene encoding heparan sulfate 2-sulfotransferase. Genes Dev 1998, 12:1894-1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miyazaki Y, Oshima K, Fogo A, Hogan BL, Ichikawa I: Bone morphogenetic protein 4 regulates the budding site and elongation of the mouse ureter. J Clin Invest 2000, 105:863-873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cho EA, Patterson LT, Brookhiser WT, Mah S, Kintner C, Dressler GR: Differential expression and function of cadherin-6 during renal epithelium development. Development 1998, 125:803-812 [DOI] [PubMed] [Google Scholar]